Abstract
The human serine protease inhibitor (serpin) gene cluster at 14q32.1 comprises 11 serpin genes, many of which are expressed specifically in hepatic cells. Previous studies identified a locus control region (LCR) upstream of the human α1-antitrypsin (α1AT) gene that is required for gene activation, chromatin remodeling, and histone acetylation throughout the proximal serpin subcluster. Here we show that the LCR interacts with multiple liver-specific transcription factors, including hepatocyte nuclear factor 3β (HNF-3β), HNF-6α, CCAAT/enhancer binding protein alpha (C/EBPα), and C/EBPβ. RNA polymerase II is also recruited to the locus through the LCR. Nongenic transcription at both the LCR and an upstream regulatory region was detected, but the deletion of the LCR abolished transcription at both sites. The deletion of HNF-3 and HNF-6 binding sites within the LCR reduced histone acetylation at both the LCR and the upstream regulatory region and decreased the transcription of the α1AT, corticosteroid binding globulin, and protein Z-dependent protease inhibitor genes. These results suggest that the LCR activates genes in the proximal serpin subcluster by recruiting liver-specific transcription factors and components of the general transcription machinery to regulatory regions upstream of the α1AT gene.
Many mammalian genes are organized into structurally related gene clusters that are expressed in a tissue- or stage-specific manner. These clusters are often controlled by complex transcriptional regulators called locus control regions (LCRs) (4). The serpin gene cluster at 14q32.1 is one such cluster (27). This ∼370-kb region contains 11 serpin genes that are organized into discrete proximal, central, and distal subclusters (27). The proximal subcluster contains four serpin genes, an α1-antitrypsin (α1AT, or SERPINA1) gene, an antitrypsin-related (SERPINA2) pseudogene, a corticosteroid binding globulin (CBG, or SERPINA6) gene, and a protein Z-dependent protease inhibitor (ZPI, or SERPINA10) gene (27). These genes are highly expressed in the liver, but they are repressed in most other cell types (23). The microcell-mediated transfer of human chromosome 14 from nonexpressing cells to expressing cells results in systematic serpin gene activation and chromatin remodeling of the entire locus into an expressing cell-typical chromatin state (34). This process provides an experimental system for studying the regulation of gene expression and chromatin structure within the serpin domain.
The activation of many hepatic genes is mediated by families of liver-specific transactivator genes, including those encoding hepatocyte nuclear factor 1 (HNF-1), HNF-3, HNF-4, HNF-6, and the CCAAT/enhancer binding protein (C/EBP). Each of these families contains several members. Furthermore, these transcription factors also regulate the expression of one another, thus forming a complex network that maintains the liver-specific transcription of albumin, α1AT, transthyretin, and α-fetoprotein genes and other hepatic genes (10). α1AT gene activation has been studied in detail previously, and HNF-1, HNF-4, HNF-3β, and HNF-6 have been shown to be involved (3, 35, 37). The data demonstrate that multiple liver-specific transcription factors are required for the activation of the α1AT gene.
More recently, homologous modifications of the chromosomal α1AT locus have identified an LCR that is required for cell-specific gene activation and chromatin remodeling throughout the proximal serpin subcluster. The deletion of five expression-associated DNase I-hypersensitive sites (DHSs) within an ∼8.0-kb genomic DNA segment upstream of the α1AT gene results in a mutant serpin allele that is completely refractory to cell-specific gene activation, DHS formation, and domain-wide histone acetylation (1, 22). Experiments with subdeletions within the 8.0-kb chromosomal segment indicated that the serpin LCR consists of multiple regulatory elements (22).
In this study, we investigated the mechanism of the serpin LCR functions. This investigation was accomplished by detailed mapping of histone acetylation and transcription factor loading at a variety of DNA sites within the proximal serpin subcluster. These sites included serpin gene promoters, intergenic regions, the serpin LCR, and an upstream regulatory region (URR) 20 to 25 kb upstream of the α1AT gene. Our results demonstrate that histones in these regions are hyperacetylated in expressing cells. However, the domain of histone acetylation is discontinuous, as hyperacetylation in the region between the α1AT and CBG genes was not observed. Significantly, the highest concentrations of liver-specific transcription factors, including HNF-3β, HNF-6α, C/EBPα, and C/EBPβ, were found not at serpin gene promoters but at the DHS of kb −7.5 (relative to the α1AT start site) of the serpin LCR. The kb −20.8 DHS of the URR was also highly enriched with HNF-6α, C/EBPα, and C/EBPβ, and the deletion of a 461-bp core element within the serpin LCR eliminated factor binding at the kb −20.8 URR site. These data suggest that the recruitment of transcription factors to the LCR and the URR is important for gene activation and chromatin remodeling in the proximal serpin domain. Interestingly, RNA polymerase II (Pol II) loading and nongenic transcription at these sites were also observed.
MATERIALS AND METHODS
Cell lines and culture conditions.
F(14n)14 and F(461wt n)9 are independent rat hepatoma microcell hybrids that contain a single, wild-type copy of human chromosome 14, and R(h14n)6 is a rat fibroblast microcell hybrid that also contains a wild-type copy of human chromosome 14 (34). F(Δ8.0)1, F(Δ8.0)6, and F(Δ8.0)15 are independent microcell hybrid clones that contain a mutant human chromosome 14 [F(Δ8.0/−8.4)] that lacks an 8.0-kb segment of genomic DNA just upstream of the hepatic α1AT gene promoter (22). The cells were grown in 1:1 Ham's F12-Dulbecco's modified Eagle's medium with 5% fetal bovine serum and 500 μg of Geneticin/ml.
Generation of F(Δ461/−7.8) mutant cells. (i) Generation of mutant allele.
The following primer pairs were used to generate the DNA homology segments for the p(Δ461/−7.8) targeting vector: a proximal arm forward primer, CTCGAGCTGCCATCAGAACAACAGGCA; a proximal arm reverse primer, CTCGAGACCAGCTTCCTTCTTTG; a distal arm forward primer, GTCGAGGATAGATGTGATATCCAAGGCATTTAAAT; and a distal arm reverse primer, GTCGAGCACTAACATCCCCACATAAGACTTTTG. Amplified arms for homology segments were initially cloned into pCR2.1-TOPOII (Invitrogen). The primers contained either a SalI or an XhoI tail (underlined) that was used for cloning into the SalI or XhoI site of the pLAHL-PGK-DipA targeting vector (22). A unique PvuI site in the vector backbone was used to linearize the recombination substrate prior to electroporation into D(h14n)F cells (22).
(ii) DNA transfections.
