Abstract
The Prp19-associated complex (NTC) is essential for pre-mRNA splicing and is associated with the spliceosome during spliceosome activation. NTC is required for specifying interactions of U5 and U6 with pre-mRNA to stabilize their association with the spliceosome after dissociation of U4. Here, we show that a novel splicing factor, Yju2, is associated with components of NTC, and that it is required for pre-mRNA splicing both in vivo and in vitro. During spliceosome assembly, Yju2 is associated with the spliceosome at nearly the same time as NTC but is destabilized after the first catalytic reaction, whereas other NTC components remain associated until the reaction is complete. Extracts depleted of Yju2 could be complemented by recombinant Yju2, suggesting that Yju2 and NTC are not entirely in association with each other. Yju2 is not required for the binding of NTC to the spliceosome or for NTC-mediated spliceosome activation. Complementation analysis of the affinity-isolated spliceosome formed in Yju2-depleted extracts demonstrated that Yju2 acts in concert with an unidentified heat-resistant factor(s) in an ATP-independent manner to promote the first catalytic reaction of pre-mRNA splicing after Prp2-mediated structural rearrangement of the spliceosome.
Splicing of nuclear precursor mRNA (pre-mRNA) is catalyzed by a large ribonucleoprotein complex, the spliceosome, which is composed of five snRNAs, U1, U2, U4, U5, and U6, in the form of snRNPs and numerous protein factors (2, 4, 17, 23, 36, 44, 47). The spliceosome is assembled by ordered interactions of snRNPs with the pre-mRNA at the 5′ splice site, the branch site, and the 3′ splice site and also with each other. After binding of the five snRNAs, a large conformational rearrangement occurs that involves the dissociation of U1 and U4 and the formation of new base pairing between U2 and U6 and between U6 and the 5′ splice site. Such structural rearrangement leads to the activation of the spliceosome, allowing catalytic reactions to proceed (2).
The DEXD/H-box RNA helicase Prp2 is required for the first catalytic reaction (26, 29). Prp2 functions as a molecular motor to restructure the spliceosome by hydrolyzing ATP, and then it leaves the spliceosome (25, 26). Another splicing factor, Spp2, originally identified as a high-copy-number suppressor of prp2-1 mutation, interacts with Prp2 and is required for the function of Prp2, possibly by mediating the binding of Prp2 to the spliceosome (28, 32, 35). After Prp2-dependent conformational rearrangement of the spliceosome, a heat-resistant heparin binding protein factor(s) of unknown identity, HP, is required to promote the step one reaction in an ATP-independent manner (26). Similarly, five protein factors have been shown to be involved in the second catalytic reaction. Hydrolysis of ATP by the DEXD/H-box RNA helicase Prp16 is required for the structural rearrangement of the spliceosome prior to the catalytic reaction (1, 19, 34). Prp17 has also been shown to act in the ATP-dependent step (22). The subsequent catalytic reaction is triggered by Slu7, Prp18, and Prp22 and does not require ATP (1, 19, 21, 33).
The Prp19-associated complex, or NTC (for “nineteen complex”), is associated with the spliceosome and plays an important role in mediating structural rearrangement of the spliceosome during its activation (37, 38). NTC is not required for the dissociation of U4, but it is required for the stabilization of U5 and U6 in the spliceosome during spliceosome activation (6). Stabilization of U5 and U6 by NTC is achieved in part through specifying interactions between U6 and the 5′ splice site and between U5 and the pre-mRNA (5, 6). A tertiary U6-pre-mRNA interaction that requires prior destabilization of Sm-like proteins from U6 also requires the presence of NTC (6). Furthermore, deficiency in NTC function affects the biogenesis of U4/U6 snRNP and, as a consequence, results in the failure of spliceosome cycling (7).
Eight NTC components have been identified, including Ntc90/Syf1, Ntc85/Cef1, Ntc77/Clf1, and Prp19, encoded by essential genes, and Ntc31/Syf2, Ntc30/Isy1, Ntc25/Snt309, and Ntc20, encoded by genes not essential for cellular growth (8-10, 41). Ntc25 has been shown to interact with Prp19 to regulate the interaction of Prp19 with other NTC components (11), whereas Ntc31, Ntc30, and Ntc20 form a stable subcomplex with Ntc90 to regulate the function of Ntc90 (9). All of these proteins are associated with the spliceosome at the same time during spliceosome assembly, suggesting that they might function as an integral particle in association with the spliceosome. Analysis of proteomic databases has revealed the possibility that other proteins are associated with NTC (16, 18, 31, 43, 47). Among these proteins, Yju2 was found to copurify with four of the identified components of NTC and other known splicing factors (16). On testing whether Yju2 is a component of NTC, we found that Yju2 is associated with NTC but has a function distinct from that of NTC. Yju2 dynamically interacts with NTC and is associated with the spliceosome at nearly the same time as NTC. Yju2 could bind to the spliceosome in association with NTC or separately from NTC after the activation of the spliceosome. Complementation analysis of the affinity-isolated spliceosome formed in Yju2-depleted extracts revealed that Yju2 functions in concert with an unidentified heat-resistant factor(s), presumably the previously described HP (26), after the ATP-dependent Prp2 step to promote the first catalytic reaction in a Mg2+-dependent and ATP-independent manner.
MATERIALS AND METHODS
Yeast strains.
Yeast strains used were BJ2168 (MATa prc1 prb1 pep4 leu2 trp1 ura3), Y187 (MATα gal4 gal80 his3 trp1 ade2 ura3 leu2 met-URA3::GAL-lacZ), PJ69-4A (MATa trp1 leu2 ura3 his3 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met::GAL7-lacZ), YSCC1 (MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP19HA), YSCC2 (MATa prc1 prb1 pep4 leu2 trp1 ura3 YJU2HA), YSCC4 (MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP4HA), YSCC12 (MATa his3 his7 ade3 ura3 prp2-1 PRP19HA), YSCC121 (MATa prc1 prb1 pep4 leu2 trp1 PRP19HA URA3::GAL-YJU2), and YSCC224 (MATa prc1 prb1 pep4 leu2 trp1 ura3 YJU2HA PRP2-4V5).
Oligonucleotides.
The following oligonucleotides were used: H6, GGCCAAGCTTAGGATCCATATGGCTAGCCATCATCATCATCATCATGGTAAACCAATTCCAAATCC; P2-1, GGCCGGATCCGGAAGGTGCTATACGAT; P2-2, CCGGAAGCTTCATGTGTTGCTATACAC; P2-3, CCGGACTAGTTCAAGTATTACATCTGAAAC; P2-4, CCGGCTCGAGGTGCTAGACGTGCCTCA; P2-5, TGTATTGATGTACAGGC; R13, GAGTGACGATTCCTATAG; Y2-1, GGCCGGATCCGAATTCATATGTCTGAAAGAAAAGC; Y2-2, GGCCGACGTCCCAGACTACGCTTGACCTAGAAAAGTAAGGAA; Y2-3, GGCCGACGTCGTATGGGTATTTGAGAGACTTTCCTCGTT; Y2-4, CCGGCTCGAGACCCGATATATCCGCT; and U6-Abio, BioTEG-TCTCTTTGTAAAACGG (QIAGEN).
Antibodies and reagents.
The antihemagglutinin (anti-HA) monoclonal antibody 8G5F was produced by immunizing mice with a keyhole limpet hemocyanin-conjugated HA peptide (data not shown). The anti-V5 antibody was purchased from Serotec, Inc. The anti-Yju2 polyclonal antibody was produced by immunizing rabbits with the full-length protein expressed in Escherichia coli. Protein A-Sepharose was from Amersham, Inc., and streptavidin-Sepharose and RNase A were from Sigma, Inc.
