Abstract
More than a dozen trithorax group (trxG) proteins are involved in activation of Drosophila HOX genes. How they act coordinately to integrate signals from distantly located enhancers is not fully understood. The female sterile (1) homeotic (fs(1)h) gene is one of the trxG genes that is most critical for Ultrabithorax (Ubx) activation. We show that one of the two double-bromodomain proteins encoded by fs(1)h acts as an essential factor in the Ubx proximal promoter. First, overexpression of the small isoform FSH-S, but not the larger one, can induce ectopic expression of HOX genes and cause body malformation. Second, FSH-S can stimulate Ubx promoter in cultured cells through a critical proximal region in a bromodomain-dependent manner. Third, purified FSH-S can bind specifically to a motif within this region that was previously known as the ZESTE site. The physiological relevance of FSH-S is ascertained using transgenic embryos containing a modified Ubx proximal promoter and chromatin immunoprecipitation. In addition, we show that FSH-S is involved in phosphorylation of itself and other regulatory factors. We suggest that FSH-S acts as a critical component of a regulatory circuitry mediating long-range effects of distant enhancers.
Drosophila HOX genes control development of body segments via highly restricted expression domains (45). These domains are first established by transiently expressed segmentation genes in early embryos and then maintained in an epigenetically heritable manner by the Polycomb group (PcG) of repressors, and the trithorax group (trxG) of activators (34, 53). Like mammalian promoters that are regulated by distant elements, transcriptional regulation of HOX genes also requires coordinated long-range interactions between the basal transcription machinery assembled around the initiation sites and factors recruited at distant regulatory elements (48). How the epigenetic inheritance imposed by PcG and trxG is integrated into the general framework of such long-range interactions remains unclear. Its elucidation should provide an important model for understanding the regulatory mechanisms of genes under strict developmental control (53).
PcG repressors form at least two types of multimeric complexes that are targeted by sequence-specific binding proteins to a core PcG response element located ∼25 kb upstream of the homeotic gene Ultrabithorax (Ubx) (20, 27). These complexes may block the access of the regulatory elements (58) or modify chromatin by associated histone deacetylase and histone methyltransferase activities (7, 8, 13, 47, 64).
In contrast to the highly targeted activities of PcG repressors, trxG activators appear to employ diverse mechanisms for chromatin remodeling and long-range interactions. For example, trithorax (trx) and absent, small or homeotic discs 1 (ash1) encode histone methyltransferases (2, 6, 44, 67) that are targeted to PcG response elements, promoters, and transcribed regions (9, 50, 51, 56). In addition to these targeted activities, brahma, moira, and osa encode subunits of an ATP-dependent chromatin remodeling complex (10, 12, 63, 68) that can modulate the nucleosome fluidity to provide an open access of regulatory sequences (62). Moreover, kohtalo and skuld encode subunits of the Mediator coactivator complex that can facilitate interactions between distal factors and basal transcription machinery (36, 66).
How signals provided by distal elements are integrated at the Ubx basal promoter remains unclear. The Ubx proximal region has several unique features. Instead of the consensus TATA box in the −30 region, Ubx contains the initiator around +1 and the downstream promoter element around +30, which are frequently found in genes lacking the TATA box in Drosophila and mammals (61). The ability of these elements to support Ubx transcription in vitro and in vivo indicates that they represent an authentic basal promoter (4, 39). However, this basal promoter fails to integrate regulatory signals from distant elements without a proximal region from −200 to −32 (9, 39), revealing a critical requirement for this region in mediating long-range interactions.
Interestingly, this critical proximal region (CPR) contains multiple binding sites for ZESTE and TRITHORAX-LIKE (TRL) proteins (4). The ZESTE sites appear to be particularly important, since CPR activity can be substantially replaced by tandem ZESTE sites (39). Consistent with the transactivating role, zeste was initially identified as required for Ubx expression through transvection, a pairing-dependent effect believed to facilitate the transutilization of the regulatory elements on one chromosome by the promoter on homologous chromosome (18). ZESTE protein can stimulate Ubx transcription in vitro (3) and is necessary for the expression of Ubx transgenes containing subsets of regulatory sequences (39). Paradoxically, zeste is not essential for normal development (22) or for expression of the endogenous Ubx promoter or a Ubx transgene with more complete regulatory sequences (38, 39). The role of zeste is further complicated by the finding that zeste may be involved in Ubx repression (29). Clearly, other factors must be required for the activating effect of the ZESTE sites in the CPR.
The maternal-effect gene female sterile (1) homeotic [fs(1)h] was identified as a transactivator of Ubx by its strong genetic interactions with Ubx, trx, and ash1 mutations (17, 21, 59). However, its direct role in homeotic gene activation has been obscured by complex phenotypes in mutant embryos (28). Sequence analysis indicates that fs(1)h encodes two putative proteins of approximately 120 and 210 kDa. The small isoform FSH-S, containing two widely spaced bromodomains (24, 63) and the extra terminal (ET) domain at its C terminus (40), is identical to the N-terminal half of the large isoform FSH-L (25). Bromodomains can bind acetylated lysine or histones (16, 30) and are frequently found in transcription or chromatin modification factors (70), whereas ET domains are found in a small family of double-bromodomain proteins (BET proteins) with no designated function (40). Several interesting properties have been shown for mammalian BET proteins. For example, human RING3 (or BRD2) is a growth-stimulated nuclear kinase acting on serine and threonine (15). Mouse BRD2-like protein can be copurified with the Mediator transcriptional coactivator complex (32). Recently, mouse BRD4 has been shown to be involved in the recruitment of positive transcription elongation factor b (31, 71).
In this report, we provide several lines of evidence to support a direct role of fs(1)h in homeotic gene activation and the idea that FSH-S is primarily responsible for this function. Furthermore, we show that FSH-S acts directly on the ZESTE site of the CPR. These results support a critical role for FSH-S in integrating signals from distal factors.
MATERIALS AND METHODS
Genetics.
