Abstract
Transcription corepressors are general regulators controlling the expression of genes involved in multiple signaling pathways and developmental programs. Repression is mediated through mechanisms including the stabilization of a repressive chromatin structure over control regions and regulation of Mediator function inhibiting RNA polymerase II activity. Using whole-genome arrays we show that the Arabidopsis thaliana corepressor LEUNIG, a member of the GroTLE transcription corepressor family, regulates the expression of multiple targets in vivo. LEUNIG has a role in the regulation of genes involved in a number of different physiological processes including disease resistance, DNA damage response, and cell signaling. We demonstrate that repression of in vivo LEUNIG targets is achieved through histone deacetylase (HDAC)-dependent and -independent mechanisms. HDAC-dependent mechanisms involve direct interaction with HDA19, a class 1 HDAC, whereas an HDAC-independent repression activity involves interactions with the putative Arabidopsis Mediator components AtMED14/SWP and AtCDK8/HEN3. We suggest that changes in chromatin structure coupled with regulation of Mediator function are likely to be utilized by LEUNIG in the repression of gene transcription.
The Arabidopsis thaliana gene LEUNIG (LUG) encodes a member of the conserved transcription corepressor family that includes Tup1 in yeast (Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Candida albicans), Groucho (Gro) in Drosophila melanogaster, and Transducin-Like Enhancer of split (TLE) in mammals. These corepressors do not possess DNA binding motifs but repress a diverse number of target genes through targeted recruitment by site-specific DNA binding transcription factors. In Arabidopsis, LUG represses target gene transcription by interacting with DNA binding transcription factors through an adaptor protein, SEUSS (SEU), in a fashion analogous to the interaction between the yeast corepressor components Tup1 and Ssn6 (27, 28). Once recruited, corepressors mediate repression through mechanisms that include stabilization of a repressive chromatin structure over control regions, inhibition of recruitment of the transcription machinery, and direct inhibition of the RNA polymerase II (Pol II) holoenzyme regulated through the associated Mediator complex. The LUG-SEU corepressor fails to repress transcription in the presence of a histone deacetylase (HDAC) inhibitor, suggesting that one mechanism of LUG repression is through the recruitment of HDAC activities (27). This observation is consistent with Arabidopsis plants that harbor mutations in HDAC-encoding genes displaying pleotropic phenotypes similar to those reported for lug mutants (31). In lug mutant flowers the class C floral homeotic MADS box gene AGAMOUS (AG) is expressed in all four floral whorls, resulting in the ectopic formation of carpels and stamens in the outer two whorls (19), suggesting that LUG is a repressor of AG. In addition, plants harboring mutations in LUG exhibit further pleotropic defects, many of which are AG independent. These defects include abnormal carpel and ovule development, reduced female and male fertility, and narrower leaves and floral organs (7, 19). Analysis of expanded leaves indicates that LUG may also act at later stages in leaf development by restricting cell expansion during leaf growth (5). Furthermore LUG is also expressed in shoot meristems, young floral primordia, leaves, and ovules (7). It therefore appears that LUG may play a wider role in plant development and signaling response.
Using genome-wide expression studies we have identified a number of novel LUG target genes in both vegetative and floral tissues, demonstrating the wider role of LUG in regulating gene expression, and show that at least two distinct mechanisms of repression are utilized to regulate a number of these targets. Analysis of the repression mechanisms employed by LUG demonstrated that LUG associates with HDA19, a class 1 HDAC. Furthermore we have shown interactions between LUG and AtMED14 (SWP) and CDK8 (HEN3), components of a putative Arabidopsis Mediator complex, suggesting that LUG may also repress transcription through the direct regulation of RNA Pol II activity.
MATERIALS AND METHODS
Plant culture.
lug-3 carries a C-to-T mutation at position 451 from the ATG in the Landsberg erecta (Ler) background that results in early termination of the protein (7). Plants were grown in Aratrays (Betatech, Belgium) under constant conditions (20°C, constant 2,400-lx globe lighting). Vegetative tissue (stage 3.9) with roots removed, rosette leaves, and flowers (stage 6.9) from Ler (wild-type) and lug-3 mutant plants were collected for protein and RNA analysis. For HDAC inhibitor studies, Ler and lug-3 mutant seeds were surface sterilized and germinated on MS agar (0.46% MS salts, 3% sucrose, and 0.8% agar, pH 5.9) under continuous light. Seedlings at the four-leaf stage were placed in MS solution (0.46% MS salts, 3% sucrose, pH 5.9) containing ethanol or 20 μM trichostatin A (TSA; Tocris Cookson Ltd., United Kingdom) for 6 h.
Plasmid constructs.
The complete AtCDK8 open reading frame (At5g63610) was amplified by PCR from the P1 clone MBK5 (24), cloned into the acceptor vector ST1-blue (Invitrogen), and verified by restriction digestion and DNA sequencing. AtCDK8 was then subcloned into yeast vector pAS64F2 (33) to obtain pAS-AtCDK8. An SWP cDNA clone (AV52360) in pBluescript II SK+ was obtained from the Kazusa DNA Research Institute (1) and verified by DNA sequence analysis before being cloned as a SalI-PciI fragment into SmaI-NcoI of the yeast vector pAS64F2 to give a LexA translational fusion (pAS-AtMED14). pJG contains the HindIII-SalI Gal4 DNA binding domain (G4BD) fragment from pGBT9 (Clontech) in HindIII-SalI sites of pJIT60 (14). pJG-LUG contains full-length LUG cDNA cloned as a translational fusion in pJG downstream of the G4BD. For the reporter vector pJC1, pJIT166 was digested with EcoICRI and EcoRV to release the CaMV35S-glucuronidase (GUS)-nopaline synthase cassette and replaced with the HindIII/EcoRI fragment GAL4 binding site-tCUP-GUS-nitric oxide synthase cassette previously excised from pCAMBIA 2300 (37). pAS-LUG has been previously described (27). For interaction assays a SalI fragment containing the LUFS+Q domains of LUG was excised from a pGBT9 construct (27) and cloned into the SalI site of pBluescript II KS+ to obtain pBS-L+Q. HDA19 (188C13T7) and HDA6 (164A11T7) clones in pBluescript II SK+ were obtained through the Nottingham Arabidopsis Stock Centre (NASC).
Repression assays in plant cells.
Isolation and transfection of Arabidopsis mesophyll protoplasts were performed as described at http://genetics.mgh.harvard.edu/sheenweb/protocols_reg.html. Protoplasts were transfected with pJG-LUG or pJG (G4BD-only control vector) vector and the reporter pJC1. Transfected protoplasts were cultured for 12 h at 24°C in the dark, and then 20 μM TSA (in ethanol) or ethanol alone was added to the cultures. Fluorometric GUS assays were performed 6 h post-TSA exposure using the substrate methylumbelliferyl-β-glucuronide as described elsewhere (15). GUS activity (U/mg protein) was normalized to protein concentration.