Construction of the YJU2-HA-tagged and PRP2-4V5-tagged strains.
Construction of the HA-tagged YJU2 strain was performed as described by Tsai et al. (40). For construction of pRS406.YJU2-HA, DNA fragments A and B were generated by PCR using primers Y2-1 and Y2-3 as well as Y2-2 and Y2-4, respectively. Following digestion with BamHI and AatII and with AatII and XhoI, respectively, fragments A and B were ligated with BamHI- and XhoI-digested pRS406. For construction of pRS406.PRP2N-4V5, DNA fragments C and D were generated by PCR using primers P2-1 and P2-2 as well as P2-3 and P2-4, respectively. Fragment C was digested with BamHI and HindIII and ligated with plasmid vector pDK85 digested with BamHI and HindIII. The resulting plasmid was digested with SpeI and XhoI and was ligated with fragment D that had been digested with SpeI and XhoI. Plasmids pRS406.YJU2-HA and pRS406.PRP2-4V5 were linearized and individually transformed into yeast strain BJ2168 to displace the wild-type allele with the tagged allele by the pop-in and pop-out gene displacement method (45).
Splicing extracts, substrates, and splicing assays.
Yeast whole-cell extracts were prepared according to the method of Cheng et al. (13), except that 240 μg/ml zymolyase was added and incubated at 25°C for 90 min for spheroplast formation of the prp2-1 mutant strain. The prp2-1 mutant extracts were heat inactivated by incubation of extracts at 37°C for 7.5 min. Actin precursors were synthesized in vitro using SP6 RNA polymerase according to the method of Cheng and Abelson (12). Biotinylated pre-mRNA was synthesized by following the procedure described by Chan et al. (6). Splicing assays were carried out according to the procedure of Cheng and Abelson (12).
Immunodepletion, immunoprecipitation, and precipitation of the spliceosome by streptavidin-Sepharose.
Immunodepletion of NTC was performed as described by Chan et al. (6). Immunodepletion of Yju2 was performed by incubation of 100 μl of splicing extracts with 100 μl of the anti-Yju2 antibody coupled to 50 μl of protein A-Sepharose. Immunoprecipitation was performed as described by Tarn et al. (39) with anti-HA, anti-Ntc20, or anti-V5 antibody. Precipitation of the spliceosome with streptavidin Sepharose was carried out according to the procedure of Chan et al. (6).
Purification of Yju2 and Prp2.
Histidine (His)-tagged full-length Yju2 was expressed in Escherichia coli under the control of the T7 promoter, and His, V5 double-tagged full-length Prp2 was expressed in yeast strain BJ2168 containing plasmid pYES2.PRP2-4V5 under the control of the GAL1 promoter. For construction of pYES2.PRP2-4V5, DNA generated by PCR using primers H6 and P2-5 and a DNA template isolated from yeast strain YSCC224 were digested with HindIII and ligated with HindIII-digested pYES2.PRP2 (14). His-tagged Yju2 and Prp2 were purified with a Ni affinity column (Novagen) according to the manufacturer's manual and Edwalds-Gilbert et al. (14), respectively. For purification of Yju2-HA from yeast, YJU2-HA extracts (600 μl) were incubated with 50 μl protein A-Sepharose conjugated with 120 μl of the anti-HA antibody at 4°C for 1 h. After unbound materials were washed off, the resin was resuspended in 50 μl of buffer DK (20 mM HEPES, pH 7.9, 60 mM KPO4, pH 7.0, 0.2 mM EDTA, 50 mM NaCl, and 20% glycerol) but without glycerol. The bound materials were then eluted by incubation at room temperature for 5 min with the HA peptide at a final concentration of 0.1 mM in buffer DK.
Dynamic interactions between Yju2 and NTC, spliceosome stability assays, UV cross-linking analysis, and primer extension analysis.
For assay of the dynamic interactions between Yju2 and NTC, 200-μl aliquots of Yju2-depleted extracts were treated with 0.1 mg/ml of RNase A at 37°C for 10 min, followed by addition of 50 ng of recombinant Yju2 and incubation at 4°C for 30 min. Immunoprecipitation of HA-tagged proteins by the anti-HA antibody was performed as described by Tarn et al. (39). Spliceosome stability assays and UV cross-linking and primer extension analyses were performed according to the method of Chan et al. (6).
Yeast two-hybrid assays.
YJU2, PRP19, NTC90, NTC85, NTC77, NTC31, NTC30, NTC25, and NTC20 were cloned into plasmid vectors pAS2 and pACT2 and individually transformed into yeast strains Y187 and PJ69-4A, respectively. Selected transformants were mated to yield diploid strains containing the pair pAS2-YJU2 and pACT2-NTC as well as pACT2-YJU2 and pAS2-NTC. Diploid cells were examined for Ade2 expression.
Preparation of ΔE and complementation of the affinity-purified spliceosome formed in Yju2-depleted or prp2-1 extracts.
Yju2-depleted extracts were incubated at 100°C for 5 min, and insoluble materials were removed by centrifugation. The supernatant was then transferred to a new Eppendorf tube, and KPO4 (pH 7.0) was added to a final concentration of 60 mM. Heat-resistant extract fractions (ΔE) were concentrated using Amicon Ultra centrifugal filter devices (Millipore) according to the recommended procedure. For complementation of the affinity-purified spliceosome formed in Yju2-depleted extracts, splicing reactions were carried out in Yju2-depleted extracts under normal conditions. Aliquots (20 μl) of the reaction mixture were precipitated with 1 μl of anti-Ntc20 antibody conjugated to 10 μl of protein A-Sepharose. After being washed, the precipitate was incubated at 25°C for 20 min with 30 μl of buffer DK without glycerol but containing 4 mM MgCl2, 0.8 U/μl RNasin, and 50 μg/ml tRNA, with or without 2 mM of ATP and in the presence or absence of 50 to 750 ng of recombinant Yju2, affinity-purified Yju2, or ΔE. The mixture was centrifuged, and supernatant and pellet fractions were collected for further analysis. The method for complementation of the affinity-purified spliceosome formed in prp2-1 extracts was the same as that described above, except that after the splicing reaction, 5 or 10 mM glucose was added and the mixture incubated for 5 min to deplete ATP. Purified Prp2-4V5 was then added and the mixture was incubated for 10 min, and the Prp2-containing spliceosome was precipitated by 1.5 μl anti-V5 antibody conjugated to protein A-Sepharose.
RESULTS
Association of Yju2 with NTC.
In recent proteomic analyses, Yju2 was found to associate with several NTC components (16, 30). To examine whether Yju2 is a component of NTC, we tagged Yju2 with the HA epitope at its carboxy terminus for immunoprecipitation analysis. Extracts prepared from the PRP19-HA or YJU2-HA strain were immunoprecipitated with the anti-HA antibody, followed by probing Western blots with antibodies against NTC components. The anti-HA antibody precipitated Prp19-HA efficiently, whereas the efficiency of precipitating Yju2-HA was poor and increased with increasing amounts of the antibody used (data not shown), presumably due to poor accessibility of the antibody to the HA epitope on Yju2. Despite the fact that five times more anti-HA antibody was used for precipitation of Yju2-HA than for precipitation of Prp19-HA, the amount of Yju2-HA protein precipitated was less than that of Prp19-HA, as shown in Fig. 1A, when the two precipitates were probed with the anti-HA antibody. Nevertheless, after being normalized to the amounts of HA-tagged proteins, all NTC components coprecipitated with Yju2 nearly as well as with Prp19, suggesting a stoichiometric association of NTC components with Yju2. Although this interpretation of stoichiometric association was later challenged by depletion analysis of Yju2 as described below, we proceeded with a functional analysis of Yju2 in the splicing reaction.