Uβ and Uβ-Z lines containing wild-type Ubx-lacZ and Ubx-lacZ with mutations were described previously (39). UAS-FSH-S or UAS-FSH-L (where UAS is upstream activation sequence) lines were established by standard germ line transformation procedures (54). Four independent lines were tested for their effects on development. For the interactions between trxG mutations with Ubx, females carrying the trxG mutations were crossed to Ubx130/ Dp(3;3)S462+In(3LR)64;84+In(3R)C, Sb males. The progeny receiving both the trxG mutation and Ubx130 were scored for transformations of dorsal metathoracic (metanotum and haltere) to mesothoracic tissues. For the interactions between trxG mutations and fs(1)h, Df(1)C128/In(1)M5 females were crossed to males carrying the trxG mutations. All progeny receiving the trxG mutation were scored for transformations.
Embryo and disc staining.
Wild-type embryos were collected at 25°C. fs(1)h1 embryos with or without transgenes (Uβ or Uβ-Z) were collected as follows: 0- to 3-h embryos laid at 21°C were aged at 21°C for about 1 h and then shifted to 29°C for 5 h. Embryos were fixed and stained with either affinity-purified S1 antibody (1:100 dilution) or digoxigenin-labeled probes by standard protocols (1). Digoxigenin-labeled DNA probes were prepared from Ubx (a 574-bp NotI fragment), cad (a 305-bp SmaI fragment), or lacZ (345- and 385-bp fragments generated by EcoRV, SacI, and BssHII). For the FSH-S probe, antisense RNA was prepared by in vitro transcription from pBT-FSH-S3 by T3 RNA polymerase. Hybridization was carried out at 45°C or 68°C for DNA or RNA probes, respectively.
Tissues from second or third instar larvae were stained with mouse anti-ANTP (11), anti-UBX (69), or a rabbit anti-Flag antibody (1:100 dilution; Sigma-Aldrich). Secondary antibodies were either Rhodamine Red-X conjugated (1:200) or Cy5 conjugated (1:500; Jackson ImmunoResearch). DNA was stained by 0.5 μM SYTOX (Molecular Probe).
Plasmid construction.
DNA fragments from previously characterized FSH-S cDNAs (25) were inserted into pUR series of vectors (55) or the pET15b vector (Novagen) to generate β-Gal/FSH-S or (His)6/FSH-S fusion constructs, respectively. pUR-S1 contains a BstXI/XbaI fragment of cDNA e3.7 (25) in pUR288. pUR-S2 contains an XbaI/PstI fragment of e1.20 in pUR292. pUR-S3 contains a EcoRI fragment of e1.15 in pUR278. FSH-S sequences were recovered from pUR constructs by restriction enzyme cleavages and reinserted into pET15b to make equivalent series. The resulting pET fusion constructs also contain approximately 50 bp of extra coding sequences 5′ to the polylinker of pUR vectors. pET-S1Δ, containing the same sequence as pET-S1 except for the 3′ part of the first bromodomain, was constructed by inserting a PCR fragment amplified from pUR-S1 into the EcoRI/HindIII sites of pET15b (5′ primer, AAGGAATTCCAGAAGATCGAATCGATGCCT; 3′ primer, GGCGTATCACGAGGCCCTTTCGTCTTCAAGAA).
The complete open reading frame (ORF) of FSH-S was assembled from the BamHI/XbaI fragment from e3.7, the XbaI/XhoI fragment was from e1.20, and the XhoI/HindIII fragment was from e1.15 into pGem-5.9. pBT-7.6-3 contains the unique part of FSH-L and C-terminal half of FSH-S. It was constructed in pBluescript (pBT; Strategene) by linking the SacI/SalI fragment from e1.20, the SalI/BsmI fragment from e5.16, and the BsmI/SalI fragment from e4.1. pAct5C-FSH-S was constructed by inserting the BamHI/EcoRI fragment from pGem-5.9 into BamHI/EcoRV sites of pAct5CPPA after repairing the EcoRI end. pAct5C-FSH-L was constructed by inserting the SacI/Asp718 fragment of pBT-7.6-3 into SacI/BglII sites of pAct5C-FSH-S after repairing Asp718 and BglII ends. For tagged FSH-S constructs, the C-terminal end of the ORF in pAct5C-FSH-S was modified by inserting in tandem two pairs of oligonucleotides corresponding to the Flag (5′-TCGACAGATCTGATGGACTACAAGGACGATGACGATAAGAA-3′) and the His6 (5′-CGCGTCCCACCATCACCACCATCACTAGGATC-3′) tags. The BamHI/EcoRI fragment from the resulting construct, pAct5C-FSH-S(FH), was inserted into BamHI/NotI sites of pMK33 to generate pMK33-FSH-S(FH) after repairing EcoRI and NotI sites. For the construction of UAS-FSH-S transgene, the BamHI/HindIII fragment of pAct5C-FSH-S(FH) was inserted into BglII/XbaI sites of pUAST after repairing HindIII and XbaI sites.
The Ubx-CAT ([UC] where CAT is chloramphenicol acetyltransferase) construct contains Ubx sequences from −3142 to +358 in pC4CAT (37). The 5′ deletion mutants of UC were created by deleting from the SalI site in the upstream linker to PstI site at −1762 (5Δ1), to HindIII site at −628 (5Δ2), to SacII site at −226 (5Δ3). The 3′ deletion mutants were constructed by cloning the Sal/BstXI (at +161; 3Δ1), or SalI/BssHII (at +36; 3Δ2) fragment of UC into the SalI/SmaI site of pC4CAT. The 3Δ21, 3Δ22, and 3Δ23 mutants were created from 3Δ2 by deleting upstream sequences from the BamHI site in the linker to the PstI, SacII, and Eco47 (at −127) sites, respectively. InΔ1 was created by cloning the PstI/BssHII fragment from UβΔ (39) to the PstI/SmaI sites of pC4CAT, thus replacing Ubx sequence from −200 to −33 by a 18-bp linker sequence.