Immunoprecipitation of LUG and associated proteins.
Nuclear proteins were isolated from seedlings treated with TSA (20 μM, 6 h) or ethanol using a CelLytic PN extraction kit in the presence of protease inhibitors (Sigma). Immunoprecipitation was performed using a polyclonal anti-LUG peptide antibody (RDLKATAQAFQAEG; AFFINITI Research Products Ltd., United Kingdom), previously purified by being blotted to immobilized LUG peptide. Equal amounts of nuclear proteins were diluted in immunoprecipitation buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% [vol/vol] Triton X-100, 1 mM dithiothreitol [DTT], 0.1% [wt/vol] sodium dodecyl sulfate [SDS], and protease inhibitor cocktail [Sigma]). One hundred microliters of purified antibody was added, and samples were incubated at 4°C overnight. Fifty microliters of activated protein G magnetic beads (QIAGEN) was added, and samples were incubated on ice for 2 h with vortexing at 15-min intervals. Immobilized samples were washed three times with immunoprecipitation buffer, and either the bead-immunocomplex was resuspended in HDAC buffer or proteins were eluted from beads for immunoblotting.
HDAC activity assay.
To assay for any HDAC activity associated with immunoimmobilized LUG, a colorimetric HDAC activity assay was used in accordance with the manufacturer's instructions (Calbiochem). Samples were resuspended in HDAC buffer and incubated with HDAC colorimetric substrate for 30 min at 37°C. The reaction was stopped with lysine developer, and the reaction mixture was incubated for a further 30 min before absorbance was read at 405 nm. HDAC activity was expressed as optical density at 405 nm (OD405)/μg protein.
Transcriptome profiling.
Three independent RNA isolations (RNeasy; QIAGEN) were made from 100 mg of pooled rosette leaves or flowers from Ler and lug-3 mutant plants. Each pooled RNA sample was hybridized independently to Affymetrix ATH1 Arabidopsis GeneChips (8). The resulting Affymetrix CEL files were analyzed using dChip (18), which implements model-based high-level expression analysis; following normalization, perfect match/mismatch difference model-based expression was used to calculate expression levels. Comparative analysis between two groups of samples (Ler versus lug-3 mutant leaves and Ler versus lug-3 mutant flowers) was used to identify genes that are reliably differentially expressed between groups, filtering criteria were set at >1.4-fold change with 90% confidence boundary limits, and the threshold for absolute difference between the two group means was set at 100. P values for t tests were set at 0.05. The complete data set has been made available through the NASC Arrays repository (8). Correlation of coclustered genes was performed using the bulk Gene Ontology annotation retrieval tool at TAIR (http://www.arabidopsis.org/tools/bulk/go/index.jsp).
Real-time PCR.
Total RNA was isolated (RNeasy Plant Mini system, with intermediate on-column DNase I digestion step; QIAGEN) from 100 mg of seedling leaves treated with TSA or ethanol and aboveground vegetative tissue, rosette leaves, and flowers collected from Ler and lug-3 mutant plants. One microgram RNA was reverse transcribed using random decamer primers (RETROscript; Ambion). The cDNA was used as a template for analyzing target gene expression (Absolute QPCR SYBR green; ABgene) using gene-specific primer pairs (Beacon Designer, Premier BioSoft; see the supplemental material for sequence information). ACTIN2 was used as an internal reference, and genomic DNA and RNA were used as positive and negative controls, respectively. Relative quantification of gene expression data was determined from threshold cycle (CT) values for each sample. Serial dilutions of cDNA were used to plot a calibration curve, and gene expression levels were quantified by plotting CT values on the curve. Expression levels were normalized with values obtained for the internal reference gene. Once normalized, expression (n-fold) of lug-3 transcript levels compared with those of the wild type was determined for each gene.
Protein interactions.
Wild-type yeast cells (FT5) transformed with pAS-AtCDK8, pAS-AtMED14, and pAS-LUG were used for purifying the LexA hybrid proteins. Transformants were grown overnight in selective liquid medium at 30°C, and whole-cell extracts were made as previously described (6). Five hundred micrograms of total protein was incubated overnight at 4°C with 15 μl of anti-LexA mouse monoclonal antibody [LexA (2-12), Santa Cruz Biotechnology] in immunoprecipitation buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1 mM DTT, and complete protease inhibitor cocktail II [Calbiochem]). Following incubation 50 μl of activated protein G magnetic beads (QIAGEN) was added to the extracts and incubated on ice for 2 h with vortexing at 15-min intervals before immobilized proteins were washed with immunoprecipitation buffer. Plasmids containing HDA19, HDA6, AtCDK8, LUG, and AtMED14 were used to direct coupled transcription-translation using T3 or T7 polymerase (TNT System; Promega). Ten microliters of [35S]methionine-labeled protein was incubated with 100 μl of bead-immobilized LexA hybrid protein in 1 ml binding buffer (20 mM Tris-HCl, pH 8, 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 0.2% Triton X-100, 0.2% bovine serum albumin, 0.2 mM EDTA, 1 mM DTT, and complete protease inhibitor cocktail II [Calbiochem]) overnight at 4°C. Following incubation the beads were washed extensively in binding buffer, eluted in SDS-polyacrylamide gel loading buffer, and analyzed by SDS-polyacrylamide gel electrophoresis for detection of 35S-labeled proteins by phosphorimaging (Pharos FX Plus; Bio-Rad) and LexA hybrid proteins by immunoblotting with the anti-LexA antibody (dilution, 1:200). The immunoreactive bands were detected using an enhanced chemiluminescence Western blotting detection system (GE Healthcare).
RESULTS
LUG functions as a repressor of transcription in vegetative and floral tissue.
Using genome-wide transcriptome microarray analysis we determined that LUG functions as a regulator of gene expression in both vegetative and floral tissue through the identification of a large number of differentially expressed genes. Statistical analysis of array data obtained using mRNA isolated from lug-3 mutant plants compared to that obtained using mRNA from wild-type plants revealed genes that were significantly up-regulated or down-regulated. Four hundred two genes were up-regulated, and 259 genes were down-regulated, at least 1.5-fold in lug-3 mutant leaves (see Table S1 in the supplemental material). In floral tissue 246 genes were up-regulated, and 436 genes were down-regulated at least 1.5-fold (see Table S2 in the supplemental material). The differential expression of a number of genes selected on the basis of their tissue expression profiles was validated by reverse transcription-PCR (RT-PCR) analysis (Table 1). Comparative analysis between vegetative and floral data sets revealed that a limited number of genes were coregulated in the two tissues. We identified only 18 genes that were up-regulated and another 18 genes that were down-regulated (including LUG) in both vegetative and floral tissue (see Table S3 in the supplemental material). Genes with down-regulated expression patterns were observed in both tissues, suggesting that some LUG targets may themselves be repressors and that LUG has an indirect upstream role in the regulation of genes where expression decreases. These microarray data reveal that LUG has a different function in floral and vegetative tissues, thus highlighting the dynamic role of this transcription regulator.