Stable and transient transfections were performed as described previously (6). Stable transfectants were selected in DT40-conditioned medium supplemented with fetal bovine serum, chicken serum, tryptone phosphate, and 1.5 mg of l-histidinol (Sigma)/ml.
(iii) Nucleic acid isolation and blot hybridization.
Genomic DNA was isolated and Southern blotting was performed as described previously (22, 32).
(iv) Excision of the histidinol selection cassette.
Cells were transfected with a bicistronic plasmid encoding Cre recombinase and enhanced green fluorescent protein. After 24 h, >104 enhanced green fluorescent protein-positive cells were harvested using a Vantage SE turbo fluorescence-activated cell sorter. The pooled cells were grown in DT40-conditioned medium.
(v) Chromosomal transfer.
The transfer of the modified human chromosome 14 from DT40 (Neor Ouas) cells to Fao-1 (Neos Ouar) cells was carried out as described previously (22). The presence of an intact chromosome 14 was confirmed by sequence-tagged site marker analysis and fluorescent in situ hybridization using a total human genomic DNA paint probe (22).
AQPCR analysis of serpin gene expression.
Total RNA (totRNA) was harvested using TRIzol per the protocol of the manufacturer (Invitrogen). totRNA was analyzed and quantitated using the Agilent 2100 bioanalyzer. Three micrograms of totRNA was treated with amplification-grade DNase I (Invitrogen) according to the manufacturer's instructions. cDNA was obtained from the treated totRNA by using the high-capacity cDNA archive kit per the protocol of the manufacturer (Applied Biosystems). Absolute quantitative PCR (AQPCR) was performed with 250 ng of cDNA by using intron-spanning primers and TaqMan MGB probes (Applied Biosystems) specific for human α1AT, CBG, and ZPI genes (see Table S1 in the supplemental material). Expression levels for each totRNA sample were normalized using GeNorm (40) and three internal control genes, the Dicer1 gene and the legumain and inositol 1,3,4-trisphosphate 5/6-kinase genes (catalog no. Hs00271599_m1 and Hs00356546_m1, respectively; Applied Biosystems).
RT assay for nongenic transcription.
Reverse transcription (RT) reactions were performed as described previously (17), with modifications. Briefly, 5 μg of RNA was treated with RNase-free DNase for 15 min at 25°C. The RNA was then reverse transcribed using Superscript II TM and random hexamer primers according to the protocol of the manufacturer (Invitrogen). The cDNA was amplified by real-time QPCR. The difference (n-fold) in the amount of a given target sequence was determined by dividing the amount of the target sequence in the cDNA sample by the amount of the target sequence in genomic DNA. The relative level of transcription of a given target sequence was then obtained by normalizing the difference (n-fold) in the amount of the sequence with the difference (n-fold) obtained for primers within Dicer1 exon 21.
ChIP assays and data analysis.
Histone acetylation and binding of Pol II, HNF-3β, and HNF-6α at specific genomic sites in vivo were studied by chromatin immunoprecipitation (ChIP) as described previously (14). Briefly, 2.5 × 107 cells were cross-linked with 1% formaldehyde for 10 min at room temperature, and chromatin was sheared to 500- to 1,000-bp fragments by sonication. The chromatin was precleared by incubation overnight with protein agarose beads at 4°C. An aliquot of precleared chromatin served as the input sample. The chromatin was subjected to reactions with specific antibodies and was precipitated using protein agarose beads. The eluted chromatin and the reserved input sample were reverse cross-linked by incubation at 65°C overnight. Purified DNA samples were analyzed by real-time QPCR using specific primers and TaqMan probes.
Differences in levels of DNA enrichment in ChIP samples were determined by real-time QPCR with 2.5% of the precipitated sample DNA and 0.02% of the input DNA samples by using an ABI Prism 7900 system (PE Applied Biosystems) (20). The threshold was set automatically to cross a point where the PCR amplification was linear, and the number of cycles required to reach the threshold was recorded and analyzed using Microsoft Excel. The difference (n-fold) in the amounts of a given target sequence precipitated by specific antibodies was determined by dividing the amount of the target sequence in the immunoprecipitated fraction by the amount of the target sequence in the input DNA. The relative level of enrichment with a given target sequence was then obtained by normalizing the difference (n-fold) in the amount of the sequence with the difference (n-fold) obtained for the primer-probe set at a DNA site 3 kb upstream of the Goosecoid gene (5′GSC) (42) or at exon 21 of the housekeeping gene Dicer1 (26).
Antibody reagents.
Anti-acH3 (06-599) and anti-acH4 (06-866) were purchased from Upstate Biotechnology, Lake Placid, NY. Anti-Pol II (sc-899), anti-HNF-3β (sc-6554), anti-HNF-6α (sc-13050), anti-C/EBPα (sc-61), and anti-C/EBPβ (sc-746) were purchased from Santa Cruz Biotechnology, Santa Cruz, CA.
Primers and TaqMan probes.
Primers and TaqMan probes selected using PE Applied Biosystems Primer Express software were obtained from Operon and PE Applied Biosystems, respectively. Amplicons were designed with an average size of 70 bp. The sequences of the primers and TaqMan and TaqMan MGB probes are given in Table S1 in the supplemental material.
RESULTS
The α1AT and CBG genes reside in distinct chromatin subdomains.
As demonstrated previously, the deletion of an ∼8.0-kb chromosomal segment containing the serpin LCR results in a mutant serpin allele that is refractory to cell-specific gene activation, fails to form expression-associated DHSs, and reduces histone acetylation throughout the proximal subcluster (1, 22). To determine whether this region comprises a continuous chromatin domain, we assayed levels of histone acetylation across the entire domain. ChIP experiments were performed using antibodies against histone H3 acetylated at lysine residues 9 and 14 and against all forms of acetyl-histone H4. The immunoprecipitated samples were tested by real-time QPCR using multiple amplicons as depicted in Fig. 1. The level of enrichment of a sample with a particular DNA sequence was normalized according to the level of enrichment with sequences 5′ of the Goosecoid gene, a nonexpressed gene located ∼380 kb from the α1AT gene toward the distal end of human chromosome 14 (27).