Internal deletion mutants of FSH-S were constructed from pAct5C-FSH-S by deleting the following DNA fragments: from BclI to the first BstXI sites (Δ1), SacI to the second BstXI sites (Δ2), and the first to second StuI sites (Δ3). The Δ12 mutant was a combination of Δ1 and Δ2. The 3′ deletion mutants were constructed by deleting the DNA fragments from SalI to the third XhoI sites (Δ4) or from the second to third XhoI sites (Δ6). The Δ5 mutant was constructed by using T4 polymerase to remove the 3′ protruding nucleotides at the SacII site.
DNA fragments used for binding assays were amplified from the Ubx proximal region by PCR and then cloned into the EcoRV site of pBT. They contain the following sequences: Ubx-5 (−230 to −98), Ubx-6 (−118 to +35), Ubx-5a (−230 to −150), Ubx-5b (−171 to −98), Ubx-6a (−118 to −35), and Ubx-6b (−56 to +35). XbaI/XhoI fragments from these plasmids were used for binding or competition assays. DNA fragments containing tandem binding sites for NTF were purified from pUβ NTF-1 plasmid after SacII/BSSHII cleavage (39).
pBT-fsh-S3 contains the 3′ untranslated region (UTR) of FSH-S (nucleotides 5092 to ∼5398) used for in situ hybridization. It was constructed by inserting a 304-bp NsiI/DraIII fragment from e1.15 into the Pst/RV sites of pBT.
pET/ZESTE was constructed in pET15b by inserting downstream of the six-His (His6) tag an SnaBI/BamHI fragment that contains the Flag tag and complete coding sequences of zeste (details will be provided upon request). The ORF was based on the work of Mansukhani et al. (41).
Antibody preparation.
β-Galactosidase fusion proteins were induced from pUR-S1, -S2, and -S3 in the TG1 strain of Escherichia coli by standard protocols, followed by 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (23). Rabbits were immunized with gel slices containing fusion proteins. For the affinity purification of antibodies, His6-tagged proteins induced from pET-S1Δ, -S2, and -S3 in the BL21(DE3) strain of E. coli were purified by Ni-nitrilotriacetic acid chromatography (Invitrogen) and coupled to Affi-gel 10 resin (Bio-Rad). Specific antibodies were eluted from appropriate resins with 0.1 M glycine (pH 2.5) according to standard protocols (23).
Protein purification.
Stable cell lines containing inducible FSH-S were established in Drosophila S2 cells with pMK-FSH-S(FH). The details for cell line establishment and protein purification followed a previously described method (26) with some modifications. Briefly, cells were grown in large T flasks, and the protein induction was carried out by adding 0.7 mM CuSO4 for overnight incubation. Whole-cell extract was prepared from ∼500 ml of cells by extracting the cell lysate with 10% (NH4)2SO4. After centrifugation for 20 min at 204,000 × g, the supernatant was precipitated by 40% (NH4)2SO4. The protein pellet was dissolved in TBS (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 0.1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin, 2 μg/ml pepstatin A), and protein solution was loaded onto a column containing 0.3 to ∼0.5 ml M2 resin (Kodak). The column was washed by 20 to ∼30 volumes of TBS and then eluted with TBS containing ∼375 μg/ml Flag peptide. To each fraction, β-mercaptoethanol was immediately added to a final concentration of 10 mM. Peak fractions were pooled and dialyzed against a buffer containing 25 mM Tris-HCl (pH 7.6), 150 mM NaCl, 0.1 mM EDTA, 10% glycerol, 10 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride.
Recombinant His6-tagged ZESTE protein was induced in the BL21(DE3) strain of E. coli by 0.5 mM isopropyl-β-d-thiogalactopyranoside for 90 min at room temperature. Proteins were purified by Ni-nitrilotriacetic acid chromatography according to the standard protocol (Invitrogen).
DNA binding assays.
For electrophoretic mobility shift assays (EMSAs), XbaI/XhoI fragments from Ubx-5, -6, -5a, -5b, -6a, and -6b plasmids were used as probes after end labeling. Binding reactions were carried out at 30°C for 20 min in 20 mM Tris-HCl (pH 7.8), 60 mM KCl, 0.1 mM EDTA, 1 mM DTT, 0.1% Tween-20, 10% glycerol, 5 μg of bovine serum albumin (BSA), and 0.25 μg of poly(dI-dC) (Pharmacia). Samples were run on a 3.5% polyacrylamide gel in Tris-acetate-EDTA buffer at 4°C for about 2 h at 180 V. The following DNA fragments were used for competition experiments: 30 ng of XbaI/XhoI fragments from Ubx-5 or Ubx-6; SacII/BSSHII fragments from pUβ GAGA, ZESTE or NTF-1 (39); AvaI-digested 123-bp ladder (Gibco); and 0.1 μg of double-stranded oligonucleotides containing TRL (5′-CGTATCTCTCCCTCTCTCCGCAT-3′) or ZESTE (5′-TTCTCGAGTGAGTGCTCGAGTT-3′) binding sites. Supershift experiments were conducted using a 10-min preincubation of purified FSH-S with 1 μl of affinity-purified antibodies before other components were added.
In-gel footprinting assays were carried out by a method using 1,10-orthophenanthroline-copper (OP-Cu) (60). Bound and free probes were eluted from gel slices. Approximately 2,000 cpm of labeled samples was separated on 8% sequencing gels.
Cell culture and transfection assay.
Haploid cell line 1182-4 was maintained in M3-D22 medium, and transfection was carried out in six-well plates as described previously (9, 14). For each well, 5 μg of total DNA was used, containing reporters (0.13 μg of pAntp-P1-CAT, 0.11 μg of pAntp-P2-CAT, 0.09 μg of pUbx-CAT, and 0.01 μg of pHsp70-CAT), effectors (0.18 μg of pAct5C-PPA or 0.25 μg of pAct5C-FSH-S), and variable amounts of pBluescript to make up the final DNA concentration. The molar ratio of the effector to the reporter was 2.4:1 for Antp and Ubx promoters and 17.7:1 for Hsp70 promoter. For the analyses of Ubx deletion mutants or FSH-S mutants, 10 ng of pAct5C-lacZ was included as an internal control. For each experiment, data from three sets of duplicates were averaged.