TABLE 1.
RT-PCR expression analysis validation of LUG-repressed genes
| Locus | Gene name | Fold change relative to Lera
|
Process (gene ontology) | |
|---|---|---|---|---|
| Leaves | Flowers | |||
| At1g26770 | AtEXP10 | 3.3 (2.1) | 1.6 (1.5) | Cell wall expansin |
| At4g21100 | DDB1B | 7.2 (10.2) | 23.9 (18.9) | DNA damage |
| At4g15260 | 8.1 (5.1) | 3.3 (4.7) | UDP-glucosyltranferase | |
| At4g16890 | SNC1 | 18.0 (16.9) | 20.2 (17.3) | Disease resistance |
| At1g22190 | RAP24 | 3.2 (3.0) | 4.3 (3.6) | Transcription |
| At3g19820 | DIMINUTO | 3.4 (2.9) | (<1.5) | Cell growth |
| At3g23250 | MYB15 | 5.4 (3.2) | (<1.5) | Transcription |
| At4g34760 | ARFX15 | 5.8 (2.4) | (<1.5) | Auxin response |
| At1g32640 | RAP1 | (<1.5) | 2.8 (2.4) | Transcription factor |
| At3g61890 | HB-12 | (<1.5) | 3.1 (3.1) | Transcription factor |
| At1g77450 | NAM | (<1.5) | 2.7 (3.6) | Transcription factor |
| At2g45660 | AGL20 | (<1.5) | 2.4 (2.4) | MADS box transcription factor |
Numbers in parentheses indicate changes identified by statistical analysis of microarray data.
Functional classification of corepressor targets revealed that they fall into several different categories including response to abiotic and biotic factors, response to stress, developmental processes, transport, and transcription (Table 1 shows examples). For example targets up-regulated in the absence of LUG included a number of genes that have functions associated with DNA damage: the UV-induced UV-damaged DNA binding protein 1-encoding gene (DDB1) (26); DWF1 (2.9-fold increase in lug-3), encoding a Ca2+-dependent calmodulin binding protein with a role in cell elongation as well as UVB defenses (9, 25); and DRT100 (2.9-fold increase in lug-3), which has a role in DNA damage repair/toleration (22). Other interesting examples of coregulated genes identified are those involved in resistance to Peronospora parasitica. Resistance genes include RPP4 and RPP5 (11), which are up-regulated 74.5- and 40.7-fold, respectively, in lug-3 mutants. However, this effect may be indirect and due to up-regulation of SNC1 (16.9- and 17.3-fold in leaves and flowers, respectively), as a gain-of-function snc1 mutation leads to the constitutive activation of a disease resistance response in Arabidopsis including RPP4 expression (39). Our analysis also identified an auxin response factor (ARFX15, Table 1) as a LUG target, supporting a role for LUG in auxin signaling. Furthermore when the expression of a second auxin response factor, ETTIN/ARF3, which has been shown to interact with SEU (23), was measured by RT-PCR, we observed that, like ARFX15, ETTIN is also differentially regulated in the lug-3 mutant (up-regulated 3.0- and 2.1-fold in leaves and flowers, respectively). Together these data suggest that LUG may have a role as an upstream regulator in auxin signaling. This genome-wide analysis of LUG function demonstrates the wider role of LUG in the regulation of gene expression in Arabidopsis, where it regulates different gene sets in floral and vegetative tissue, and highlights a number of novel processes under the regulation of the corepressor.
LUG repression involves HDAC-dependent and -independent mechanisms.
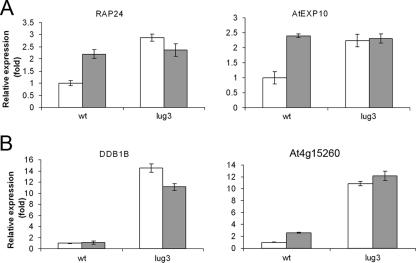
When recruited to an artificial promoter via SEU, LUG can repress transcription, a function that is reduced upon exposure to the HDAC inhibitor TSA, indicating that the corepressor represses transcription through the recruitment of an HDAC activity (27). In order to determine whether this mechanism was utilized for the repression of in vivo LUG targets, we tested the effect of TSA on a number of LUG-regulated genes identified in our microarray analysis and validated by RT-PCR. Genes were selected on the basis that they were derepressed in both floral and vegetative tissue and were therefore likely to be derepressed in other tissues and at other developmental stages, making them suitable targets for the seedling-based assay used for investigating the response to HDAC treatment. Expression levels of several genes derepressed in lug-3 mutant plants were measured by RT-PCR in wild-type and lug-3 mutant plants grown to the seedling stage. Seedlings were treated with TSA before being harvested, and RNA was isolated in order to determine the expression levels of target genes. When wild-type seedlings were treated with TSA, the expression level of AtEXP10 and RAP24 was increased to the same level measured in lug-3 mutants with or without exposure to TSA (Fig. 1A). This suggests that the mechanism utilized by LUG to repress transcription of AtEXP10 and RAP24 is largely HDAC dependent. For DDB1B and At4g15260, treatment of wild-type plants with TSA resulted in relatively little derepression of transcription compared to lug-3 mutants (Fig. 1B). These effects were not due to changes in the expression of LUG since LUG levels remained unchanged after HDAC inactivation (data not shown). It therefore appears that both HDAC-dependent and -independent mechanisms are involved in the regulation of DDB1B and At4g15260. Alternatively it is possible that secondary, downstream effects resulting from the constitutive loss of LUG or HDAC activity are responsible for changes in gene expression.
FIG. 1.
HDAC-dependent and -independent regulation of in vivo LUG targets. TSA treatment reveals HDAC-dependent (A) and HDAC-independent (B) LUG target genes. Shown are results of real-time PCR analysis of reverse-transcribed mRNA from Ler and lug-3 mutant seedlings 6 h post-TSA treatment. Relative quantification of gene expression data was carried out using CT values for each sample normalized to ACTIN2 expression levels. Normalized values for lug-3 expression levels are given as change (n-fold) relative to values for Ler seedlings.
Transcription repression by LUG involves direct interaction with HDACs.