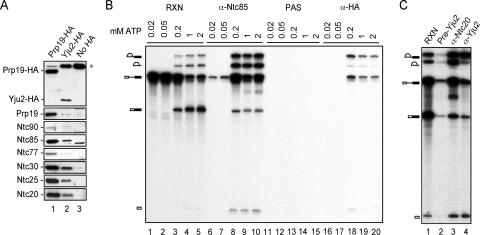
FIG. 1.
Yju2 is associated with components of NTC and is associated with the spliceosome at nearly the same time as NTC. (A) Extracts prepared from Prp19-HA (lane 1), Yju2-HA (lane 2), and untagged (No HA; lane 3) strains were immunoprecipitated with the anti-HA antibody, followed by Western blotting using antibodies against the HA epitope and components of NTC. The asterisk indicates a nonspecific protein recognized by the anti-HA antibody. (B) Splicing reactions were carried out in Yju2-HA extracts at 2 mM (lanes 5, 10, 15, and 20), 1 mM (lanes 4, 9, 14, and 19), 0.2 mM (lanes 3, 8, 13, and 18), 0.05 mM (lanes 2, 7, 12, and 17), and 0.02 mM (lanes 1, 6, 11, and 16) of ATP, and reaction mixtures were subjected to immunoprecipitation in the absence of antibodies (lanes 11 to 15) or with anti-Ntc85 (lanes 6 to 10) and anti-HA (lanes 16 to 20) antibodies. Lanes 1 to 5 represent 1 μl of the reaction mixture. Lanes 6 to 20 represent precipitates from 10 μl of reaction mixtures. (C) Splicing reactions were carried out at 2 mM of ATP, and reaction mixtures were subjected to immunoprecipitation with preimmune (lane 2), anti-Ntc20 (lane 3), or anti-Yju2 (lane 4) antibody. Lane 1 represents 0.5 μl of the reaction mixture. Lane 3 represents the precipitate from 5 μl of reaction mixtures, and lanes 2 and 4 represent precipitate from 20 μl of reaction mixtures. RXN, reaction mixture; PAS, protein A-Sepharose; Pre-Yju2, preimmune serum of Yju2.
To see whether Yju2 is also associated with the spliceosome during spliceosome assembly and whether the association occurs at the same time as it does for NTC, splicing reactions were carried out in Yju2-HA extracts at various ATP concentrations, and the reaction mixtures were then precipitated with either anti-HA or anti-Ntc85 antibody. NTC has been shown to associate with the spliceosome during spliceosome activation and not to associate with spliceosomes assembled at the lower ATP concentrations that prevent spliceosome activation (6, 38). Figure 1B shows that although the efficiency of Yju2-HA precipitation by the anti-HA antibody was low (lanes 16 to 20), it is clear that spliceosomes coprecipitated with Yju2-HA in the presence of ATP at concentrations of 0.2 mM or higher (lanes 18 to 20) but not at 0.02 or 0.05 mM (lanes 16 and 17), similar to what was seen for spliceosome precipitation with the anti-Ntc85 antibody (lanes 6 to 10). However, unlike with Ntc85, which remained stably associated after binding to the spliceosome (lanes 8 to 10), the amount of the spliceosome coprecipitated with Yju2-HA was largest at 0.2 mM ATP and decreased at higher ATP concentrations (lanes 18 to 20). Furthermore, the spliced product intron-lariat was coprecipitated to a lesser extent than splicing intermediates, suggesting that Yju2-HA might be destabilized when the reaction was complete. Precipitation with polyclonal antibodies raised against recombinant Yju2 protein also precipitated less intron-lariat (Fig. 1C, lane 4), excluding the possibility of epitope blocking during or after catalytic reactions. These results indicate that Yju2 is associated with the spliceosome at nearly the same time as NTC, but it is destabilized when the reaction progresses. Therefore, Yju2 may not function with NTC in a concerted manner during the splicing reaction.
Yju2 is essential for pre-mRNA splicing in vivo.
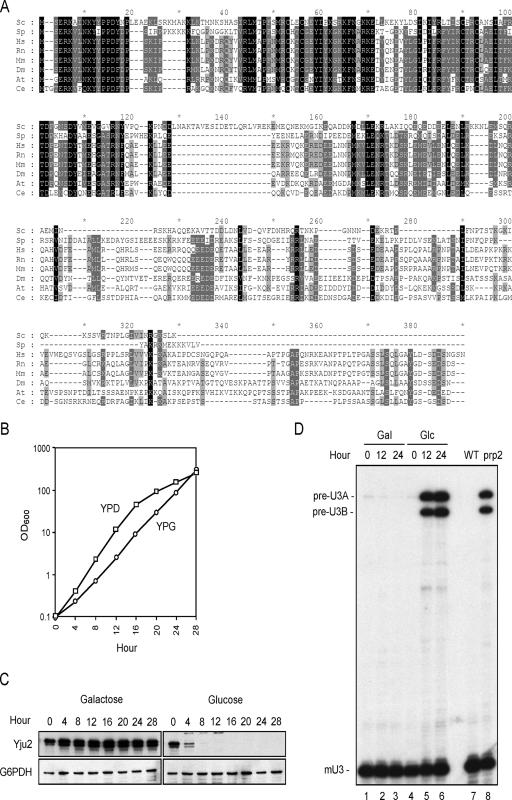
Sequence alignment of YJU2 orthologs shown in Fig. 2A reveals that although the amino-terminal half of the protein is conserved, the carboxy-terminal half is quite divergent. YJU2 is reported to be essential for yeast vegetative growth (15). The sequence does not have an obvious motif to indicate the possible function of the protein, except for a coiled-coil domain in the middle region of the protein. To see whether Yju2 is essential for splicing in vivo, we constructed a yeast strain in which the YJU2 gene was under the control of an inducible galactose promoter. A shift of cells to glucose-containing medium depleted the Yju2 protein, as shown in Fig. 2C, and led to growth arrest after 16 h, as shown in Fig. 2B. To see whether splicing was affected upon depletion of Yju2, RNA was isolated from cell cultures grown in galactose- or glucose-supplemented medium and subjected to primer extension analysis using a 5′-end-labeled primer complementary to a region in the second exon of the U3 gene (40). Figure 2D shows that two extension products representing pre-mRNA of U3A and U3B accumulated at 12 or 24 h after growth in glucose medium (lanes 5 and 6) but not in galactose medium (lanes 1 to 3) or in the wild-type control (lane 7). Similar extension products were also seen in the prp2-1 temperature-sensitive mutant after growth at restrictive temperatures for 2 h (lane 8). This indicates that in vivo depletion of Yju2 leads to a defect in pre-mRNA splicing and that Yju2 is required for splicing in vivo.
FIG. 2.
Yju2 is required for pre-mRNA splicing in vivo. (A) Protein sequence alignment of Yju2 orthologs using MAFFT and GeneDoc. Putative Yju2 orthologs were identified by using published KEGG (Kyoto Encyclopedia of Genes and Genomes) databases (24). Conserved residues are shaded black for 100% conservation, gray for 80% conservation, and light gray for 60% conservation. Abbreviations, protein accession numbers (from NCBI), and percentages of identity with Yju2 are the following: Sc, Saccharomyces cerevisiae, NP_012828, 100%; Sp, Schizosaccharomyces pombe, NP_001018234, 40%; Hs, Homo sapiens, NP_060544, 36.32%; Rn, Rattus norvegicus, XP_576700, 36.32%; Mm, Mus musculus, NP_082657, 36.32%; Dm, Drosophila melano-gaster, NP_611092, 35.38%; At, Arabidopsis thaliana, NP_173156, 33.99%; and Ce, Caenorhabditis elegans, NP_498576, 33.33%. (B) Growth curves of GAL-YJU2 cells in glucose medium (YPD) and galactose medium (YPG). Cells were grown in galactose medium to mid-log phase and then either continuously maintained in YPG or shifted to YPD. Cells were collected at 0, 4, 8, 12, 16, 20, 24, and 28 h after the shift for measurements of the optical density at 600 nm (OD600). (C) Protein extracted from collected cells was analyzed by probing Western blots with the anti-Yju2 antibody. Anti-G6PDH antibody was used as an internal control. (D) Total RNA extracted from collected cells was analyzed by primer extension using R13 as the primer. The prp2 mutant was grown at 37°C for 2 h before harvest. YPD, yeast extract-peptone-dextrose; YPG, yeast extract-peptone-galactose; Gal, galactose; Glc, glucose.