Chromatin immunoprecipitation assay.
Fifty fs(1)h17 male third instar larvae were dissected in Drosophila M3 medium and fixed by 1% formaldehyde for 20 min, followed by addition of glycine to 0.125 M. After three washes with a buffer containing 10 mM Tris-HCl (pH 8.0) and 150 mM NaCl, imaginal discs and ganglia were suspended in 0.5 ml of lysis buffer (Upstate) and sonicated to shear chromatin to the size range of 300 to ∼800 bp. A 0.1-ml chromatin fraction was used for immunoprecipitation by S3 or L3 antibody or preimmune serum according to the manufacturer's protocol (Upstate). Two percent of the precipitate was used for 35 cycles of amplification with primers that cover following regions of Ubx (GenBank accession number U31961): P1 (positions 243702 to 243264), P2 (243277 to 242767), P3 (242900 to 242486), P4 (242524 to 241898), and P5 (241947 to 241452). Primers for cad cover the region from 20769994 to 20770550 (GenBank accession number NT_033779).
In vitro phosphorylation assays.
In vitro phosphorylation was conducted at 30°C for 1 h in reaction mixtures containing 30 mM HEPES (pH. 8.0), 30 mM MgCl2, 0.5 mM EDTA, 2 mM DTT, 0.1% NP-40, 10 μM ATP, and 3 μCi of [γ-32P]ATP. Alternatively, 0.1 mM ATP was used (see Fig. 8D). For dephosphorylation, FSH-S was treated with 2 units of calf intestine phosphatase as described by the manufacturer (New England Biolab). Phosphoamino acids were determined by acid hydrolysis of labeled proteins, followed by high-voltage two-dimensional electrophoresis in an HTLE 7000 chamber (CBS Scientific). For cross-linking assays, 1 μl of FSH-S was incubated with 1.25 mM 5′-p-flurosulfonylbenzoyl adenosine (FSBA) for 20 min at 30°C. For competition assays, 7.5 mM ATP was added. FSBA-conjugated proteins were detected by an anti-FSBA antibody (Roche Applied Science).
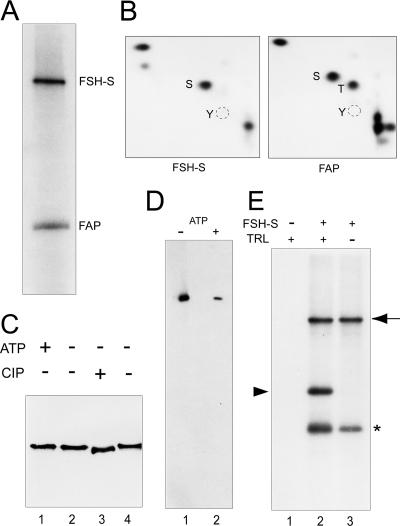
FIG. 8.
Characterization of a kinase activity of FSH-S. (A) Kinase assays. The kinase reaction was carried out with purified FSH-S and [γ-32P]ATP. Phosphorylated FSH-S and FAP56 are indicated. (B) Phosphoamino acid analysis. Following the transfer to the polyvinylidene difluoride membrane, labeled FSH-S and FAP56 proteins were excised and subjected to phosphoamino acid analysis. The positions of phosphoserine (S), phosphothreonine (T) or phosphotyrosine (Y) are indicated. (C) FSH-S as a phosphoprotein. Purified FSH-S was either phosphorylated with (lane 1) or without 0.1 mM ATP (lane 2) or dephosphorylated with (lane 3) or without (lane 4) 2 units of calf intestine phosphatase. Following 5% SDS-PAGE, FSH-S was detected by the anti-Flag antibody. (D) ATP binding assays. Purified FSH-S was incubated with 1.25 mM FSBA in the absence (lane 1) or presence (lane 2) of 7.5 mM ATP, followed by immunoblotting with an anti-FSBA antibody. (E) TRL as a kinase substrate. Kinase reactions were performed with purified FSH-S (lanes 2 and 3) and 0.1 μg of purified recombinant TRL (lanes 1 and 2). Phosphorylated FSH-S (arrow), TRL (arrowhead), and FAP56 (asterisk) are indicated.
RESULTS
Requirement of fs(l)h for active Ubx expression.
While many trxG mutations were identified by their suppressing effects on specific homeotic phenotypes caused by PcG mutations (36), their contributions to regulation of individual HOX genes have not been systematically examined. To address this issue, we examined the effect of mutations of 18 trxG genes on homeotic phenotypes caused by reduced Ubx expression, i.e., transformation of the third to second thoracic segment in adult flies. Interestingly, only fs(1)h [i.e., Df(1)C128], trx, and ash1 mutations showed strong enhancement on Ubx130 phenotypes (increased from <1% to ∼10%) (Table 1). Other trxG mutations showed weak or no effects on Ubx130 mutation, despite that many could suppress PcG phenotypes as strongly as trx mutations (36). Thus, Ubx activation appeared to be highly sensitive to the dosages of fs(1)h, trx, and ash1. This selective effect was further supported by genetic interactions between fs(1)h and other trxG mutations. Again, fs(1)h showed strong synergistic effects with trx (>20%) and ash1 mutations (∼10%) on Ubx phenotypes (Table 1). By contrast, it showed weaker or no interactions with other trxG mutations. These results strongly suggest that fs(1)h, trx, and ash1 share some common role in certain critical steps of Ubx activation.
TABLE 1.