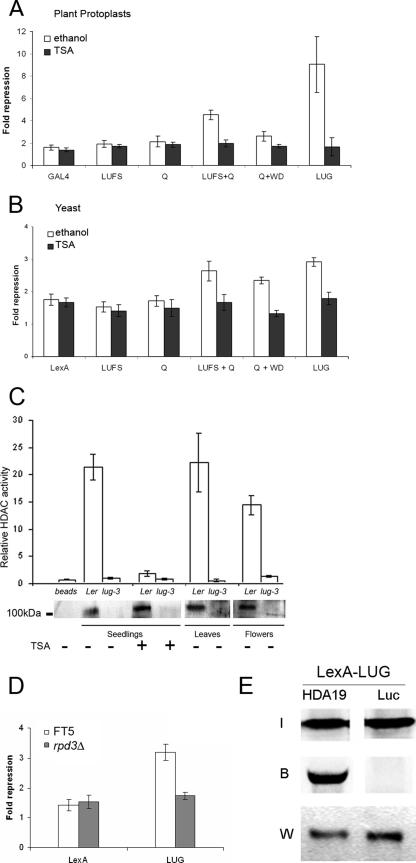
Having demonstrated that one mechanism utilized for the repression of in vivo LUG targets requires HDAC activity, we sought to further investigate this HDAC dependency. In Arabidopsis, the repression activity of LUG, when recruited to a test promoter via SEU, can be suppressed by treatment with the HDAC inhibitor TSA (27). To determine whether this loss of repression activity was due to a direct effect of TSA on LUG activity and not due to an indirect effect which may disrupt the interaction between LUG and SEU, we tested the effect of TSA directly on LUG repression function. Repression assays in Arabidopsis protoplasts transformed with full-length LUG and truncated LUG derivatives (27) revealed that LUG, and the two derivatives LUFS+Q and Q+WD, previously shown to possess repression activity, failed to repress transcription in the presence of TSA when directly recruited to the test promoter pJC1 (Fig. 2A). Similarly the loss of repression activity due to TSA exposure was observed in a heterologous yeast repression assay for full-length LUG and the LUFS+Q and Q+WD repression domains (Fig. 2B). These results support a mechanism in which HDAC activity is directly involved in repression by LUG, in what appears to be a highly conserved process.
FIG. 2.
LUG interacts functionally and physically with HDACs to repress transcription. (A) LUG repression activity in plant protoplasts is abolished by TSA treatment. Arabidopsis leaf protoplasts were transfected with GAL4-LUG, GAL4-LUG derivatives, or GAL4-only effector plasmids plus reporter plasmid pJC1 and treated with 20 μM TSA (dark bars) or ethanol (white bars) for 6 h before fluorescence levels were determined relative to untransformed controls (blank). (B) LUG repression activity in yeast is abolished by TSA treatment. Yeast strain FT5 was transformed with the reporter vector pJK1621 (16) together with the indicated LexA-LUG derivatives (27) or LexA only. Individual transformants were treated with 20 μM TSA (dark bars) or ethanol (white bars) and grown overnight in liquid medium to early log phase (OD600 of <1) before β-galactosidase activity was determined. (C) LUG copurifies with an HDAC activity. Nuclear proteins isolated from Ler and lug-3 mutant leaves, flowers, and seedlings (with or without 6 h of TSA treatment) were incubated with immobilized anti-LUG antibody to immunopurify LUG-associated proteins, and the immunoprecipitate was either analyzed for HDAC activity relative to the negative bead-only control (top) or immunoblotted and probed with anti-LUG antibody (bottom). (D) LUG requires RPD3 to repress transcription. Yeast strains FT5 and FT5::rpd3Δ were transformed with the reporter vector pJK1621 (16) together with full-length LexA-LUG (27) or LexA only. Individual transformants were assayed as described for panel B. (E) LUG interacts with HDA19 in vitro. LexA-LUG was immunoprecipitated from yeast whole-cell extracts and incubated with [35S]methionine-labeled HDA19 or Luciferase (Luc). Input (I) and bound (B) 35S-labeled proteins were visualized using a phosphorimager. LUG was detected using an anti-LexA antibody (W).
Inhibition of LUG repression activity by the HDAC inhibitor TSA suggested that LUG would be directly associated with HDAC(s) in vivo. In order to provide direct evidence of such an interaction, LUG was immunoprecipitated from plant nuclear extracts and assayed for HDAC activity. Nuclear extracts were isolated from wild-type or lug-3 mutant vegetative tissue, flowers, or seedlings grown either in the presence or in the absence of TSA. LUG was then immunoprecipitated from these samples using an anti-LUG antibody, and the presence of LUG in the immunoprecipitate was verified by protein blotting (Fig. 2C, bottom). A band of approximately 100 kDa corresponding to LUG protein was detected in all samples prepared from wild-type plants but was absent in lug-3 mutant-derived samples or in the negative (bead-antibody) control. Immunoprecipitated complexes were then assayed directly for HDAC activity (Fig. 2C, top). In wild-type vegetative tissue, flowers, and untreated seedlings high levels of HDAC activity were detected, demonstrating that HDACs were directly associated with LUG, an association that required functional LUG, as samples prepared from lug-3 mutant tissue displayed no HDAC activity. Such an activity was also absent from TSA-treated wild-type seedlings, suggesting that the inhibitor copurified with the LUG complex and remained associated with HDACs during the assay (Fig. 2C, top).
Eukaryotic transcription corepressors including Sin3, Groucho, and Tup1 have been shown to interact with class 1 (Rpd3-like) HDACs to repress transcription. The Arabidopsis genome contains four class 1 HDACs (HDA6, -7, -9, and -19) predicted to be involved in transcription repression (36). In order to determine whether transcription repression by LUG involved class 1 HDACs, we tested the requirement for Rpd3 in a heterologous repression assay (27). When transformed into yeast cells harboring a deletion in RPD3, the repression function of LUG was lost completely, demonstrating the requirement for this class of HDAC by LUG (Fig. 2D).
Recent reports have highlighted the involvement of HDA19 in biological processes that have also been associated with LUG, and hda19 mutants display phenotypic similarities to lug mutants (13, 20, 30, 31). We argued that these observations could be due to a functional interaction between LUG and HDA19 and that HDA19 could account for the LUG-associated HDAC activity. We therefore tested whether LUG interacted directly with HDAC19 in vitro. Epitope-tagged LUG was immunoprecipitated from whole-cell extracts, and the bead-immobilized protein was incubated with [35S]methionine-labeled HDA19. After extensive washing HDA19 remained associated with LUG, demonstrating a direct physical interaction between these two proteins (Fig. 2E), although we cannot rule out the possibility that this interaction could be stabilized by copurifying proteins. This result strongly supports the argument that HDA19 will be utilized by LUG to repress transcription. In order to determine the specificity of this interaction, we also tested whether LUG interacted with a second HDAC which did not appear to have any function overlapping with LUG. HDA6 has been described as a putative HDAC with a role in rRNA gene silencing in nucleolar dominance (10). Although we were able to observe a weak interaction with LUG, this interaction was unstable and was lost under conditions in which the HDA19-LUG interaction remained stable (not shown). Together these data demonstrate the specificity of the interaction between HDA19 and LUG, indicating that the HDA19 is likely to be a predominant LUG partner, and effectively rule out HDA6 as a specific LUG partner.