Yju2 functions in splicing independently of NTC.
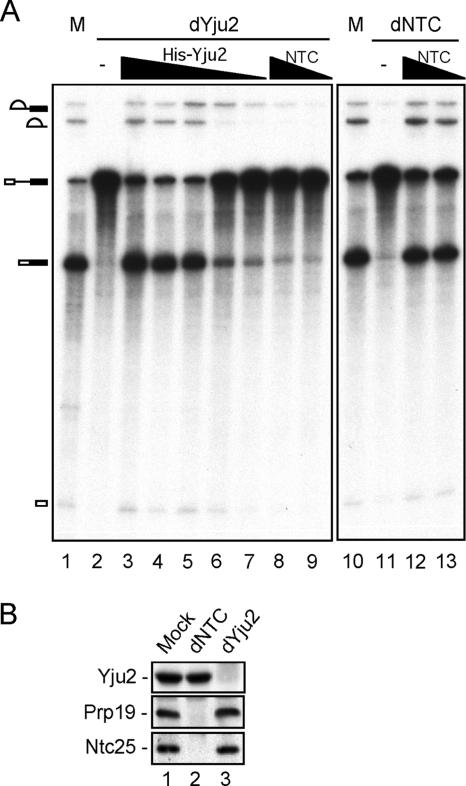
To see whether Yju2 is required for the same step of spliceosome assembly as NTC, Yju2 was depleted from extracts with the anti-Yju2 antibody. As shown in Fig. 3A, depletion of Yju2 completely abolished the splicing activity (lane 2). Surprisingly, addition of affinity-purified NTC only marginally restored the splicing activity (lanes 8 and 9), despite the same amount of NTC being able to fully recover splicing of NTC-depleted extracts (lanes 10 to 13). Furthermore, purified recombinant Yju2 with a His tag at its amino terminus (His-Yju2) efficiently restored the splicing activity of Yju2-depleted extracts (lanes 3 to 7). These results suggest that although immunoprecipitation analysis revealed the association of Yju2 and NTC components, it is likely that only a small fraction of the Yju2 protein is associated with NTC and vice versa. Moreover, Yju2 might have a function independent of NTC in the splicing reaction. Indeed, Western blotting confirmed that the level of Prp19 or Ntc25 was not significantly affected after depletion of Yju2 (Fig. 3B, lane 3), and that of Yju2 was not affected to a great extent upon depletion of NTC (Fig. 3B, lane 2).
FIG. 3.
Complementation of Yju2-depleted extracts by recombinant Yju2. (A) Splicing reactions were carried out in mock-depleted (lanes 1 and 10), Yju2-depleted (lanes 2 to 9), or NTC-depleted (lanes 11 to 13) extract with the addition of recombinant His-Yju2 (lane 3, 150 ng; lane 4, 50 ng; lane 5, 15 ng; lane 6, 5 ng; and lane 7, 2 ng) or affinity-purified NTC (lanes 8 and 12, 2 μl; lanes 9 and 13, 1 μl). (B) Western blot of mock-depleted (lane 1), NTC-depleted (lane 2), or Yju2-depleted (lane 3) extract probed with anti-Yju2, anti-HA, or anti-Ntc25 antibody. M, mock depleted; dYju2, Yju2 depleted; dNTC, NTC depleted.
Dynamic interactions between Yju2 and NTC.
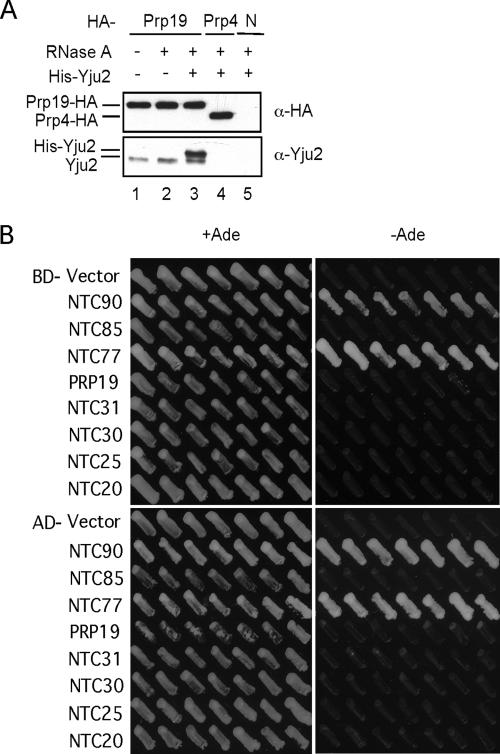
The fact that only small fractions of Yju2 and NTC are associated with each other raises the possibility that Yju2 might interact with NTC in a dynamic manner, perhaps for one to recruit the other to the spliceosome. To examine the possibility of dynamic interactions, recombinant His-Yju2 was added to splicing extracts, followed by immunoprecipitation of NTC to see whether His-Yju2 had become associated with NTC. Figure 4A shows that immunoprecipitation of Prp19-HA extracts with the anti-HA antibody coprecipitated both endogenous Yju2 and recombinant His-Yju2 (lane 3). Association of Yju2 or His-Yju2 with NTC was not mediated through binding to RNA, because pretreatment of extracts with RNase A did not affect the association (lanes 1 to 3). Precipitation of Prp4 with Prp4-HA extracts did not coprecipitate endogenous or recombinant Yju2 (lane 4), further demonstrating the specificity of the Yju2-NTC association. These results strongly suggest that Yju2 interacts dynamically with NTC.
FIG. 4.
Interactions of Yju2 with NTC and NTC components. (A) His-Yju2 (50 ng) was added to Prp19-HA (lanes 1 to 3), Prp4-HA (lane 4), or untagged extract (lane 5) and immunoprecipitated with the anti-HA antibody, followed by probing Western blots with anti-HA and anti-Yju2 antibodies. Extracts were pretreated with RNase A prior to precipitation (lanes 2 to 5). N, untagged extracts. (B) Yju2 and NTC components were fused to the GAL4-DNA binding domain (BD) and the GAL4 activation domain (AD), and interactions were assayed for activation of ADE2 under the control of the GAL2 promoter on plates with or without adenine.
To identify the component(s) mediating the Yju2-NTC association, interactions of Yju2 with NTC components were examined by two-hybrid assays. Yju2 was fused to the GAL4-DNA binding domain and NTC components were fused to the GAL4 activation domain and vice versa, and interactions were assayed for the activation of ADE2 under the control of the GAL2 promoter on plates supplemented with adenine or left unsupplemented. As shown in Fig. 4B, Yju2 interacted only with Ntc90 and Ntc77 but not with Ntc85, Prp19, Ntc31, Ntc30, Ntc25, or Ntc20. Association of Yju2 with NTC is therefore mediated through its interaction with Ntc90 and Ntc77.
Yju2 is required for splicing after spliceosome activation.