Genetic interactions between trxG mutations, Ubx, and fs(1)h
| trxG mutation | Frequency (%) of T3 to T2 transformation (no. of transformations/no. of scored flies)a
|
|
|---|---|---|
| Ubx130 | Df(1)C128 | |
| Oregon R | <1 (1/1,456) | <1 (1/1,170) |
| Df(1)C128 | 14 (49/352) | Lethal |
| trxE2 | 10 (20/201) | 36 (109/300) |
| trxB22 | 13 (28/209) | 24 (49/207) |
| Df(3)trxE12 | 11 (29/260) | 24 (51/210) |
| ash16 | 4 (9/219) | 7 (19/273) |
| ash1B1 | 15 (32/211) | 9 (21/224) |
| ash21 | 0 (0/291) | 0 (0/323) |
| ash22 | 0 (0/220) | 0 (0/202) |
| brm1 | 1 (2/262) | 2 (6/329) |
| brm2 | 2 (8/533) | 2 (10/621) |
| brm5Df(3L)vtd12 | 2 (5/210) | 1 (2/243) |
| mor1 | 1 (4/603) | 0 (0/255) |
| mor2 | 2 (4/213) | 1 (2/210) |
| mor4 | 0 (0/203) | 0 (0/206) |
| osa1 | 1 (2/213) | 0 (0/249) |
| osa2 | <1 (1/243) | 0 (0/220) |
| osa4 | 0 (0/223) | <1 (1/220) |
| kis1 | 0 (0/239) | 0 (0/223) |
| kis2 | 0 (0/209) | 0 (0/232) |
| kto1 | 0 (0/239) | 0 (0/216) |
| kto3 | 1 (2/209) | 0 (0/205) |
| skd1 | 1 (2/212) | 0 (0/227) |
| skd2 | 3 (11/319) | 0 (0/238) |
| Df(3R)urd | 0 (0/217) | 0 (0/218) |
| urd2 | <1 (1/215) | <1 (1/268) |
| btldev1 | 0 (0/216) | 0 (0/251) |
| btldev2 | <1 (1/216) | <1 (1/414) |
| sls1 | 0 (0/212) | 0 (0/248) |
| Trl3Df(3L)vtd2 | <1 (1/212) | 0 (0/217) |
| Df(3L)vtd3 | 0 (0/226) | 0 (0/211) |
| Vha5512 | 1 (2/223) | 1 (2/267) |
| Vha5516 | 1 (2/236) | 1 (4/468) |
| tna1 | <1 (1/234) | 0 (0/317) |
| tna5 | 0 (0/245) | 0 (0/305) |
| tara2 | 0 (0/227) | 0 (0/316) |
| tara20 | 0 (0/224) | <1 (1/300) |
For Ubx interactions, trxG−/+ females were crossed to Ubx130/+ males. For fs(1)h interactions, Df(1)C128/+ females [fs(1)h deletion] were crossed to trxG−/+ males. T3, third thoracic segment; T2, second thoracic segment.
Loss of fs(1)h function results in complex defects in early embryos, leading to severe body distortion and lethality (19, 28). These defects hamper the analysis of the role of fs(1)h in Ubx activation. To circumvent these problems, we inactivated fs(1)h function by shifting heat-sensitive fs(1)h1 mutant embryos from the permissive temperature (21°C) to the restrictive temperature (29°C) during the onset of gastrulation. In wild-type embryos, high levels of Ubx transcripts can be detected in the ventral nerve cord (VNC) in a domain encompassing parasegments (PS) 5 to 12 (Fig. 1A). In mutant embryos, a marked reduction of Ubx transcripts was seen (Fig. 1B). By contrast, no change was observed for caudal (cad), a HOX gene controlling the development of most posterior segments (46). Thus, fs(1)h appeared to be required for a subset of HOX genes.
FIG. 1.
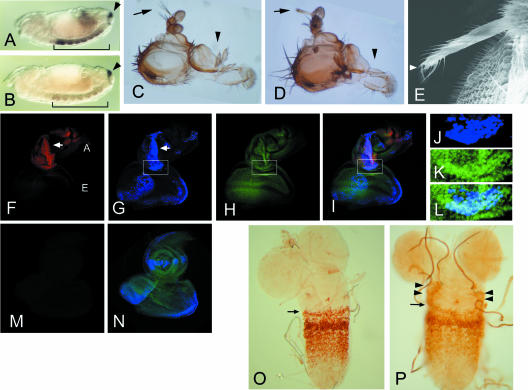
Essential role of fs(1)h1 in HOX gene activation. (A and B) Whole-mount in situ hybridization to reveal Ubx (bracket) and cad (arrowhead) transcripts in the ventral nerve cord of stage 13 embryos. (A) A wild-type embryo showing high levels of Ubx transcripts in PS5 to PS12. (B) An fs(1)h1 embryo showing severely reduced Ubx transcripts. No difference was found for cad transcripts between these embryos. (C to E) Head defects caused by targeted expression of FSH-S driven by dpp-Gal4. A wild-type adult head showing normal aristae (arrow) and maxillary palpus (C, arrowhead). Targeted FSH-S expression resulted in aristae-to-leg transformation and loss of maxillary palpus (D). No abnormality was observed when FSH-L was induced. Anterior is to the top. Scanning electron microscopy of ectopic leg shows a claw indicated by an arrowhead (E). (F to N) ANTP expression in antennal discs with dpp-driven FSH-S (F to L) or FSH-L (M and N). The eye-antennal discs from third instar larvae were stained by anti-ANTP (red in panels F, I, and M), anti-Flag antibodies (blue in panels G, I, J, L, and N), or a DNA dye SYTOX (green in panels H, I, K, L, and N). The lower portion of the antennal disc (boxes in G to I) is shown with higher magnification to reveal the staining of FSH-S (J) and DNA (K). The merged images of multiple staining are shown (I, L, and N). Ectopic ANTP signals were seen in antennal discs with induced FSH-S but not in wild-type discs or discs with induced FSH-L. The antennal (A) and eye (E) discs are indicated. The overlap between ANTP and FSH-S signals in the antennal disc is marked by arrows (F and G). Note that FSH-S induction also caused distortion of antennal tissue. No distortion was seen by FSH-L induction. (O and P) Ectopic UBX expression by en-driven FSH-S. Ganglia from second instar larvae were stained with an anti-UBX antibody. Normal UBX expression pattern is shown with its anterior boundary in PS5 indicated (O, arrow). Induction of FSH-S by en-Gal4 resulted in ectopic UBX signals at lateral parts of more anterior tissue (P, arrowheads). en-Gal4 is expressed in a transverse row of cells in each PS.