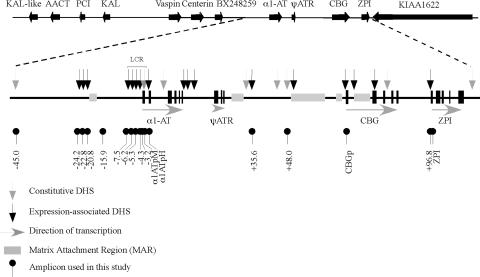
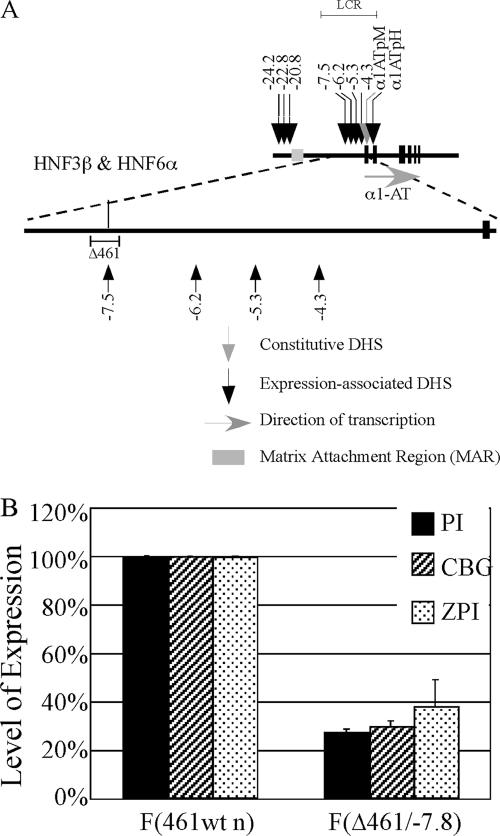
FIG. 1.
Organization of the human serpin gene cluster on chromosome 14q32.1. The serpin cluster in humans on chromosome 14q32.1 is composed of 11 genes (2 pseudogenes and 9 expressed genes). Previous work demonstrated the presence of 5 matrix attachment regions and 17 expression-associated and 8 constitutive DHSs in rat hepatoma cells. The positions of the ChIP and nongenic transcript PCR amplicons used in this study are indicated. KAL, kallistatin gene; AACT, α1-antichymotrypsin gene; PCI, protein C inhibitor gene; ψATR, antitrypsin-related pseudogene; 1ATpM, macrophage α1AT gene promoter; α1ATpH, hepatic α1AT gene promoter; CBGp, CBG gene promoter.
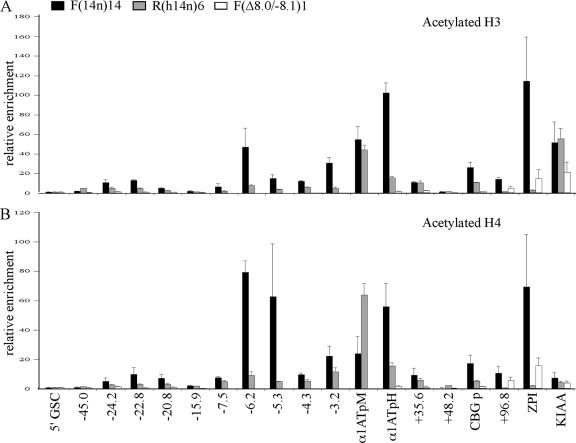
Averages of results from three independent experiments are shown in Fig. 2. In expressing F(14n)14 cells, peaks of histone H3 and H4 hyperacetylation at the kb −6.2 DHS of the serpin LCR, at the macrophage and hepatic promoters of the α1AT gene, and at the CBG and ZPI gene promoters were seen (Fig. 2), consistent with our previous data (1). Lower levels of histone hyperacetylation at the kb −7.5, −5.3, and −4.3 DHSs within the LCR were detected, but the entire LCR was hyperacetylated to some degree. Notably, DNA sequences at kb −3.2, which is between the LCR core (kb −8.1 to −5.8) (22) and the α1AT gene promoters, were also hyperacetylated. This finding suggests that histone hyperacetylation in the region between the serpin LCR and the α1AT gene, which is the first gene of the proximal serpin subcluster (Fig. 1), is continuous. Histones at DHSs in the region from kb −20 to −24 were moderately (5- to 12-fold) acetylated. In contrast, histones in the kb −15.9 region were hypoacetylated in both expressing [F(14n)14] and nonexpressing [R(h14n)6] cells. This finding suggests that the URR at kb −24 to −20 and the serpin LCR at kb −8.4 to −0.3 are separated by a hypoacetylated chromatin zone.
FIG. 2.
Histone acetylation throughout the proximal serpin locus is dependent on the LCR. ChIP was performed with F(14n)14, R(14n)6, and F(Δ8.0/−8.1)1 cells with antibodies to acetylated histone H3 (A) and H4 (B). The averages (± standard deviations) of results from three independent experiments are shown. The relative enrichment (n-fold) with each amplicon was normalized according to the level of the 5′GSC amplicon (see Materials and Methods). Enrichment data for the LCR are absent in F(Δ8.0/−8.1)1 cells as this is the region deleted by the Δ8.0/−8.1 modification. α1ATpM, macrophage α1AT gene promoter; α1ATpH, hepatic α1AT gene promoter; CBG p, CBG gene promoter.
The level of histone acetylation throughout the region in nonexpressing R(h14n)6 cells was generally lower than that in expressing F(14n)14 cells, which demonstrates the tissue specificity of the acetylated chromatin domain. One exception to this rule was the macrophage promoter of the α1AT gene, which showed constitutive hyperacetylation in both cell types. It is noteworthy that the macrophage promoter in both F(14n)14 and R(h14n)6 cells is also constitutively sensitive to DNase I (34). Levels of H3 and H4 acetylation in the KIAA 1622 gene (Fig. 2), which is expressed in both cell types, were also similar.
To more extensively map the histone hyperacetylation domain, we designed and analyzed amplicons throughout the ∼150-kb region (Fig. 1). The most distal amplicon, near the kb −45 DHS, was not hyperacetylated in either cell type (Fig. 2). This finding suggests that the acetylated chromatin domain of the proximal serpin subcluster does not extend to the central subcluster. Histones are hypoacetylated at kb +48 (Fig. 2), a constitutive DHS that lies between the α1AT and CBG genes (Fig. 1). This hypoacetylation suggests that the histone hyperacetylation domain within the proximal serpin subcluster is discontinuous. However, histone hyperacetylation throughout the domain requires the serpin LCR, as hyperacetylation in the region from kb −24 to +100 in F(Δ8.0)1 (Fig. 2) and F(Δ8.0)6 and F(Δ8.0)15 (data not shown) cells, which lack the serpin LCR, was not observed. This finding confirms the dominant function of the LCR in establishing an active chromatin domain in this ∼125-kb region.
Pol II is recruited to the LCR.