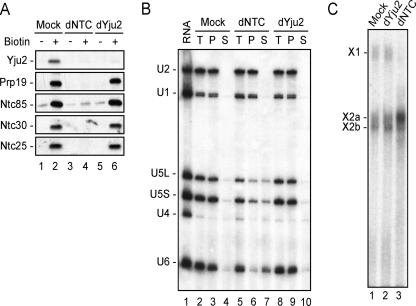
To determine whether Yju2 acts in spliceosome assembly before or after NTC, the spliceosome was assembled in NTC- or Yju2-depleted extracts by using biotinylated pre-mRNA, and components of the spliceosome were then isolated by precipitation with streptavidin-Sepharose and examined by Western blotting. Figure 5A shows that NTC components, exemplified by Prp19, Ntc85, Ntc30, and Ntc25, were present on the spliceosome formed in Yju2-depleted extracts (lane 6), while Yju2 was not present on the spliceosome formed in NTC-depleted extracts (lane 4). These results suggest that Yju2 is not required for the binding of NTC to the spliceosome, whereas NTC is required for the binding of Yju2 to the spliceosome. Thus, Yju2 is associated with the spliceosome after or concomitantly with the binding of NTC.
FIG. 5.
Yju2 was not required for spliceosome activation. (A) Splicing reactions were carried out in mock-depleted (lanes 1 and 2), NTC-depleted (lanes 3 and 4), or Yju2-depleted (lanes 5 and 6) extract using biotinylated (lanes 2, 4, and 6) or nonbiotinylated (lanes 1, 3, and 5) ACAC pre-mRNA. The spliceosome was isolated by precipitation with streptavidin-Sepharose, followed by Western blotting using antibody against Yju2, Prp19, Ntc85, Ntc30, or Ntc25. (B) Splicing reactions were carried out in mock-depleted (lanes 2 to 4), NTC-depleted (lanes 5 to 7), or Yju2-depleted (lanes 8 to 10) extract using biotinylated ACAC pre-mRNA as the substrate, and the spliceosome was precipitated with streptavidin-Sepharose. After the unbound materials were washed off, the pellet was separated into two fractions: one was the total precipitate (T; lanes 2, 5, and 8), and the other was added to the splicing buffer and incubated at room temperature for 20 min. After separating the supernatant (S; lanes 4, 7, and 10) and pellet fractions (P; lanes 3, 6, and 9), RNA was extracted and analyzed by Northern blotting. Lane 1 is total RNA from 2 μl of extracts. (C) Splicing reactions were carried out in mock-depleted (lane 1), Yju2-depleted (lane 2), or NTC-depleted extract (lane 3) using Ac/Cla pre-mRNA as the substrate. The reaction mixtures were precipitated with the anti-Smd1 antibody, followed by UV irradiation. RNA was then isolated and selected with 5′-end-biotinylated oligonucleotide U6-Abio through precipitation with streptavidin-Sepharose and was then further analyzed. The cross-linked products X1, X2a, and X2b are as described by Chan and Cheng (5). dYju2, Yju2-depleted; dNTC, NTC-depleted.
Since NTC is required for spliceosome activation after the dissociation of U1 and U4, Yju2 could also be required for spliceosome activation either in concert with NTC or after the action of NTC. Alternatively, it could be required only for catalytic steps. We have demonstrated that depletion of NTC results in destabilization of U5 and U6 due to failure to form specific interactions between U6 and the intron and between U5 and the exon sequence near splice junctions (6). We thus examined whether U5 and U6 remained stably associated with the spliceosome post-U4 dissociation in the absence of Yju2. Splicing reactions were carried out in Yju2-depleted extracts by using biotinylated pre-mRNA, and the spliceosome was isolated by precipitation with streptavidin-Sepharose. The isolated spliceosome, containing U2, U5, and U6, was reincubated in the splicing buffer at 25°C for 20 min (Fig. 5B, lanes 2, 5, and 8) (6). With NTC-depleted extracts, the majority of U6 and more than half of U5 were dissociated from the spliceosome, while U2 remained stably bound (lanes 5 to 7) (6). In contrast, U5 and U6 remained stably associated with the spliceosome with Yju2-depleted extracts (lane 8 to 10) and with mock-depleted extracts (lanes 2 to 4). UV cross-linking analysis, as shown in Fig. 5C, also revealed cross-linking products of U6 identical to those of pre-mRNA in both Yju2-depleted extracts and mock-depleted extracts (lanes 1 and 2), and they were quite distinct from those of NTC-depleted extracts (lane 3). These results suggest that spliceosome activation is not affected by depletion of Yju2, and Yju2 might be involved in the first catalytic reaction after spliceosome activation.
Yju2 and heat-resistant factors are required for an ATP-independent step of the first catalytic reaction.
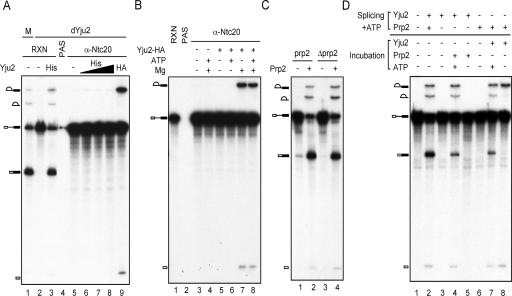
To examine how Yju2 is involved in the first catalytic reaction, the spliceosome was formed in Yju2-depleted extracts and was isolated by precipitation with the anti-Ntc20 antibody. Recombinant Yju2 (His-Yju2) was added to the precipitated spliceosome to see whether Yju2 was sufficient to promote the first catalytic reaction on the spliceosome formed in the absence of Yju2. Figure 6A shows that although small amounts of His-Yju2 could restore the splicing activity of the Yju2-depleted extract (lane 3), larger amounts of His-Yju2 failed to promote catalytic reactions on the affinity-isolated spliceosome (lanes 6 to 8). This indicates that Yju2 alone was not sufficient to promote splicing catalysis of the affinity-isolated activated spliceosome formed in Yju2-depleted extracts and that splicing might require another factor(s) to act in concert. Interestingly, affinity-purified Yju2 from Yju2-HA extracts using the anti-HA antibody did support the first catalytic reaction, albeit at a low activity level (lane 9), suggesting that this unidentified factor(s) might have been copurified with Yju2-HA from splicing extracts.
FIG. 6.
Yju2 is required for the first catalytic reaction after the Prp2 step. (A) Splicing was carried out in Yju2-depleted extracts and reaction mixtures (lane 2) precipitated with the anti-Ntc20 antibody (lanes 5 to 9). After the unbound materials were washed off, the pellet was incubated in the presence of ATP and Mg2+ at 25°C for 20 min alone (lane 5) or with the addition of recombinant His-Yju2 (lane 6, 50 ng; lane 7, 250 ng; lane 8, 750 ng) or affinity-purified Yju2-HA from 150 μl yeast extracts (lane 9). His, His-Yju2; HA, affinity-purified Yju2-HA from yeast; RXN, reaction; PAS, protein A-Sepharose. (B) Splicing was carried out in Yju2-depleted extracts and reaction mixtures (lane 1) precipitated with the anti-Ntc20 antibody (lanes 3 to 8). The precipitates were incubated alone (lanes 3 and 4) or with the addition of affinity-purified Yju2-HA (lanes 5 to 8) alone or in the presence of ATP or Mg2+. (C) Splicing in untreated (lanes 1 and 2) or heat-treated prp2-1 mutant extracts (lanes 3 and 4) alone (lanes 1 and 3) or with (lanes 2 and 4) the addition of 100 ng of recombinant Prp2 protein (lanes 2 and 4). (D) Splicing was carried out in Yju2-depleted prp2-1 extracts with the addition of recombinant Yju2 (lanes 2 to 5) or Prp2 (lanes 2 and 6 to 8) protein. Glucose was then added to a final concentration of 5 mM, and the mixture was incubated for 5 min to deplete ATP. Recombinant Yju2, Prp2, and/or 10 mM ATP was then added and further incubated at 25°C for 10 min.