fs(1)h encodes two double-bromodomain proteins, FSH-S and FSH-L. To define their roles in HOX activation, we employed the Gal4/UAS binary system (5) to induce high levels of FSH-S or FSH-L and examine their effects on HOX expression. UAS transgenes containing epitope-tagged FSH-S or FSH-L were driven by dpp-Gal4 in small subsets of imaginal cells. As shown in Fig. 1D, targeted expression of FSH-S caused striking defects in the adult. Frequently, the adults' heads lacked maxillary palpi, and their aristae were transformed into distal legs with claws (Fig. 1E). Severe defects were also found in thoracic legs, including bifurcation of tibial segments and deletion of tarsal segments (data not shown). Surprisingly, no discernible defect was seen in adults with targeted FSH-L expression, suggesting that FSH-L and FSH-S act differently.
Antenna-to-leg transformations can be induced by ectopic expression of the HOX gene Antennapedia (Antp) in antennal discs (33, 57). To determine whether extra legs induced by FSH-S might be related to ectopic Antp expression, we stained eye-antennal discs from third instar larvae with an anti-ANTP antibody. Whereas ANTP is normally not expressed in these discs, strong ANTP signals were seen in antennal discs of transgenic animals. Using an anti-Flag antibody to mark tagged FSH-S, we found extensive overlaps between FSH-S and ANTP signals (Fig. 1F to I), suggesting that FSH-S is directly involved in ANTP induction. By contrast, no ectopic ANTP was induced by FSH-L (Fig. 1M and N), which was consistent with the normal appearance of adult flies. These effects further distinguished the role of FSH-S and FSH-L in HOX activation. Curiously, very little ANTP expression was induced by FSH-S in eye discs, despite its comparable levels in antennal and eye discs. The nature of this tissue-dependent response is unclear. Furthermore, no ectopic UBX signal was found in eye-antennal and other discs. To avoid problems caused by induction timing or tissue dependence, we used an en-Gal4 line to drive FSH-S expression. Under such conditions, most larvae died before the third instar, while rare adult escapers (less than 1%) showed partial deletion of thoracic segments. In second instar larvae, ectopic UBX signals could be detected in ventral ganglions. In addition to the transverse rows normally found within PS5 to PS12, UBX signals appeared in small clusters of cells near the lateral margins of PS4, PS3, and PS2 at anteriorly diminishing frequencies. Occasionally, ectopic UBX was found in PS4 extending to PS2 on both sides of the ganglion (compare Fig. 1O and P). These results strongly suggested that FSH-S can induce HOX genes.
FSH-S is a ubiquitous nuclear factor activating Ubx via proximal elements.
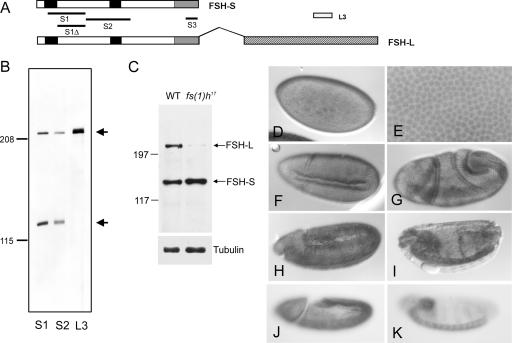
Previous analysis predicted that fs(1)h protein products might be membrane associated, implicating a role in signal transduction (25). To further characterize FSH-S, we raised antibodies against three regions (S1, S2, and S3) common to both FSH-S and FSH-L. Using affinity-purified antibodies, we detected two common bands in embryonic extracts (Fig. 2B and C). The sizes of these two bands were consistent with predicted sizes of FSH proteins (∼210 and ∼120 kDa). In addition, an antibody specific for FSH-L (i.e., L3) reacted only with the larger protein. The authenticity of these proteins was further confirmed by the analysis of a larval-lethal mutant, fs(1)h17, which results from an insertion of a copia element in the intron following the FSH-S coding sequences. Unlike many other fs(1)h mutations, this mutation did not cause homeotic effects (17). Interestingly, the larger protein was severely diminished in mutant larvae at third instar, while the small one was unaffected (Fig. 1C). These results indicate that these proteins represent the two specific FSH isoforms and, more importantly, that FSH-L is not essential for the homeotic effect.
FIG. 2.
FSH-S is a ubiquitous nuclear protein in embryos. (A) Organization of the two ORFs of the fs(1)h gene. FSH-S contains 1,110 amino acids, while FSH-L contains an additional 932 amino acids by the alternative splicing as indicated by the overhang. Splicing junctions in other regions are omitted. The double-bromodomain, the ET domain, and the unique region of FSH-L are indicated by black, gray, and hatched boxes, respectively. S1, S2, S3, and L3 correspond to regions used for immunization. A smaller region (S1Δ) lacking the first bromodomain was used for affinity purification of S1 antibody. (B) Analysis of FSH proteins. Embryonic extracts were immunoblotted with affinity-purified S1, S2, and L3 antibodies. S1 and S2 antibodies detected both a ∼220-kDa and a ∼120-kDa protein, while the L3 antibody detected only the ∼220-kDa protein. (C) Differential effect of fs(1)h17 mutation on fs(1)h products. Extracts from wild-type or fs(1)h17/Y third instar larvae were immunoblotted with affinity-purified S3 antibody. FSH-L was severely reduced in an fs(1)h17 mutant. α-Tubulin control is shown in the bottom panel. (D to I) Embryonic patterns of FSH proteins. Wild-type embryos were stained with affinity-purified S1 antibody. FSH proteins were detected ubiquitously at blastoderm stage (D and E), beginning of gastrulation (F and G), fully extended germ-band stage (H), and dorsal closure stage (I). An enlarged view showing nuclear staining at the blastoderm stage in shown in panel E. (J and K) Distribution of FSH-S-specific transcripts. Wild-type embryos were hybridized with an antisense RNA probe from the 3′ UTR of FSH-S. A uniform distribution was seen at fully extended germ band (J) and dorsal closure (K) stages.