LUG repression function is associated with the Arabidopsis Mediator components AtMED14 (SWP) and AtCDK8 (HEN3).
For some of the in vivo LUG targets tested, repression by LUG appeared to be largely independent of HDAC activity (Fig. 1B). We therefore sought to establish the molecular nature of this HDAC-independent mechanism. Previously it has been shown that transcription corepressors require components of the Mediator complex including Cdk8 (Srb10), Med14 (Rgr1), and Med16 (Sin4) to repress transcription (6, 17), and we reasoned that such a mechanism could also be utilized by Arabidopsis corepressors.
In order to investigate whether LUG could function through Mediator to repress transcription, we first determined whether Mediator was likely to be present in Arabidopsis. Comparative analysis with yeast, Drosophila, mouse, and human Mediator components using full-length protein sequences identified several putative Arabidopsis Mediator components including subunits of the head, middle, and Cdk8 modules of Mediator, as well as MED14, which forms the bridge to the tail module (Table 2), and were named in accordance with adopted convention (4). The homology of MED14 is limited to a highly conserved region at the N terminus of the protein (2). These sequences are supported by expressed sequence tag and full-length cDNA sequences, suggesting that a Mediator complex containing the components identified here is present in Arabidopsis. This analysis correlates well with previous highly conserved homology block analysis (3), which has been further annotated here to show the similarity in molecular weights between Arabidopsis and yeast Mediator orthologues. However, the similarity for the tail component MED15 was poor, and MED1, -2, -3, -8, -9, -16, and -19 were not identified, suggesting that an Arabidopsis Mediator will display a degree of structural diversity. Of the Mediator components previously shown to function in transcription repression, two of these, CDK8 and MED14, were identified by comparative analysis, whereas for a third component, MED16, no orthologues were found. We therefore focused on investigating the functions of MED14 and CDK8.
TABLE 2.
Arabidopsis Mediator homologues identified through comparative analysis with eukaryotic Mediator components using BLASTp
| Mediator componenta,b | Module | Arabidopsis gene name | BLAST score (% similarity) | Protein mol wt (103)
|
|
|---|---|---|---|---|---|
| Arabidopsis | Yeast | ||||
| MED6 | Head | At3g21350 | 5e-8 (62) | 28.8 | 32.8 |
| MED7 | Middle | At5g03220 | 2e-14 (50) | 19.4 | 25.6 |
| MED10 | Middle | At5g41910 | 1e-8 (50) | 20.7 | 17.9 |
| MED14 | Tail | At3g04740 (SWP) (2) | 1e-24 (52)e | 185.5 | 123.4 |
| MED21 | Middle | At4g04780 | 2e-14 (50) | 42.7 | 16.1 |
| MED31 | At5g19910 | 3e-12 (69) | 22.8 | 14.7 | |
| CDK8 | CDK8 | At5g63610 (HEN3) (35) | 1e-69 (63) | 52.8 | 62.8 |
| MED4 | Middle | At5g02850 | bsd | 46.2 | 32.2 |
| MED11 | Head | At5g63480 | bs | 19.8 | 15.2 |
| MED12 | CDK8 | At4g00450 (CRPc) | bs | 237.5 | 166.9 |
| MED13 | CDK8 | At1g55325 | bs | 208.5 | 160.0 |
| MED17 | Head | At5g20170 | bs | 73.5 | 78.5 |
| MED20 | Head | At4g09070 | bs | 25.1 | 22.9 |
| MED22 | Head | At1g16430 | bs | 16.2 | 13.9 |
Components in bold are essential for viability in yeast.
MED1, -2, -3, -5, -8, -9, -15, -16, and -19 were not identified (search criteria for alignment score and predicted molecular weight not met).
CRP, CRYPTIC PRECOCIOUS, GenBank annotation.
bs, peptide sequence block identified in reference 3.
N-terminal region amino acids 39 to 284.
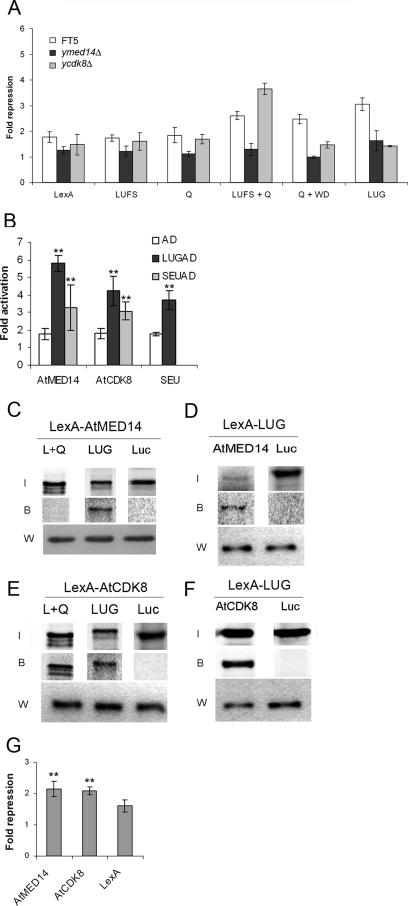
MED14 is an invariant component of Mediator, which in yeast is required for the repression of certain genes (6). Due to the conserved repression function of LUG in heterologous yeast repression assays we tested whether there was a requirement for yMED14 (RGR1) in LUG-mediated repression. In a yeast strain harboring a partial deletion of yMED14 it was found that LUG repression activity was impaired (Fig. 3A), showing that a plant transcription repressor can function through a Mediator-dependent mechanism. Both the N- and C-terminal repression domains of LUG (LUFS+Q and Q+WD, respectively) required yMED14 for their repression activity, suggesting that the interaction between LUG and yMED14 occurs through multiple domains of the LUG protein (Fig. 3A). In order to determine whether this requirement for MED14 was due to a physical interaction with LUG, a yeast two-hybrid assay was conducted between LUG and the Arabidopsis MED14 sequence orthologue AtMED14. AtMED14 was obtained as a full-length cDNA clone from the Kazusa DNA Research Institute and cloned into a yeast artificial recruitment vector as a LexA hybrid. When a LUG-activation domain (AD) fusion and AtMED14 were cotransformed into a wild-type yeast strain (FT5) together with the reporter plasmid JK103, a strong interaction between LUG and AtMED14 was observed (Fig. 3B). In the same assay we also recorded an interaction between SEU and AtMED14 (Fig. 3B), suggesting that AtMED14 is likely to form a complex with LUG-SEU. To verify this in vivo interaction, epitope-tagged AtMED14 was immunoprecipitated from whole-cell extracts and incubated with [35S]methionine-labeled LUG and LUFS+Q proteins. Subsequent analysis demonstrated that AtMED14 interacts with full-length LUG but not the N-terminal repression domain (Fig. 3C). This observation supports the direct interaction between LUG and AtMED14. The lack of an interaction between LUFS+Q and AtMED14 suggests that the direct interaction between these two proteins occurs through the C-terminal repression domain of LUG. In the reciprocal experiment epitope-tagged LUG was found to interact with the conserved N-terminal region of SWP (amino acids 1 to 959), indicating that interaction between these two proteins is likely to be through this highly conserved region of MED14 (Fig. 3D). Having established that LUG interacted both functionally and physically with MED14, we tested whether AtMED14 displayed any inherent repression activity that could contribute to the LUG-mediated repression process. When recruited to the constitutively active promoter of reporter plasmid pJK1621 (16), LexA-AtMED14 exhibited repression activity in a heterologous yeast system (Fig. 3G), demonstrating that, like its yeast counterpart, it has a negative effect on transcription.