Since ATP is required for both catalytic steps, we used the same complementation experiment to examine whether the function of Yju2 requires ATP. Figure 6B shows that complementation by affinity-purified Yju2-HA required Mg2+ but not ATP (lanes 5 to 8). The step one reaction requires Prp2 to drive the conformational rearrangement of the spliceosome, utilizing the energy from ATP hydrolysis (25, 26). The fact that Yju2 could promote the first catalytic reaction independently of ATP suggests that Yju2 might act after Prp2. To confirm the order of Yju2 and Prp2, we used prp2 mutant extracts to block the function of Prp2. Figure 6C shows that the extract prepared from the prp2 temperature-sensitive mutant had a low level of splicing activity (lane 1), but this could be complemented by the addition of Prp2 purified from Prp2-overexpressed extracts (26), whether or not the extract was pretreated with heat (lanes 2 and 4). Splicing was carried out in Yju2-depleted prp2 heat-inactivated extracts in the presence or absence of purified Yju2 or Prp2. After depletion of ATP by the addition of 5 mM glucose, followed by incubation for 5 min, Yju2 or Prp2 was added alone or with 10 mM ATP to see whether the catalytic reaction could proceed (Fig. 6D). Yju2 added either before Prp2 (lane 4) or after Prp2 (lanes 7 and 8) could promote the catalytic reaction and did not require ATP (lane 8), whereas Prp2 added after depletion of ATP required the addition of ATP to drive the catalytic reaction (lanes 4 and 5). This result confirms that Yju2 could function after the ATP-dependent action of Prp2 in mediating the step one reaction.
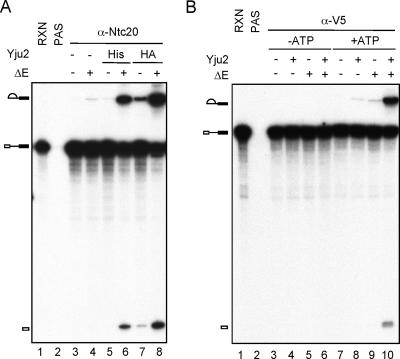
A heat-resistant protein factor, HP, was previously shown to be required for the first catalytic reaction after the Prp2 step (26). While HP has been partially purified, its identity has not yet been revealed. Judging from the order of its involvement in the first step, we speculated that Yju2 might be HP or might cooperate with HP to function in the first catalytic reaction after Prp2. To clarify the relationship between Yju2 and HP, extracts depleted of Yju2 were heat treated by boiling for 5 min, and the soluble materials (ΔE) were used to complement the affinity-isolated spliceosome formed in Yju2-depleted extracts. As shown in Fig. 7A, heat-treated Yju2-depleted extracts (lane 4) or recombinant His-Yju2 (lane 5) only barely promoted the catalytic reaction. However, the heat-treated extract was able to promote the reaction when it was in combination with His-Yju2 (lane 6), and it greatly enhanced the activity of Yju2-HA affinity purified from yeast extracts (lanes 7 and 8). This suggests that Yju2 is distinct from HP and that HP is likely a factor copurified with Yju2-HA from splicing extracts that cooperates with Yju2 to promote catalysis following Prp2 action, although the association of Yju2 and HP was not quantitative.
FIG. 7.
Yju2 and a heat-resistant factor(s) are required for the first catalytic reaction after the Prp2 step. (A) Splicing was carried out in Yju2-depleted extracts and reaction mixtures (lane 1) precipitated with the anti-Ntc20 antibody (lanes 3 to 8). Precipitates were incubated in the presence of Mg2+ at 25°C for 20 min alone (lane 3) or with the addition of recombinant His-Yju2 (lanes 5 and 6, 50 ng) or affinity-purified Yju2-HA (lanes 7 and 8) from 150 μl yeast extracts and either in combination with 4 μl heat-treated extracts prepared from Yju2-depleted extracts (lanes 4, 6, and 8) or alone (lanes 3, 5, and 7). (B) Splicing was carried out in Yju2-depleted heat-inactivated prp2-1 extracts, followed by addition of glucose (final concentration, 5 mM) to deplete ATP. Affinity-purified V5-tagged Prp2 was then added and incubated for 10 min, and the Prp2-bound spliceosome was precipitated with the anti-V5 antibody. Affinity-purified Yju2-HA and/or heat-treated extracts were then added to precipitates for incubation in the presence (lanes 7 to 10) or absence (lanes 3 to 6) of 2 mM ATP at 25°C for 20 min. ΔE, heat-treated extracts; RXN, reaction; PAS, protein A-Sepharose.
The requirement of HP for the first catalytic reaction after Prp2 was further demonstrated by isolating the Prp2-containing spliceosome for complementation. Splicing reactions were carried out in heat-treated Yju2-depleted prp2 mutant extracts, followed by incubation of the reaction mixtures in the presence of 5 mM glucose to deplete ATP. Affinity-purified Prp2 (His-V5 tagged) was then added to the reaction mixture, and the Prp2-bound spliceosome was precipitated with the anti-V5 antibody, as shown in Fig. 7B (lanes 3 to 10). When Yju2 or ΔE was added to the isolated spliceosome, a minute amount of splicing could be detected, but only in the presence of ATP (lanes 8 and 9). The splicing activity was greatly enhanced when both Yju2 and ΔE were added (lane 10). This confirms that Prp2 binds to the spliceosome prior to the function of Yju2 and HP and that it allows Yju2 and HP to promote the catalytic reaction only after ATP hydrolysis.
DISCUSSION
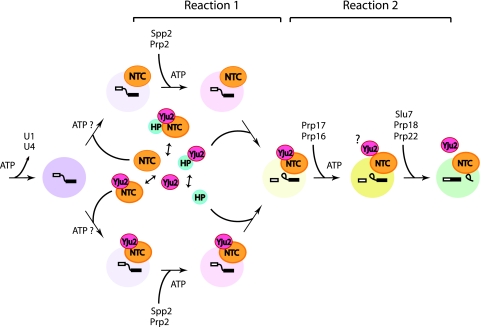
In this study, we have identified a novel splicing factor, Yju2, essential for the first catalytic step of the pre-mRNA splicing reaction. Yju2 is required for pre-mRNA splicing both in vivo and in vitro. It is dynamically associated with NTC but does not participate in NTC-dependent spliceosome activation. We provide evidence that Yju2 functions after Prp2 and acts in concert with an unidentified heat-resistant factor(s) to promote the first catalytic reaction in a Mg2+-dependent but ATP-independent manner. The requirement for Mg2+ might simply reflect the involvement of Mg2+ in the transesterification reaction. A diagram summarizing the interaction of Yju2 with NTC, with HP, and with the spliceosome and its function in splicing is shown in Fig. 8.
FIG. 8.
Diagram illustrating the interaction of Yju2 with NTC, with HP, and with the spliceosome; the diagram also illustrates the function of Yju2 in the splicing reaction. Yju2 interacts with NTC and with HP in dynamic manners. Double arrowheads indicate equilibrium between the association and dissociation forms. After U1 and U4 are dissociated from the spliceosome, NTC, either in association with Yju2 or with no association, binds to activate the spliceosome. The first catalytic reaction involves an ATP-dependent step, which requires Prp2 and Spp2, and an ATP-independent step, which requires HP and Yju2. The second catalytic reaction also involves an ATP-dependent step, which requires Prp17 and Prp16, and an ATP-independent step, which requires Slu7, Prp18, and Prp22. The requirement for ATP in each step is indicated. Yju2 is destabilized from the spliceosome after the reaction is complete, but its association with the spliceosome after the Prp16 step remains undetermined and is indicated by a question mark. A change in the color of the spliceosome represents a structural change of the spliceosome during each transition.