The developmental profiles and subcellular localization of FSH proteins in embryos were analyzed by immunostaining with affinity-purified S1 antibody. As shown in Fig. 2D and E, nuclear staining was clearly seen in syncytial embryos. Although S1 antibody reacted with both FSH-S and FSH-L, we attribute this nuclear staining to FSH-S, since FSH-L is primarily a centrosomal protein at this stage (unpublished data). In addition, tagged FSH-S was localized in the nuclei in both transgenic lines (Fig. 1J to L) and transfected S2 cells (data not shown). The staining intensity appeared to be uniform throughout all developmental stages except in those cells located near invaginating furrows (Fig. 2F to H) or in VNC (Fig. 2I). To further confirm the distribution pattern of FSH-S, we performed whole-mount in situ hybridization using a probe from the 3′ UTR of FSH-S mRNA, which is absent in FSH-L mRNA. Again, a ubiquitous distribution of FSH-S transcripts was observed (Fig. 2J and K).
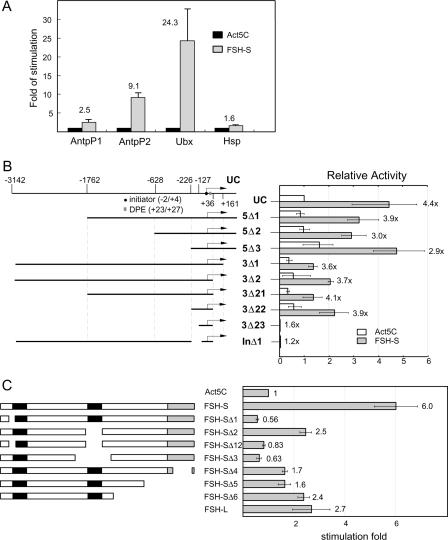
The genetic interactions, induction of HOX gene expression, nuclear localization, and the presence of a double bromodomain raised a strong possibility that FSH-S might directly affect HOX promoters. To test this, we examined the ability of FSH-S to stimulate reporter constructs containing various promoters by cotransfection experiments in a Drosophila haploid cell line which was shown to recapitulate Ubx regulation by trx and Pc (14). Reporter activities from constructs containing the P1 or P2 promoters of Antp or promoters of Ubx and the Heat shock protein 70 (Hsp70) were assayed following cotransfection of an Act5C-FSH-S effector or an Act5C control vector. As shown in Fig. 3A, the activities of the Antp-P2 and Ubx promoters were stimulated approximately 10-fold and 20-fold, respectively, while the Antp-P1 and Hsp70 promoters were only weakly affected. Thus, the effect of FSH-S appeared to be highly selective.
FIG. 3.
Transactivation of the Ubx promoter by FSH-S. (A) Activation of specific homeotic promoters by FSH-S. CAT reporter constructs containing Antp-P1 (−6 kb to +793), Antp-P2 (−10 kb to +200), Ubx (−3154 to +358), or Hsp-70 (−1.5 kb to +90) promoter were cotransfected with either Act5C control vector or Act5C-FSH-S into Drosophila cells. The CAT activities obtained from cells transfected with Act5C-FSH-S were normalized to those with Act5C vector to determine relative stimulation. Note that much less Hsp70-CAT was used because of its strong promoter activity. (B) Mapping of FREs. Diagrams of UC and deletion constructs are shown in the left panel. The transcription start site, initiator, and downstream promoter element (DPE) are marked by a bent arrow, a circle, and a box, respectively. The end point of each construct is indicated in the top diagram. Each deletion construct was cotransfected with either Act5C or Act5C-FSH-S in the presence of an internal control, Act5C-lacZ. The CAT activity from each construct was first normalized with the β-galactosidase activity. The relative activity obtained from the UC construct with Act5C vector was then used for normalization of other constructs. Relative stimulation is indicated for each construct. (C) Identification of functional domains of FSH-S. Diagrams of full-length FSH-S and deletion constructs are shown in the left panel. The bromodomain and ET domain are marked by black and gray boxes, respectively. Sequences deleted in mutant constructs are the following: amino acid residues 24 to 82 (Δ1), 490 to 621 (Δ2), 24 to 82 and 490 to 621 (Δ12), 371 to 698 (Δ3), 1024 to 1104 (Δ4), 815 to 1110 (Δ5), and 681 to 1110 (Δ6). Act5C, Act5C-FSH-S, or Act5C-FSH-L was cotransfected with UC and an internal control, Act5C-lacZ. For each effector construct, the CAT activity was first normalized to the β-galactosidase activity. The relative activity from Act5C was then used as a standard to derive relative stimulation. Note that the weaker effects of FSH-S on Ubx in experiments B and C might result from the interference from using Act5C-lacZ as the internal control in these experiments.