FIG. 3.
The Arabidopsis Mediator components AtMED14/SWP and AtCDK8/HEN3 are involved in LUG-mediated repression. (A) LUG requires yeast yMED14 and yCDK8 for repression function. Yeast strains FT5 (white bars), FT5::med14Δ (dark bars; viable partial deletion), and FT5::cdk8Δ (light gray bars) were transformed with the reporter vector pJK1621 (4× LexA operator) or the control vector pLG312S (0× LexA operator) together with the indicated LexA-LUG derivatives (27) or LexA only. Individual transformants were grown in liquid medium to an early log phase (OD600 of <1), and β-galactosidase activity was determined. Error bars show standard deviations. (B) LUG interacts with AtMED14 and AtCDK8 in vivo. Interactions were determined using yeast two-hybrid assays. FT5 was transformed with the reporter plasmid JK103 (4× LexA operator sequences-CYC1 minimal promoter-LacZ) plus LexA-AtMED14 or LexA-AtCDK8 together with either AD (AD only; white bars), LUG-AD (dark bars), or SEU-AD (light gray bars) (27). LexA-SEU was used as a positive control for LUG interaction. Error bars show standard deviations. Unpaired t tests comparing bait vector alone with bait plus prey vectors were used to test significance (**, P < 0.01). (C and E) LUG interacts with AtMED14 (C) and AtCDK8 (E) in vitro. LexA-MED14 or LexA-AtCDK8 was immunoprecipitated from whole-cell extracts and incubated with [35S]methionine-labeled LUG, LUFS+Q (L+Q), or luciferase (Luc) proteins. 35S-labeled proteins were visualized using a phosphorimager. LUG was detected using an anti-LexA antibody (W). (D and F) Like panels C and E but using LexA-LUG immunoprecipitated from yeast whole-cell extracts incubated with [35S]methionine-labeled AtMED14 (conserved N-terminal domain) (D) and AtCDK8 (F) or luciferase (Luc). I, input; B, bound. (G) AtMED14 and AtCDK8 display inherent repression activity. Repression activity of LexA-AtMED14 or LexA-AtCDK8 was determined by comparing LacZ activity in FT5 transformed with pJK1621 and activity in FT5 transformed with pLG312S, and significance was determined using unpaired t tests between pJK1621 and pLG312S values (**, P < 0.01). Error bars show standard deviations.
The MED14 homologue identified through our analysis has been previously described as STRUWWELPETER (SWP) (2). In an swp mutant the shoot apical meristems are severely disorganized as a result of the ectopic expression of WUSCHEL (WUS) and a reduction in the expression of SHOOTMERISTEMLESS (STM) (2). Due to the functional interaction between LUG and AtMED14, we tested whether WUS and STM expression levels were altered in lug-3 mutant plants compared to the wild type. In leaf tissue we observed significant changes in expression for both genes that mirrored those of the swp mutant (WUS, 4.5-fold increase; STM, 2.0-fold decrease), and when the genes were tested in flowers significant changes were again observed (WUS, 4.1-fold increase; STM, 2.0-fold increase). AtMED14 levels remained unchanged in the lug-3 mutant, showing that the effect of the lug-3 mutation is not due to the down-regulation of AtMED14 expression (not shown). The identification of common biological targets for LUG and AtMED14 further supports the notion that these two proteins may function in the same molecular complex to repress transcription.
The cyclin-dependent kinase CDK8 has been extensively studied in yeast and mammalian systems, where it has a negative role in transcription (6, 17, 29) and has been shown to associate with specific corepressors (38). An orthologue of CDK8 was identified in our search for Arabidopsis Mediator components. This gene (AtCDK8, At5g63610) has previously been described as HEN3, a weak regulator of AG which is a known target of LUG (35), and a repressor of WUS expression in flowers. The predicted protein sequence of AtCDK8 has a high degree of homology to Srb10 in yeast including the catalytic center (D921 in AtCDK8), suggesting that the kinase activity associated with the protein (35) will function through this conserved region. Based on the functional overlap between AtCDK8 and LUG in regulating AG and WUS and the negative role played by CDK8 in the regulation of transcription, we examined whether there was a requirement for CDK8 in LUG-mediated repression. When assayed in a yeast strain deleted for yCDK8, the repression activity of full-length LUG and the C-terminal repression domain (Q+WD) was found to be impaired; however, for the N-terminal repression domain (LUFS+Q) repression activity was not reduced but enhanced slightly (Fig. 3A). This loss of LUG repression activity in the absence of yCDK8 demonstrates the involvement of a second Mediator component in the function of a plant transcription repressor, and our data suggest that this involvement occurs specifically through the Q+WD repression domain. Based on the functional interaction between LUG and CDK8 a two-hybrid assay was used to establish if AtCDK8 interacted with LUG (Fig. 3B). AtCDK8 was amplified by PCR and cloned into a yeast artificial recruitment vector as a translational fusion with the LexA DNA binding domain. Wild-type yeast cells were transformed with LexA-AtCDK8, LUG-AD, or SEU-AD and the reporter pJK103 and assayed. An interaction was observed between LUG and AtCDK8 (Fig. 3B) and between SEU and AtCDK8 (Fig. 3B), suggesting that the LUG-SEU corepressor may also interact with a plant Mediator through AtCDK8. To confirm the in vivo interaction with LUG, epitope-tagged AtCDK8 was immunoprecipitated from whole-cell extracts and incubated with [35S]methionine-labeled LUG and LUFS+Q proteins. AtCDK8 was found to interact with both full-length LUG and the N-terminal repression domain, demonstrating the likelihood of a direct interaction between the corepressor and the kinase (Fig. 3E). A similar interaction was observed in the reciprocal experiment using epitope-tagged LUG and [35S]methionine-labeled AtCDK8 (Fig. 3F). When tested for any intrinsic repression activity, LexA-AtCDK8 displayed a significant reduction in reporter gene expression (Fig. 3G), which is consistent with its predicted role as a negative regulator of transcription.