Proteomic study has shown that Yju2 copurifies with four identified components of NTC and other known splicing factors (16). Yju2 has also been described as Cwc16 for its association with Cefl/Ntc85 (30). Consistent with previous results, we have shown that Yju2 is associated with all identified components of NTC. Nevertheless, depletion of NTC did not codeplete a significant fraction of Yju2 and vice versa, indicating that only a small fraction of Yju2 is associated with NTC, and a similarly small fraction of Prp19 is associated with Yju2. The finding that affinity-purified NTC had only a low level of activity in complementing Yju2-depleted extracts supports this notion. Interestingly, the relative amounts of NTC components that coprecipitated with Prp19 and with Yju2 were similar (Fig. 1A). This suggests that, like Yju2, not all Prp19 is associated with other NTC components. Indeed, the level of Prp19 or of Ntc25 was not affected to a great extent upon depletion of Ntc20 (data not shown), suggesting that interactions between NTC components might also be dynamic to some extent, as is the case between Yju2 and NTC. Our two-hybrid analysis revealed that Yju2 does not directly interact with Prp19 or Ntc25, and the association of Yju2 with NTC is likely mediated through its interactions with Ntc90 and Ntc77. Both Ntc90 and Ntc77 contain multiple copies of TPR repeats and interact with all identified NTC components, except Prp19 and Ntc25 (9). They are speculated to act as the scaffold of the Prp19-associated complex. Their interactions with Yju2 suggest that they might also serve to recruit other splicing factors for later functional steps. It will be interesting to see whether binding of HP is also mediated through interactions with Ntc90 and Ntc77.
Although Yju2 can be associated with NTC, Yju2 and NTC are not functionally linked. NTC is required for stabilization of U5 and U6 on the spliceosome through specifying interactions between U6 and the 5′ splice site and between U5 and pre-mRNA during spliceosome activation (5, 6). Yju2 was determined to function after NTC, since depletion of Yju2 did not affect NTC-mediated spliceosome activation, as judged by assays for the stable association of U5 and U6 with the spliceosome, specific interactions between U6 and the 5′ splice site (Fig. 5C), and destabilization of Sm-like proteins from U6 (data not shown). Complementation analysis of the affinity-isolated spliceosome formed in Yju2-depleted extracts further showed that Yju2 is involved in the first catalytic reaction and can function after the ATP-dependent Prp2 step. It is not unusual for splicing factors to be able to bind to the spliceosome early in the splicing reaction but to function only in later steps. Prp18 was shown to associate with U5 snRNP and presumably can be associated with the spliceosome upon binding of tri-snRNP, but it is involved only in the second step of pre-mRNA splicing (20). The NTR complex, which mediates spliceosome disassembly, is weakly associated with NTC, and it could bind to the spliceosome in association with NTC during spliceosome activation (40).
Unlike the other two identified step-one splicing factors, Spp2 and Prp2 (25, 27, 32), Yju2 appears to be more specifically associated with splicing intermediates. Precipitation of Yju2 from splicing reaction mixtures coprecipitated predominantly splicing intermediates as well as a smaller fraction of pre-mRNA and lariat-intron, suggesting that Yju2 becomes associated with the spliceosome prior to the catalytic step, more stably associated after the first catalytic reaction, and destabilized from the spliceosome as the second catalytic reaction proceeds. Thus, not only is Yju2 not functionally linked to NTC, but also it is distinct from NTC in its spliceosome binding pattern; NTC becomes associated with the spliceosome during spliceosome activation and remains stably associated until the reaction is complete.
Yju2 is involved in the step one reaction, but it interacts with neither Spp2 nor Prp2 in our yeast two-hybrid assays (data not shown). Considering that Prp2 and Spp2 are only transiently associated with the spliceosome during spliceosome assembly, it is unlikely that they are involved in recruiting Yju2 to the spliceosome by interacting with Yju2. In contrast, NTC is stably associated with the spliceosome and interacts with Yju2 in a dynamic manner. Such dynamic interaction might serve to mediate the recruitment of Yju2 to the spliceosome, adding another role for NTC besides its functional role in spliceosome activation. In fact, a component of NTC, Ntc30/Isy1, has also been shown to interact with Prp16, a DEXD/H-box RNA helicase involved in the second catalytic reaction, to regulate the fidelity of pre-mRNA splicing (42). This reveals multiple roles for NTC in the splicing reaction during its association with the spliceosome.
We have demonstrated that Yju2 can function after the Prp2 step, although there is no evidence that Prp2 has to function before Yju2. Yju2 alone is not sufficient to promote the catalytic reaction after the action of Prp2. A heat-resistant factor, analogous to the previously described HP (26), is also required. Yju2 is distinct from HP but might interact with it, since affinity-purified Yju2 from yeast extracts had a low level of activity in complementing the affinity-isolated spliceosome formed in Yju2-depleted extracts. The complementation activity was enhanced by the addition of a soluble fraction of the heat-treated extract, indicating that the amount of HP associated with Yju2 was limiting. It has been speculated that HP might interact with Yju2 in a dynamic manner, which results in the association of HP with a small fraction of Yju2 and vice versa. Thus, it is possible that HP also can bind to the spliceosome early in its association with Yju2. In fact, it has been suggested that HP could be associated with the spliceosome prior to Prp2, since a Prp2-bound spliceosome isolated from a low-salt glycerol gradient can perform the first catalytic reaction when it is incubated with ATP without the addition of any other protein factors (25). Consistent with this view, we found that affinity-purified NTC is also barely able to complement the affinity-isolated spliceosome formed in Yju2-depleted extracts (data not shown). These results strongly suggest that dynamic interactions between Yju2, HP, and NTC (Fig. 8), leading to the formation of a complex containing Yju2, HP, and NTC, play important roles in the recruitment of Yju2 and HP to the spliceosome for the first catalytic reaction.
The function of Yju2 in step one of splicing is not known. Prp2 is presumed to mediate a conformational change in the spliceosome upon hydrolyzing ATP prior to the catalytic reaction. Like HP, Yju2 functions after the Prp2 step and does not require ATP for its action. Yju2 and HP might be required for further structural changes to the spliceosome to juxtapose the 5′ splice site and the branch site so that the catalytic reaction can take place. Since both proteins can bind to the spliceosome in association with NTC in a manner independent of their later action, it cannot be distinguished whether they function sequentially or must act in concert in mediating the final structural rearrangement of the spliceosome to allow the catalytic reaction.
The second catalytic reaction also involves ATP-dependent and ATP-independent steps. Prp17 and Prp16 function in an ATP-dependent manner, followed by the ATP-independent, sequential action of Slu7, Prp18, and Prp22 (21, 22). However, precursor mRNAs in which the 3′ splice site is close to the branch point can be spliced efficiently in the absence of Slu7, Prp18, or Prp22 (3, 33, 46). When the distance from the branch point to the 3′ splice site is greater than 12 nucleotides, Slu7 and Prp18 are required for splicing of such pre-mRNA. Prp22 is required only when the distance is greater than 21 nucleotides (3, 33, 46). These proteins are thus thought to play a role in bringing a distant 3′ splice site into the catalytically active site of the spliceosome during the second catalytic reaction. However, the exact part of the pre-mRNA with which each step one factor interacts is unknown. Whether the requirement of Yju2 and HP for the ATP-independent step is dispensable for splicing of selected classes of pre-mRNA remains to be investigated.
Acknowledgments
We thank R.-J. Lin for providing plasmid pYES2.PRP2. We also thank P. Lin for reading the manuscript and Harry Wilson for English editing.
This work was supported by Academia Sinica and National Science Council (Taiwan) Grant NSC94-2321-B-001-021.
Footnotes
Published ahead of print on 21 May 2007.