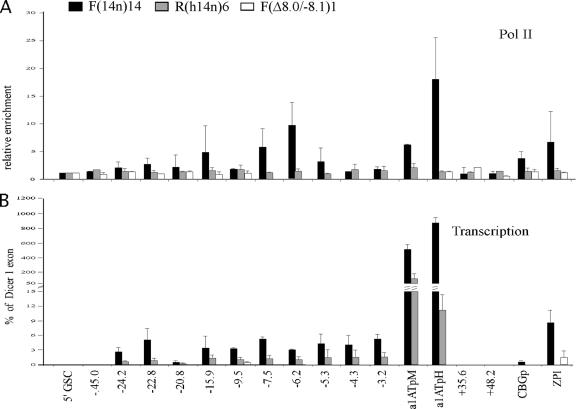
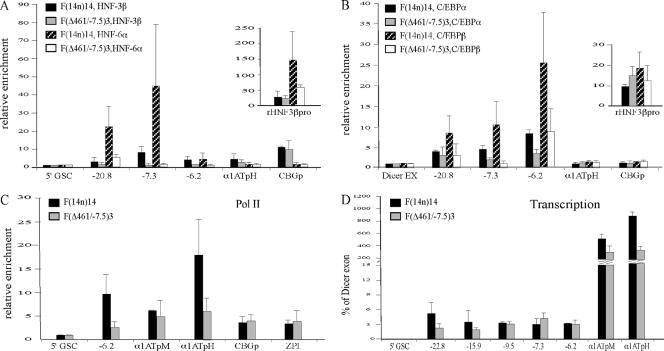
ChIP experiments were performed to determine the distribution of Pol II across the proximal serpin subcluster. As expected, no significant Pol II enrichment in nonexpressing R(h14n)6 cells was detected (Fig. 3A). In contrast, the α1AT gene promoter in F(14n)14 cells was highly enriched (∼20-fold) with Pol II, consistent with the high level of expression of α1AT in this cell type (Fig. 3A). Moderate levels (∼5-fold) of Pol II enrichment at the promoters of the CBG and ZPI genes, which are expressed at levels 50- to 100-fold lower than the α1AT gene, were also observed. Significantly, both the kb −7.5 and the kb −6.2 DHSs of the serpin LCR were enriched ∼5- to 10-fold with Pol II (Fig. 3A). However, Pol II enrichment in the region upstream of the α1AT gene was discontinuous, as the α1AT gene promoter and the serpin LCR were enriched with Pol II but the regions between them were not (Fig. 3A, kb −5.3 and −4.3 amplicons). Similarly, the region just upstream of the LCR (Fig. 3A, kb −9.5 amplicon) was not enriched with Pol II. These results suggest that Pol II is specifically recruited to the kb −7.5 and −6.2 DHSs of the serpin LCR, which is consistent with the observation that core LCR activity lies within the 2.3-kb DNA segment between kb −8.1 and −5.8 (22). It is also possible that these sites may be enriched with Pol II by reduced rates of transcriptional elongation of nongenic transcripts (see below) in these regions. Pol II is also found at the macrophage promoter of the α1AT gene, which is active at low levels in this cell type (Fig. 3A).
FIG. 3.
Pol II distribution and nongenic transcription across the proximal serpin locus. (A) ChIP experiments were performed as described in the legend to Fig. 2, except that antibody to Pol II was used. The relative enrichment is expressed as n-fold. (B) RT-PCR was performed to detect the level of nongenic transcription. The level of nongenic transcription at each of the amplicons is depicted as the percentage of the transcription from the constitutively expressed Dicer1 exon 21 amplicon. Enrichment data for the LCR are absent in F(Δ8.0/−8.1)1 cells as this is the region deleted by the Δ8.0/−8.1 modification. α1ATpM, macrophage α1AT gene promoter; α1ATpH, hepatic α1AT gene promoter; CBGp, CBG gene promoter.
We demonstrated previously that α1AT and CBG expression decreases dramatically when the LCR is deleted (22). To test whether the deletion of the LCR affected the recruitment of Pol II to serpin gene promoters, ChIP assays with F(Δ8.0/−8.4)1 cells, which lack the LCR, were performed. In these cells, Pol II recruitment to the α1AT, CBG, and ZPI gene promoters was abolished (Fig. 3A). Thus, the serpin LCR is important not only for recruitment to the neighboring α1AT gene promoter, but also for Pol II loading at the CBG and ZPI gene promoters, which are ∼67 and 100 kb away, respectively.
Nongenic transcription occurs at the LCR and upstream regions.
To determine whether RNA transcripts were generated at the LCR and other regions of Pol II enrichment, RT-PCR experiments were performed. The enrichment of cDNA samples with specific target sequences was tested by real-time QPCR, and the levels of enrichment were normalized according to the levels of enrichment with sequences at exon 21 of the human Dicer gene, a constitutively expressed locus which is ∼700 kb toward the distal end of human chromosome 14. Normalization with levels of other gene sequences yielded similar results. In F(14n)14 cells, transcription at the LCR core was detected, with levels ∼6 to 10% of those at Dicer exon 21 (Fig. 3B, −7.5 and −6.2). Notably, nongenic transcripts at the kb −4.3 DHS and kb −3.2 were also detected (Fig. 3B), although these regions were not significantly enriched with Pol II (Fig. 3A). This finding suggests that there may be progressive transcription between the LCR core and the α1AT gene promoter. Nongenic transcripts at the kb −24.2, −22.8, −15.9, and −9.5 sites were also detected (Fig. 3B). This finding suggests that nongenic transcripts in F(14n)14 cells are generated over a broad region. Nongenic transcription in R(h14n)6 cells was also detected, but the levels were ∼3- to 5-fold lower than those in F(14n)14 cells. Nongenic transcripts in F(Δ8.0/−8.4)1 cells were not detected (Fig. 3B), indicating that the nongenic transcription we observed is LCR dependent.
The serpin LCR functions as a recruitment center for transcription factors.
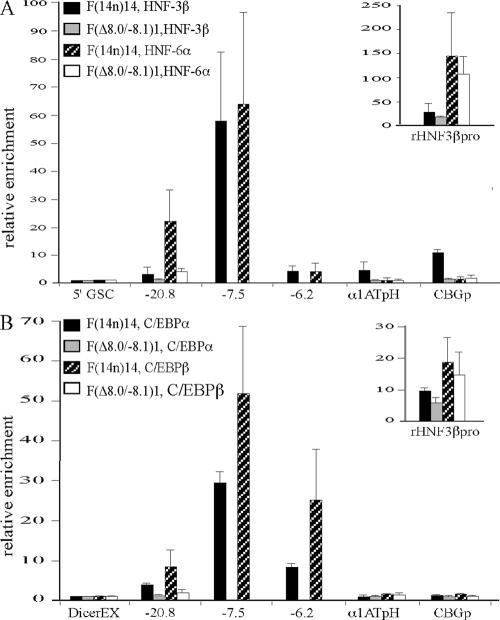
Hepatic genes are regulated by a set of liver-specific transcription factors. For example, HNF-3 and HNF-6 bind to the transthyretin, HNF-3β, and α1AT gene promoters in vitro, and this binding activates linked reporter gene expression in transfection assays (3, 35). Transcriptional activation of the α1AT gene also requires the binding of HNF-1α and HNF-4α (13, 33) to the α1AT gene promoter/enhancer (37). ChIP studies have shown that HNF-6 and C/EBPα and C/EBPαβ are bound at the mouse HNF-3β gene promoter in vivo (44).