The above observations suggest that LUG is likely to utilize a second, previously undescribed repression mechanism that involves direct interaction with components of a putative plant Mediator complex including the negative regulators AtMED14 and AtCDK8. Furthermore, this mechanism may account for the HDAC-independent repression function of LUG observed in this study.
DISCUSSION
In this study comparative analysis of genome-wide transcriptome array data obtained from Arabidopsis plants harboring a mutation in LUG clearly indicates that LUG functions as a transcription corepressor. These observations demonstrate that the role of LUG extends beyond the regulation of AG in floral development. Our data indicate that LUG regulates a number of target genes and is likely to have a role in the response to abiotic and biotic stresses, as well as in plant development and signaling. A specific involvement for LUG in auxin signaling is now emerging. Arabidopsis lug mutants display increased cell expansion (5), plants harboring mutations in lug and seu have altered petal vascular development (12), and the Antirrhinum LUG orthologue STYLOSA shows hypersensitivity towards auxin and polar auxin inhibitors, suggesting reduced auxin transport in these mutants (21). This involvement is consistent with our observations showing a function for LUG in the regulation of the auxin response factors ARFX15 and ETTIN. The regulatory network involving ETTIN appears complex, as ETTIN, which has been shown to interact with SEU (23), was found to repress transcription through auxin response elements in transient protoplast assays. This suggests that ETTIN may recruit SEU to target genes to repress transcription (32, 34). Such a model would require the concomitant recruitment of LUG, as we have previously shown that SEU cannot repress transcription in the absence of LUG (27). Our observation that ETTIN, like ARFX15, is also differentially regulated in the lug-3 mutant suggests that these auxin response factors may function in a negative feedback loop regulating their own expression.
The repression function of LUG appears to be mediated by both HDAC-dependent and -independent mechanisms: inhibition of HDAC function completely derepresses one group of LUG-regulated genes, yet fails to derepress a second group of LUG-regulated genes. The HDAC-dependent repression activity of LUG is due to the association of LUG with an HDAC activity. It appears likely that this activity is provided by HDA19, as LUG interacts with HDA19, but not with a second HDAC, HDA6, when tested in vitro. In plants class 1 HD1/Rpd3-like and also HD2-type HDACs are emerging as important determinants in growth and development (13, 20, 31, 37). While we have demonstrated that HDACs are involved in the regulation of biologically relevant targets of LUG and have shown a specific interaction with HDA19, we cannot rule out that, for the regulation of other LUG target genes, the corepressor may recruit different HD1 or HD2-type HDACs. HDAC-independent LUG repression is likely to function through interactions with AtMED14 and AtCDK8, components of a putative Arabidopsis Mediator complex. These plant proteins, like their orthologues in other eukaryotic systems, have negative roles in transcription (6). The likelihood of an important role for a Mediator complex in Arabidopsis is becoming more evident, revealed here and in other studies by the functional analysis of AtCDK8 and AtMED14 (2, 35) and their interactions with the corepressor LUG. While we have yet to complete the biochemical purification of Mediator from plants using Tap-tagged AtCDK8 and AtMED14, the importance of the functional interactions between LUG, MED14, and CDK8 is clear. If AtMED14 and AtCDK8 do not turn out to be components of a plant Mediator, the regulatory complex that they do form with LUG will be equally important given the role of LUG in several different biological processes.
Through our studies, and those involving other eukaryotic corepressors, it is apparent that a dynamic interaction between two distinct repression mechanisms used by transcription corepressors is likely to exist. One scenario for this interaction would be a temporal pathway whereby transcription is paused by blocking polymerase function via Mediator, which is then followed by stabilization of local chromatin architecture by HDACs. A second would be an additive pathway whereby inhibition of RNA Pol II function through regulating Mediator would result in a partial reduction in target gene expression, with the gene becoming completely shut off following HDAC recruitment by the corepressor. The first scenario would allow for rapid and complete shutoff of transcription, while the second would allow for a more gradual reduction in gene expression.
The observation that the Arabidopsis corepressor LUG interacts with components involved in two distinct mechanisms of transcription repression is an important step towards a more complete understanding of transcription repression in plants. As LUG appears to play a role in several distinct signaling and developmental pathways, the precise characterization of its function in coordinating gene regulation will remain of significant interest.
Supplementary Material
Acknowledgments
We thank Zhongchi Liu and Anuj Bhatt for critical reading of the manuscript, Zhongchi Liu for lug-3 mutant seed, Nurul Hamidi for yeast strain construction, and Oguz Özkeser for SWP cloning. Microarray hybridization was undertaken at the BBSRC GARNET facility at NASC, Nottingham, United Kingdom (8).
This work was supported by Biotechnology and Biology Sciences Research Council grant 58/G16919 to R.S.C.