REFERENCES
- 1.Ansari, A., and B. Schwer. 1995. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 14:4001-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333-360. [DOI] [PubMed] [Google Scholar]
- 3.Brys, A., and B. Schwer. 1996. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA 2:707-717. [PMC free article] [PubMed] [Google Scholar]
- 4.Burge, C. B., T. H. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosome, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), RNA world II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 5.Chan, S.-P., and S.-C. Cheng. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 280:31190-31199. [DOI] [PubMed] [Google Scholar]
- 6.Chan, S.-P., D.-I. Kao, W.-Y. Tsai, and S.-C. Cheng. 2003. The Prp19p-associated complex in spliceosome activation. Science 302:279-282. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C.-H., D.-I. Kao, S.-P. Chan, T.-C. Kao, J.-Y. Lin, and S.-C. Cheng. 2006. Functional links between the Prp19-associated complex, U4/U6 biogenesis and spliceosome recycling. RNA 12:765-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C.-H., W.-Y. Tsai, H.-R. Chen, C.-H. Wang, and S.-C. Cheng. 2001. Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J. Biol. Chem. 276:488-494. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C.-H., W.-C. Yu, T. Y. Tsao, L.-Y. Wang, H.-R. Chen, J.-Y. Lin, W.-Y. Tsai, and S.-C. Cheng. 2002. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 30:1029-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H.-R., S.-P. Jan, T. Y. Tsao, Y.-J. Sheu, J. Banroques, and S.-C. Cheng. 1998. Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol. Cell. Biol. 18:2196-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, H.-R., T. Y. Tsao, C.-H. Chen, W.-Y. Tsai, L.-S. Her, M.-T. Hsu, and S.-C. Cheng. 1999. Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 96:5406-5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, S.-C., and J. Abelson. 1987. Spliceosome assembly in yeast. Genes Dev. 1:1014-1027. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, S.-C., A. Newman, R.-J. Lin, G. D. McFarland, and J. N. Abelson. 1990. Preparation and fractionation of yeast splicing extract. Methods Enzymol. 181:89-96. [DOI] [PubMed] [Google Scholar]
- 14.Edwalds-Gilbert, G., D.-H. Kim, S.-H. Kim, Y.-H. Tseng, Y. Yu, and R.-J. Lin. 2000. Dominant negative mutants of the yeast splicing factor Prp2 map to a putative cleft region in the helicase domain of DExD/H-box proteins. RNA 6:1106-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrova, H., J. Kolarov, M. Ghislain, and A. Goffeau. 1992. Sequence of the novel essential gene YJU2 and two flanking reading frames located within a 3.2 kb EcoRI fragment from chromosome X of Saccharomyces cerevisiae. Yeast 8:419-422. [DOI] [PubMed] [Google Scholar]
- 16.Gavin, A., M. Bösche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. Michon, C. Cruciat, M. Remor, C. Höfert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. Heartier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 17.Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel, M. Wilm, and R. Lührmann. 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 99:16719-16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazbun, T. R., L. Malmström, S. Anderson, B. J. Graczyk, B. Fox, M. Riffle, B. A. Sundin, D. Aranda, W. H. McDonald, C.-H. Chiu, B. E. Snydsman, P. Bradley, E. G. D. Muller, S. Fields, D. Baker, J. R. I. Yates, and T. N. Davis. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12:1353-1365. [DOI] [PubMed] [Google Scholar]
- 19.Horowitz, D. S., and J. Abelson. 1993. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 7:320-329. [DOI] [PubMed] [Google Scholar]
- 20.Horowitz, D. S., and J. Abelson. 1993. A U5 small nuclear ribonucleoprotein particle protein involved only in the second step of pre-mRNA splicing in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:2959-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James, S., W. Tirmer, and B. Schwer. 2002. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8:1068-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, M. H., D. N. Frank, and C. Guthrie. 1995. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc. Natl. Acad. Sci. USA 92:9687-9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurica, M. S., L. J. Licklider, S. R. Gygi, N. Grigorieff, and M. J. Moore. 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8:426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa, M., S. Goto, S. Kawashima, and A. Nakaya. 2002. The KEGG databases at GenomeNet. Nucleic Acids Res. 30:42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, S.-H., and R.-J. Lin. 1993. Pre-mRNA splicing within an assembled yeast spliceosome requires an RNA-dependent ATPase and ATP hydrolysis. Proc. Natl. Acad. Sci. USA 90:888-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S.-H., and R.-J. Lin. 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 16:6810-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King, D. S., and J. D. Beggs. 1990. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 18:6559-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Last, R. L., J. R. Maddock, and J. L. J. Woolford. 1987. Evidence for related functions of the RNA genes of Saccharomyces cerevisiae. Genetics 117:619-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, R.-J., A. J. Lustig, and J. Abelson. 1987. Splicing of yeast nuclear pre-mRNA in vitro requires a functional 40S spliceosome and several extrinsic factors. Genes Dev. 1:7-18. [DOI] [PubMed] [Google Scholar]
- 30.Ohi, M. D., and K. L. Gould. 2002. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8:798-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohi, M. D., A. J. Link, L. Ren, J. L. Jennings, W. H. McDonald, and K. L. Gould. 2002. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22:2011-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy, J., K. Kim, J. R. Maddock, J. G. Anthony, and J. Woolford. 1995. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1:375-390. [PMC free article] [PubMed] [Google Scholar]
- 33.Schwer, B., and C. H. Gross. 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwer, B., and C. Guthrie. 1992. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 11:5033-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverman, E. J., A. Maeda, J. Wei, P. Smith, J. D. Beggs, and R.-J. Lin. 2004. Interaction between a G-patch protein and a spliceosome DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 24:10101-10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 37.Tarn, W.-Y., C.-H. Hsu, K.-T. Huang, H.-R. Chen, H.-Y. Kao, K.-R. Lee, and S.-C. Cheng. 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarn, W.-Y., K.-R. Lee, and S.-C. Cheng. 1993. Yeast precursor mRNA processing protein PRP19 associates with the spliceosome concomitant with or just after dissociation of U4 small nuclear RNA. Proc. Natl. Acad. Sci. USA 90:10821-10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarn, W.-Y., K.-R. Lee, and S.-C. Cheng. 1993. The yeast PRP19 protein is not tightly associated with small nuclear RNAs but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol. 13:1883-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai, R.-T., R.-H. Fu, F.-L. Yeh, C.-K. Tseng, Y.-C. Lin, Y.-H. Huang, and S.-C. Cheng. 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 19:2991-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai, W.-Y., Y.-T. Chow, H.-R. Chen, K.-T. Huang, R.-I. Hong, S.-P. Jan, N.-Y. Kuo, T. Y. Tsao, C.-H. Chen, and S.-C. Cheng. 1999. Cef1p is a component of the Prp19p-asociated complex and essential for pre-mRNA splicing. J. Biol. Chem. 274:9455-9462. [DOI] [PubMed] [Google Scholar]
- 42.Villa, T., and C. Guthrie. 2005. The Isy1p component of the NineTeen Complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 19:1894-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, Q., K. Hobbs, B. Lynn, and B. C. Rymond. 2003. The Clf1p splicing factor promotes spliceosome assembly through N-terminal tetratricopeptide repeat contacts. J. Biol. Chem. 278:7875-7883. [DOI] [PubMed] [Google Scholar]
- 44.Will, C. L., and R. Lührmann. 1997. Protein functions in pre-mRNA splicing. Curr. Opin. Biol. 9:320-328. [DOI] [PubMed] [Google Scholar]
- 45.Winston, F., F. Chumley, and G. R. Fink. 1983. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 101:211-228. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, X., and B. Schwer. 1997. Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucleic Acids Res. 25:2146-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]