LCRs and enhancers generally activate target genes by recruiting transcriptional activators. To determine whether the serpin LCR functions in this way, we assayed the binding of different liver-specific transactivators to the locus by using ChIP. In these experiments, amplicons 5′ of the Goosecoid gene or within Dicer1 exon 21 served as negative controls, and an amplicon within the rat HNF-3β gene promoter was the positive control. Results from three independent experiments are summarized in Fig. 4. As expected, the rat HNF-3β gene promoter in F(14n)14 cells was highly enriched with HNF-3β, HNF-6α, C/EBPα, and C/EBPβ (Fig. 4, insets). Within the human serpin locus, the kb −7.5 DHS of the LCR and the CBG gene promoter were highly enriched with HNF-3β (Fig. 4A). Low-level HNF-3β enrichment at the kb −20.8 DHS, the kb −6.2 DHS, and the hepatic α1AT gene promoter was also found (Fig. 4A). In contrast, HNF-6α was found only at the kb −7.5 and −20.8 DHSs (Fig. 4A). The kb −7.5 and −6.2 DHSs of the serpin LCR were highly enriched with C/EBPα and C/EBPβ, and the kb −20.8 DHS was less so (Fig. 4B). These results suggest that the LCR is a center for the recruitment of liver-specific transcription factors. Consistent with this view, the deletion of the LCR in F(Δ8.0/−8.1)1 cells abolished the binding of all four transcription factors at other sites in the proximal subcluster (Fig. 4).
FIG. 4.
The LCR is enriched with transcription factors. (A) ChIP experiments were performed as described in the legend to Fig. 2, except that antibodies to HNF-3β and HNF-6α were used. (B) ChIP experiments were performed as described in the legend to Fig. 2, except that antibodies to C/EBPα and C/EBPβ were used. The relative enrichment was normalized to the level of the Dicer1 exon 21 amplicon. Enrichment data for the LCR are absent in F(Δ8.0/−8.1)1 cells as this is the region deleted by the Δ8.0/−8.1 modification. The relative enrichment is expressed as n-fold. α1ATpH, hepatic α1AT gene promoter; CBGp, CBG gene promoter; rHNF3βpro, rat HNF-3β gene promoter; DicerEX, Dicer1 exon 21.
Establishment of the histone acetylation domain requires HNF-3β and HNF-6α binding to the LCR.
To test the function of HNF-3β and HNF-6α bound at the kb −7.5 DHS of the LCR, we generated a mutant serpin allele (Δ461/−7.8) in which the core region of the kb −7.5 DHS was deleted specifically (Fig. 5A). Four independent F(Δ461/−7.8) clones carrying this mutant allele showed reduced levels of expression of the α1AT (∼4-fold), CBG (∼4-fold), and ZPI (∼3-fold) genes as assessed by AQPCR (Fig. 5B).
FIG. 5.
Excision of the HNF3β-HNF6α binding region at the kb −7.5 DHS results in decreased expression of the proximal cluster. (A) Illustration of the α1AT gene and the URR, including DHSs and the HNF-6α binding site at kb −7.5. The region removed by the Δ461/−7.8 modification is indicated. α1ATpM, macrophage α1AT gene promoter; α1ATpH, hepatic α1AT gene promoter. (B) The expression of the α1AT (PI), CBG, and ZPI genes in F(461wt n) and F(Δ461/−7.8) cells was measured via AQPCR. Levels of expression were normalized according to that in F(461wt n) cells and displayed as a percentage of that in the control hepatoma cell line. The average levels of expression (± standard deviations) from three independent AQPCR experiments are shown.
ChIP experiments were carried out to define the chromatin landscape of the Δ461/−7.8 mutant allele. As the amplicon at kb −7.5 was deleted in this mutant allele, a new primer-probe set at kb −7.3 was used to monitor this region. The binding of HNF-3β and HNF-6α at this site in wild-type cells was readily detected (Fig. 6A), but the binding of both HNF-3β and HNF-6α at kb −7.3 in the F(Δ461/−7.8) mutant was abolished (Fig. 6A). However, the binding of HNF-3β to the CBG gene promoter was not affected by the Δ461/−7.8 deletion, suggesting that HNF-3β can be recruited to the CBG promoter independently of its binding to the kb −7.5 site of the LCR (Fig. 6A). In contrast, the binding of HNF-6α to the kb −20.8 DHS in the URR was dramatically reduced by the 461-bp deletion. The binding of C/EBPα and C/EBPβ to the LCR and the upstream region was modestly affected (Fig. 6B).
FIG. 6.
The region excised by the Δ461/−7.8 modification is crucial for the recruitment of transcription factors. ChIP experiments were performed (as described in the legend to Fig. 2) with F(14n)14 and F(Δ461/−7.8) cells with antibodies to HNF-3β and HNF-6α (A), C/EBPα and C/EBPβ (B), and Pol II (C). The relative enrichment is expressed as n-fold. (D) Levels of nongenic transcription in F(14n)14 and F(Δ461/−7.8) cells were measured (as described in the legend to Fig. 3). α1ATpH, hepatic α1AT gene promoter; α1ATpM, macrophage α1AT gene promoter; CBGp, CBG gene promoter; rHNF3βpro, rat HNF-3β gene promoter; Dicer EX, Dicer1 exon 21.
Pol II loading at the hepatic α1AT gene promoter and the kb −6.2 DHS of the LCR was decreased two- to threefold in the F(Δ461/−7.8) mutant relative to that in the wild type, but Pol II loading at the macrophage α1AT gene promoter or the CBG or ZPI gene promoter was not affected (Fig. 6C). This finding suggests that the recruitment of Pol II to the LCR core requires HNF-3β and HNF-6α binding at this site. Nongenic transcription in the F(Δ461/−7.8) mutant was largely unaffected (Fig. 6D).