Footnotes
Published ahead of print on 25 May 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Asamizu, E., Y. Nakamura, S. Sato, and S. Tabata. 2000. A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 7:175-180. [DOI] [PubMed] [Google Scholar]
- 2.Autran, D., C. Jonak, K. Belcram, G. T. Beemster, J. Kronenberger, O. Grandjean, D. Inze, and J. Traas. 2002. Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J. 21:6036-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boube, M., L. Joulia, D. L. Cribbs, and H. M. Bourbon. 2002. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. [DOI] [PubMed] [Google Scholar]
- 4.Bourbon, H. M., A. Aguilera, A. Z. Ansari, F. J. Asturias, A. J. Berk, S. Bjorklund, T. K. Blackwell, T. Borggrefe, M. Carey, M. Carlson, J. W. Conaway, R. C. Conaway, S. W. Emmons, J. D. Fondell, L. P. Freedman, T. Fukasawa, C. M. Gustafsson, M. Han, X. He, P. K. Herman, A. G. Hinnebusch, S. Holmberg, F. C. Holstege, J. A. Jaehning, Y. J. Kim, L. Kuras, A. Leutz, J. T. Lis, M. Meisterernest, A. M. Naar, K. Nasmyth, J. D. Parvin, M. Ptashne, D. Reinberg, H. Ronne, I. Sadowski, H. Sakurai, M. Sipiczki, P. W. Sternberg, D. J. Stillman, R. Strich, K. Struhl, J. Q. Svejstrup, S. Tuck, F. Winston, R. G. Roeder, and R. D. Kornberg. 2004. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14:553-557. [DOI] [PubMed] [Google Scholar]
- 5.Cnops, G., S. Jover-Gil, J. L. Peters, P. Neyt, S. De Block, P. Robles, M. R. Ponce, T. Gerats, J. L. Micol, and M. Van Lijsebettens. 2004. The rotunda2 mutants identify a role for the LEUNIG gene in vegetative leaf morphogenesis. J. Exp. Bot. 55:1529-1539. [DOI] [PubMed] [Google Scholar]
- 6.Conlan, R. S., N. Gounalaki, P. Hatzis, and D. Tzamarias. 1999. The Tup1-Cyc8 protein complex can shift from a transcriptional co-repressor to a transcriptional co-activator. J. Biol. Chem. 274:205-210. [DOI] [PubMed] [Google Scholar]
- 7.Conner, J., and Z. Liu. 2000. LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. USA 97:12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craigon, D. J., N. James, J. Okyere, J. Higgins, J. Jotham, and S. May. 2004. NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 32:D575-D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du, L., and B. W. Poovaiah. 2005. Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437:741-745. [DOI] [PubMed] [Google Scholar]
- 10.Earley, K., R. J. Lawrence, O. Pontes, R. Reuther, A. J. Enciso, M. Silva, N. Neves, M. Gross, W. Viegas, and C. S. Pikaard. 2006. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 20:1283-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eulgem, T., V. J. Weigman, H. S. Chang, J. M. McDowell, E. B. Holub, J. Glazebrook, T. Zhu, and J. L. Dangl. 2004. Gene expression signatures from three genetically separable resistance gene signaling pathways for downy mildew resistance. Plant Physiol. 135:1129-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franks, R. G., Z. Liu, and R. L. Fischer. 2006. SEUSS and LEUNIG regulate cell proliferation, vascular development and organ polarity in Arabidopsis petals. Planta 224:801-811. [DOI] [PubMed] [Google Scholar]
- 13.Gao, M. J., I. Parkin, D. Lydiate, and A. Hannoufa. 2004. An auxin-responsive SCARECROW-like transcriptional activator interacts with histone deacetylase. Plant Mol. Biol. 55:417-431. [DOI] [PubMed] [Google Scholar]
- 14.Guerineau, F., A. Lucy, and P. Mullineaux. 1992. Effect of two consensus sequences preceding the translation initiator codon on gene expression in plant protoplasts. Plant Mol. Biol. 18:815-818. [DOI] [PubMed] [Google Scholar]
- 15.Jefferson, R., T. Kavanagh, and M. Bevan. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keleher, C. A., M. J. Redd, J. Schultz, M. Carlson, and A. D. Johnson. 1992. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68:709-719. [DOI] [PubMed] [Google Scholar]
- 17.Kuchin, S., and M. Carlson. 1998. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol. Cell. Biol. 18:1163-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Z., and E. Meyerowitz. 1995. LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121:975-991. [DOI] [PubMed] [Google Scholar]
- 20.Long, J. A., C. Ohno, Z. R. Smith, and E. M. Meyerowitz. 2006. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312:1520-1523. [DOI] [PubMed] [Google Scholar]
- 21.Navarro, C., N. Efremova, J. F. Golz, R. Rubiera, M. Kuckenberg, R. Castillo, O. Tietz, H. Saedler, and Z. Schwarz-Sommer. 2004. Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131:3649-3659. [DOI] [PubMed] [Google Scholar]
- 22.Pang, Q., J. B. Hays, and I. Rajagopal. 1992. A plant cDNA that partially complements Escherichia coli recA mutations predicts a polypeptide not strongly homologous to RecA proteins. Proc. Natl. Acad. Sci. USA 89:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfluger, J., and P. Zambryski. 2004. The role of SEUSS in auxin response and floral organ patterning. Development 131:4697-4707. [DOI] [PubMed] [Google Scholar]
- 24.Sato, S., H. Kotani, Y. Nakamura, T. Kaneko, E. Asamizu, M. Fukami, N. Miyajima, and S. Tabata. 1997. Structural analysis of Arabidopsis thaliana chromosome 5. I. Sequence features of the 1.6 Mb regions covered by twenty physically assigned P1 clones. DNA Res. 4:215-230. [DOI] [PubMed] [Google Scholar]
- 25.Savenstrand, H., M. Brosche, and A. Strid. 2004. Ultraviolet-B signalling: Arabidopsis brassinosteroid mutants are defective in UV-B regulated defence gene expression. Plant Physiol. Biochem. 42:687-694. [DOI] [PubMed] [Google Scholar]
- 26.Schroeder, D. F., M. Gahrtz, B. B. Maxwell, R. K. Cook, J. M. Kan, J. M. Alonso, J. R. Ecker, and J. Chory. 2002. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12:1462-1472. [DOI] [PubMed] [Google Scholar]
- 27.Sridhar, V. V., A. Surendrarao, D. Gonzalez, R. S. Conlan, and Z. Liu. 2004. Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 101:11494-11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sridhar, V. V., A. Surendrarao, and Z. Liu. 2006. APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133:3159-3166. [DOI] [PubMed] [Google Scholar]
- 29.Sun, X., Y. Zhang, H. Cho, P. Rickert, E. Lees, W. Lane, and D. Reinberg. 1998. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell 2:213-222. [DOI] [PubMed] [Google Scholar]
- 30.Tian, L., and Z. J. Chen. 2001. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98:200-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian, L., J. Wang, M. P. Fong, M. Chen, H. Cao, S. B. Gelvin, and Z. J. Chen. 2003. Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics 165:399-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tiwari, S. B., G. Hagen, and T. Guilfoyle. 2003. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzamarias, D., and K. Struhl. 1994. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature 369:758-761. [DOI] [PubMed] [Google Scholar]
- 34.Ulmasov, T., G. Hagen, and T. J. Guilfoyle. 1999. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96:5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, W., and X. Chen. 2004. HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131:3147-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu, K., K. Malik, L. Tian, D. Brown, and B. Miki. 2000. Functional analysis of a RPD3 histone deacetylase homologue in Arabidopsis thaliana. Plant Mol. Biol. 44:167-176. [DOI] [PubMed] [Google Scholar]
- 37.Wu, K., L. Tian, K. Malik, D. Brown, and B. Miki. 2000. Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22:19-27. [DOI] [PubMed] [Google Scholar]
- 38.Zaman, Z., A. Z. Ansari, S. S. Koh, R. Young, and M. Ptashne. 2001. Interaction of a transcriptional repressor with the RNA polymerase II holoenzyme plays a crucial role in repression. Proc. Natl. Acad. Sci. USA 98:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, Y., and X. Li. 2005. A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1, constitutive 1. Plant Cell 17:1306-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.