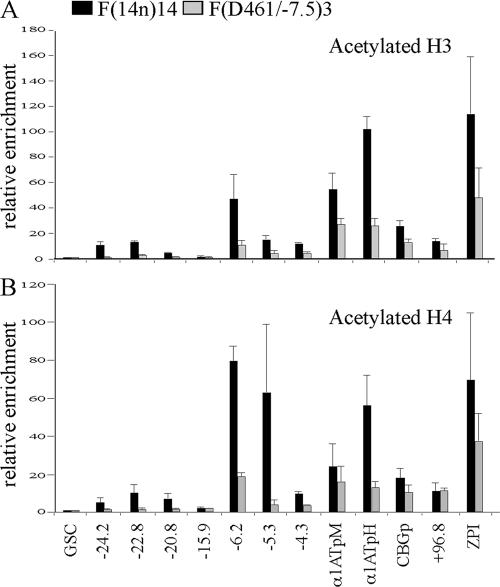
It has been shown previously that HNF-6, C/EBPα, and CBP form a complex in mouse liver and that increased levels of both C/EBPα and HNF-6 proteins are required for CBP association with the mouse HNF-3β gene promoter (44). This finding implies a role for HNF-6 in the recruitment of histone acetyltransferases. To test this possibility, we investigated patterns of histone H3 and H4 acetylation in the F(Δ461/−7.8) mutant. Histone acetylation across the entire proximal serpin subcluster was significantly decreased (Fig. 7). These data demonstrate that the establishment of a domain of histone hyperacetylation within the proximal subcluster requires HNF-3β and HNF-6α binding to the kb −7.5 DHS of the serpin LCR.
FIG. 7.
The Δ461/−7.8 modification results in reduced histone acetylation levels across the proximal subcluster. ChIP analysis of the levels of acetylated H3 (A) and H4 (B) in F(14n)14 and F(Δ461/−7.8) cells was performed. The relative enrichment (n-fold) with each amplicon was normalized to the level of the 5′GSC amplicon (see Materials and Methods). α1ATpM, macrophage α1AT gene promoter; α1ATpH, hepatic α1AT gene promoter; CBGp, CBG gene promoter.
DISCUSSION
LCRs were originally defined based on their ability to induce integration site-independent and copy number-dependent expression of linked genes in transgenic mice (18). These gene activation phenomena are generally correlated with alterations in chromatin structure, for example, increased nuclease accessibility and posttranslational histone modifications (25). It has therefore been assumed that LCRs provide a dominant chromatin-opening activity (4) that is required for gene expression. Consistent with this view, the deletion of one DHS in the human growth hormone (hGH) LCR results in decreased transgene expression and the loss of histone acetylation in an ∼32-kb domain (11). Moreover, the deletion of the 5′ HS3 core region of the β-globin LCR affects DNase I accessibility and reduces histone acetylation throughout the locus in transgenic mice (7). However, deletions of the mouse chromosomal β-globin LCR reduce β-globin gene expression, but the chromatin structure of the locus is largely unaffected (2, 36). These results suggest that the β-globin LCR affects gene expression by a mechanism that functions downstream of activation-specific alterations in chromatin structure, possibly at the level of RNA elongation (36). In marked contrast, chromosomal deletions in the serpin LCR abolish gene expression, DHS formation, and histone acetylation across the entire proximal serpin subcluster, a genomic region of ∼130 kb (1, 22). These results suggest that LCRs function in a variety of different ways in vivo.
Although the α1AT, CBG, and ZPI genes are expressed at different levels, the transcription of all three genes is LCR dependent. In liver cells, α1AT mRNA is abundant, constituting ∼1% of Pol II transcripts (5), while the CBG and ZPI genes are expressed at much lower levels (unpublished data). Levels of histone acetylation also vary across the locus, with the highest levels at the hepatic α1AT gene promoter, the serpin LCR, and the ZPI gene promoter. However, as shown both in a previous study (1) and in the present study, the zone of histone hyperacetylation across the proximal serpin subcluster is discontinuous, as there are areas of hypoacetylation between genes. This finding suggests that the differential transcription of the α1AT gene versus the CBG and ZPI genes may be due, at least in part, to the residence of these genes in different chromatin subdomains. Further studies of the region using high-density probe arrays may help to resolve this issue. Whether proximity to the LCR and/or the presence of intervening matrix attachment regions also affects gene activity in the region is presently unknown. In any event, the activation domain(s) that includes the proximal serpin subcluster does not extend to the kb −45 DHS upstream of the locus or to the KIAA 1622 gene downstream.
There is increasing evidence that the activation of target genes through distal enhancer and/or LCR elements is mediated by the recruitment of transcriptional machinery components. For example, an enhancer 25 kb away from the pDβ1 gene in the T-cell receptor β locus binds Pol II (38). Moreover, Pol II is recruited to the upstream enhancer of the androgen-responsive prostate-specific antigen gene in a hormone-dependent but promoter-independent manner (21). Pol II is also found at DHSs within the β-globin LCR in both mouse and human (15, 41). More recently, it was shown that the β-globin genes localize to active Pol II foci in expressing cells and that the β-globin LCR is required for colocalization (28, 30). These findings are consistent with the notion that during transcription, genes are localized to discrete nuclear sites of concentrated RNA polymerase. In the present study, we found that not only serpin gene promoters but also the serpin LCR is enriched with Pol II. Furthermore, deletions of the LCR abolished Pol II binding to the hepatic α1AT gene promoter, suggesting that the promoter alone is not sufficient for the recruitment of Pol II at its native chromosomal site. LCR deletions also abolished Pol II binding near the CBG and ZPI gene promoters, which are 67 and 98 kb away, respectively. These results suggest that the serpin LCR plays a fundamental role in recruiting Pol II to the locus, and genes both nearby and at a distance require Pol II binding at the LCR for their subsequent activation. It will be interesting to determine whether the LCR can recruit Pol II by itself or whether it requires serpin promoters for this activity, possibly via physical interactions between transcriptional complexes at the sites that interact at a distance.
Nongenic transcription in the LCRs of several loci, including β-globin (19), hGH (12), and major histocompatibility class II (24) genes, has been detected previously. However, the functional role of these nongenic transcripts in long-range gene activation remains obscure. For β-globin transgenes, nongenic transcription across the locus has been detected (29), and levels of nongenic transcripts correlate with DNase I sensitivity (8). These results suggest that nongenic transcription and chromatin remodeling may be linked events. However, nongenic transcription and H4 acetylation profiles of the human β- globin locus do not correlate in any simple way (9). Moreover, placing a transcription terminator downstream of the hGH LCR reduces the expression of the hGH normal gene, but it does not alter H3 or H4 acetylation patterns (12). Thus, the functions of nongenic transcription, if any, remain to be determined.
In the proximal serpin subcluster, nongenic transcription in the URR and LCR, where the chromatin is hyperacetylated, was detected. Curiously, nongenic transcription at kb −15.9 was also apparent, although histones at this site were hypoacetylated. In contrast, histones were modestly acetylated at the kb +35.6 DHS, but nongenic transcription in this region was not detected. Furthermore, histone acetylation at the LCR in the mutant with a 461-bp deletion was significantly reduced, but nongenic transcription was not affected. Thus, the histone acetylation domain(s) at the proximal serpin cluster is not related strictly to nongenic transcription. These results suggest that the establishment of histone acetylation domains is not simply a result of Pol II elongation.
The core region of the serpin LCR is a 2.3-kb DNA segment that extends from kb −8.1 to −5.8 (22). This region includes two expression-associated DHSs, at kb −7.5 and −6.2. Deletions in this region affect serpin gene expression and DHS formation across the locus (22), and histone hyperacetylation is also affected by the 2.3-kb deletion (unpublished data). ChIP experiments demonstrated that the kb −6.2 DHS within this region was highly enriched with acetylated histones and that C/EBPα and C/EBPβ were also bound at this site. Significantly, the kb −7.5 DHS was highly enriched with HNF-3β, HNF-6α, C/EBPα, and C/EBPβ. The deletion of a 461-bp core element of the kb −7.5 DHS abolished HNF-3β and HNF-6α binding in this region and reduced histone H3 and H4 acetylation across the locus. These results suggest that DNA sequences in the deleted 461-bp segment may serve as a platform for the recruitment of histone acetyltransferases to the locus through HNF-3β and HNF-6α.
The formation of expression-associated DHSs in the region from kb −25 to −20 upstream of the α1AT gene is strongly affected by LCR deletions (22). Here we show that factor recruitment in this region also requires the serpin LCR. For example, the kb −20.8 DHS is enriched with C/EBPα, C/EBPβ, and HNF-6α, but both DHS formation and factor binding at this site are abolished by the 8.0-kb LCR deletion. This finding raises the possibility that the binding of these factors to the kb −20.8 DHS may actually reflect a physical interaction between the LCR and this site by the formation of a chromatin loop (39). Alternatively, it may be due simply to the inaccessibility of sequences in this upstream region to trans-acting factors in the absence of the LCR. An analysis of the 461-bp-deletion mutant allowed us to distinguish between these possibilities. When the kb −7.5 core region, the region most highly enriched with HNF-3β and HNF-6α, is deleted, the kb −20.8 DHS still forms (unpublished data) and this region is enriched with C/EBPα but HNF-6α is no longer bound to this site. Thus, HNF-6α binding at kb −20.8 requires HNF-6 and/or HNF-3 binding at the kb −7.5 DHS of the LCR. Furthermore, there are no HNF-6 binding sites at kb −20.8 that can be recognized in silico, although there are candidate C/EBP binding sites at both kb −20.8 and kb −7.5. As C/EBPα and C/EBPβ can form homo- and heterodimers through leucine zipper sequences at their C termini (16), the serpin URR and LCR may contact each other through dimer formation between C/EBP factors.
The deletion of the URR significantly decreases the expression of the CBG gene but only modestly affects α1AT gene expression (unpublished data), suggesting that the differential activation of α1AT and CBG genes requires distinct combinations of regulatory elements. Interestingly, we found that the URR and the CBG promoter are enriched with HNF-6α and HNF-3β, respectively. It has been shown previously that HNF-3β and HNF-6 can physically interact (31). This finding suggests that the URR may contact the CBG gene promoter through this interaction, which may be an intermediate step during CBG gene activation by the LCR.
Our findings that Pol II is recruited to the LCR and that nongenic transcripts are generated in the LCR are consistent with predictions of the transcription factory model (43). As proposed, the model suggests that if transcribed regions are being sequestered in Pol II foci, both regulatory elements and target genes will be in proximity to one another. Our observation that transcription factors bound at the LCR may also bind to the region from kb −24 to −20 implies that these two regions may interact physically. Studies of interactions between regulatory elements and target serpin genes, as well as colocalization studies of these DNA sequences, should provide information that will be important for understanding long-range gene control.
Supplementary Material
Acknowledgments
We thank Stephanie Namciu and Ann Dean for their comments on the manuscript.
These studies were supported by grant GM26449 from the National Institute of General Medical Sciences.
Footnotes
Published ahead of print on 25 May 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Baxter, E. W., W. J. Cummings, and R. E. Fournier. 2005. Formation of a large, complex domain of histone hyperacetylation at human 14q32.1 requires the serpin locus control region. Nucleic Acids Res. 33:3313-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender, M. A., M. Bulger, J. Close, and M. Groudine. 2000. Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol. Cell 5:387-393. [DOI] [PubMed] [Google Scholar]
- 3.Costa, R. H., D. R. Grayson, and J. E. Darnell, Jr. 1989. Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol. Cell. Biol. 9:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dean, A. 2006. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 22:38-45. [DOI] [PubMed] [Google Scholar]
- 5.Derman, E., K. Krauter, L. Walling, C. Weinberger, M. Ray, and J. E. Darnell, Jr. 1981. Transcriptional control in the production of liver-specific mRNAs. Cell 23:731-739. [DOI] [PubMed] [Google Scholar]
- 6.Dieken, E. S., E. M. Epner, S. Fiering, R. E. Fournier, and M. Groudine. 1996. Efficient modification of human chromosomal alleles using recombination-proficient chicken/human microcell hybrids. Nat. Genet. 12:174-182. [DOI] [PubMed] [Google Scholar]
- 7.Fang, X., J. Sun, P. Xiang, M. Yu, P. A. Navas, K. R. Peterson, G. Stamatoyannopoulos, and Q. Li. 2005. Synergistic and additive properties of the beta-globin locus control region (LCR) revealed by 5′HS3 deletion mutations: implication for LCR chromatin architecture. Mol. Cell. Biol. 25:7033-7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gribnau, J., K. Diderich, S. Pruzina, R. Calzolari, and P. Fraser. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol. Cell 5:377-386. [DOI] [PubMed] [Google Scholar]
- 9.Haussecker, D., and N. J. Proudfoot. 2005. Dicer-dependent turnover of intergenic transcripts from the human beta-globin gene cluster. Mol. Cell. Biol. 25:9724-9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi, Y., W. Wang, T. Ninomiya, H. Nagano, K. Ohta, and H. Itoh. 1999. Liver enriched transcription factors and differentiation of hepatocellular carcinoma. Mol. Pathol. 52:19-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, Y., F. Elefant, N. Cooke, and S. Liebhaber. 2002. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9:291-302. [DOI] [PubMed] [Google Scholar]
- 12.Ho, Y., F. Elefant, S. A. Liebhaber, and N. E. Cooke. 2006. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell 23:365-375. [DOI] [PubMed] [Google Scholar]
- 13.Hu, C., and D. H. Perlmutter. 1999. Regulation of alpha1-antitrypsin gene expression in human intestinal epithelial cell line caco-2 by HNF-1alpha and HNF-4. Am. J. Physiol. 276:G1181-G1194. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, K. D., and E. H. Bresnick. 2002. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods (Duluth) 26:27-36. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, K. D., J. A. Grass, C. Park, H. Im, K. Choi, and E. H. Bresnick. 2003. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol. Cell. Biol. 23:6484-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, P. F. 2005. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J. Cell Sci. 118:2545-2555. [DOI] [PubMed] [Google Scholar]
- 17.Kim, A., and A. Dean. 2004. Developmental stage differences in chromatin subdomains of the beta-globin locus. Proc. Natl. Acad. Sci. USA 101:7028-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Q., K. R. Peterson, X. Fang, and G. Stamatoyannopoulos. 2002. Locus control regions. Blood 100:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ling, J., B. Baibakov, W. Pi, B. M. Emerson, and D. Tuan. 2005. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J. Mol. Biol. 350:883-896. [DOI] [PubMed] [Google Scholar]
- 20.Litt, M. D., M. Simpson, F. Recillas-Targa, M. N. Prioleau, and G. Felsenfeld. 2001. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 20:2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louie, M. C., H. Q. Yang, A. H. Ma, W. Xu, J. X. Zou, H. J. Kung, and H. W. Chen. 2003. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc. Natl. Acad. Sci. USA 100:2226-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsden, M. D., and R. E. Fournier. 2003. Chromosomal elements regulate gene activity and chromatin structure of the human serpin gene cluster at 14q32.1. Mol. Cell. Biol. 23:3516-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsden, M. D., and R. E. Fournier. 2005. Organization and expression of the human serpin gene cluster at 14q32.1. Front. Biosci. 10:1768-1778. [DOI] [PubMed] [Google Scholar]
- 24.Masternak, K., N. Peyraud, M. Krawczyk, E. Barras, and W. Reith. 2003. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 25.Maston, G. A., S. K. Evans, and M. R. Green. 2006. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 7:29-59. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda, S., Y. Ichigotani, T. Okuda, T. Irimura, S. Nakatsugawa, and M. Hamaguchi. 2000. Molecular cloning and characterization of a novel human gene (HERNA) which encodes a putative RNA-helicase. Biochim. Biophys. Acta 1490:163-169. [DOI] [PubMed] [Google Scholar]
- 27.Namciu, S. J., R. D. Friedman, M. D. Marsden, L. M. Sarausad, C. L. Jasoni, and R. E. Fournier. 2004. Sequence organization and matrix attachment regions of the human serine protease inhibitor gene cluster at 14q32.1. Mamm. Genome 15:162-178. [DOI] [PubMed] [Google Scholar]
- 28.Osborne, C. S., L. Chakalova, K. E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065-1071. [DOI] [PubMed] [Google Scholar]
- 29.Plant, K. E., S. J. Routledge, and N. J. Proudfoot. 2001. Intergenic transcription in the human beta-globin gene cluster. Mol. Cell. Biol. 21:6507-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragoczy, T., M. A. Bender, A. Telling, R. Byron, and M. Groudine. 2006. The locus control region is required for association of the murine beta-globin locus with engaged transcription factories during erythroid maturation. Genes Dev. 20:1447-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rausa, F. M., Y. Tan, and R. H. Costa. 2003. Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol. Cell. Biol. 23:437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rollini, P., and R. E. Fournier. 1997. A 370-kb cosmid contig of the serpin gene cluster on human chromosome 14q32.1: molecular linkage of the genes encoding alpha 1-antichymotrypsin, protein C inhibitor, kallistatin, alpha 1-antitrypsin, and corticosteroid-binding globulin. Genomics 46:409-415. [DOI] [PubMed] [Google Scholar]
- 33.Rollini, P., and R. E. Fournier. 1999. The HNF-4/HNF-1alpha transactivation cascade regulates gene activity and chromatin structure of the human serine protease inhibitor gene cluster at 14q32.1. Proc. Natl. Acad. Sci. USA 96:10308-10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rollini, P., and R. E. Fournier. 1999. Long-range chromatin reorganization of the human serpin gene cluster at 14q32.1 accompanies gene activation and extinction in microcell hybrids. Genomics 56:22-30. [DOI] [PubMed] [Google Scholar]
- 35.Samadani, U., and R. H. Costa. 1996. The transcriptional activator hepatocyte nuclear factor 6 regulates liver gene expression. Mol. Cell. Biol. 16:6273-6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubeler, D., M. Groudine, and M. A. Bender. 2001. The murine beta-globin locus control region regulates the rate of transcription but not the hyperacetylation of histones at the active genes. Proc. Natl. Acad. Sci. USA 98:11432-11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 38.Spicuglia, S., S. Kumar, J. H. Yeh, E. Vachez, L. Chasson, S. Gorbatch, J. Cautres, and P. Ferrier. 2002. Promoter activation by enhancer-dependent and -independent loading of activator and coactivator complexes. Mol. Cell 10:1479-1487. [DOI] [PubMed] [Google Scholar]
- 39.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 40.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: RESEARCH0034. [DOI] [PMC free article] [PubMed]
- 41.Vieira, K. F., P. P. Levings, M. A. Hill, V. J. Crusselle, S. H. Kang, J. D. Engel, and J. Bungert. 2004. Recruitment of transcription complexes to the beta-globin gene locus in vivo and in vitro. J. Biol. Chem. 279:50350-50357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wakamiya, M., J. A. Rivera-Perez, A. Baldini, and R. R. Behringer. 1997. Goosecoid and goosecoid-related genes in mouse embryogenesis. Cold Spring Harbor Symp. Quant. Biol. 62:145-149. [PubMed] [Google Scholar]
- 43.West, A. G., and P. Fraser. 2005. Remote control of gene transcription. Hum. Mol. Genet. 14(Spec. no. 1):R101-R111. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida, Y., D. E. Hughes, F. M. Rausa III, I. M. Kim, Y. Tan, G. J. Darlington, and R. H. Costa. 2006. C/EBPalpha and HNF6 protein complex formation stimulates HNF6-dependent transcription by CBP coactivator recruitment in HepG2 cells. Hepatology 43:276-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.