Several trxG genes have been shown to act on regulatory sequences located about 20 kb upstream of the initiation site (9, 65). Since the UC construct used here only contained sequences from −3142 to +360 (Fig. 3B), FSH-S appeared to act via distinct sequences near the basal promoter. To identify the FSH-S response elements (FRE), we analyzed the effect of FSH-S on a series of Ubx deletion mutants. As shown in Fig. 3B, sequential deletion of 5′ sequences from −3142 to −1762 (5Δ1), −628 (5Δ2), or −226 (5Δ3) did not alter the ability of the Ubx promoter to respond to cotransfected FSH-S. Deletion from +360 to +161 (3Δ1) resulted in a general reduction of the promoter activity by about twofold, regardless of the presence or absence of cotransfected FSH-S. Since the stimulatory effect of FSH-S was not affected, this downstream region most likely contains a positive element that is unrelated to FRE. No further effect was observed when sequences from +161 to +36 (3Δ2) were deleted. These results indicated that the FRE is not present in the regions upstream of −226 or downstream of +36. Consistently, a construct containing sequences from −226 to + 36 (3Δ22) was sufficient to respond to FSH-S. Conversely, an internal deletion of sequence from −200 to −32 (InΔ1; In is initiator) almost completely abolished the promoter activity. Since the initiator (ACATTC from −2 to +4) and downstream promoter elements (GGATA from +23 to +27) were intact in InΔ1 construct, the inactivation of the Ubx promoter should reflect the removal of regulatory elements. These results led us to conclude that the FRE is located between −200 and −32, which corresponds to the CPR determined previously (39). Further refinement of the boundaries of the FRE was unsuccessful, since deletions from −226 to −127 (3Δ23) or from −127 to −32 inactivated the promoter.
We also examined whether any specific domain of FSH-S is required for transactivation. Mutant constructs carrying deletions of the N-terminal half of the first bromodomain (Δ1), the entire second bromodomain (Δ2) and its flanking sequences (Δ3), or both bromodomains (Δ12) or the C-terminal sequences including the ET domain (Δ4-6) were tested in transfection assays. It appeared that deletion of the first bromodomain results in a complete inactivation of FSH-S(Fig. 3C, Δ1), suggesting a critical requirement of this domain. However, the full activity of FSH-S was also dependent on the second bromodomain and ET domain, since deletion of these domains resulted in partial inactivation. Interestingly, although FSH-L contains the entire FSH-S sequence, it appeared to be much less active than FSH-S (Fig. 3C, compare the second and last bars). These results are consistent with our observation that FSH-L could not induce HOX genes in imaginal tissues and support further that FSH-S is primarily, if not exclusively, responsible for the transactivation function of fs(1)h.
Identification of the ZESTE site as the FRE.
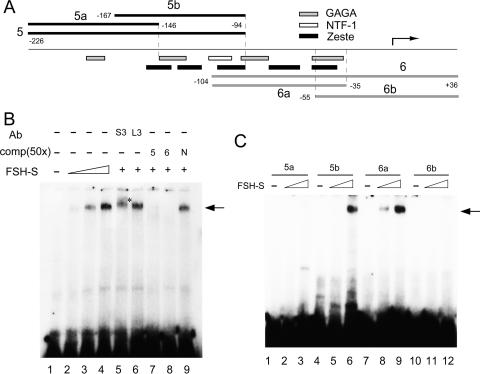
We next examined whether FSH-S could bind any specific sequences in the CPR. An inducible S2 cell line containing the metallothionein promoter-driven Flag-tagged FSH-S was established. Tagged FSH-S was purified by immunoaffinity chromatography from whole-cell extracts after (NH4)2SO4 enrichment. In addition to the major band corresponding to FSH-S, several less abundant proteins were also copurified (Fig. 4A). Although fs(1)h mutant showed strong genetic interactions with trx or ash1 mutants, FSH-S was not copurified with these proteins or OSA (Fig. 4B). In addition, FSH-S was not associated with ZESTE protein, which was shown to bind the CPR (3) (Fig. 4C). The ability of purified FSH-S to bind specific sequences of the CPR was demonstrated by EMSAs. Upon addition of increasing amounts of FSH-S to labeled Ubx-5 probe (Fig. 5A), a slower-migrating band appeared near the top of 3.5% native polyacrylamide gels (Fig. 5B), indicating the formation of protein-DNA complexes. The exceedingly slow mobility of this band suggested that a multisubunit protein complex is involved. FSH-S is a constituent of this putative complex, since a small but significant supershift was observed when an antibody against FSH-S was briefly incubated with FSH-S protein. A supershift was not observed when an antibody to FSH-L was used instead. Furthermore, this binding was sequence specific, since it could be completely blocked by the addition of excess amounts of unlabeled Ubx-5 or Ubx-6 but not by a random DNA fragment (Fig. 5B, N). Similar results were also obtained when Ubx-6 was used as the probe (data not shown). To further narrow the binding region, we used four smaller probes from the CPR for EMSA. As shown in Fig. 5C, specific binding was observed with probes Ubx-5b (−167 to ∼−94) and Ubx-6a (−104 to ∼−35) but not Ubx-5a (−226 to ∼−146) or Ubx-6b (−55 to ∼+36), indicating that the FRE is located between −167 and −35.
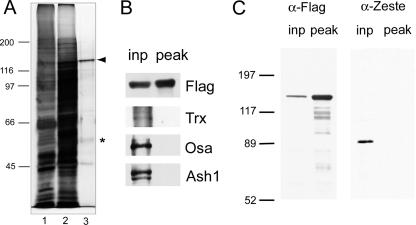
FIG. 4.
Characterization of purified FSH-S. (A) FSH-S purification. FSH-S was purified by an immunoaffinity method from a transformed S-2 cell line containing Flag-tagged FSH-S. A total of 3.5 μl of whole-cell extracts (lane 1) or protein fractions before (lane 2) or after (lane 3) immunoaffinity purification were separated by 7.5% SDS-PAGE and visualized by silver staining. FSH-S and a 56-kDa phosphoprotein are indicated by an arrowhead and asterisk, respectively. The sizes of molecular mass markers are indicated. (B) Lack of other trxG in FSH-S preparation. Aliquots of proteins before (inp) and after (peak) immunoaffinity purification were blotted and probed with antibodies against Flag, Trx, Osa, or Ash1. (C) Lack of ZESTE protein in FSH-S preparation. Aliquots of proteins before (inp) and after (peak) immunoaffinity purification were blotted and probed with antibodies against Flag or ZESTE. α, anti.
FIG. 5.
FSH-S binds specific Ubx proximal sequences. (A) Diagram of the Ubx proximal promoter. The region from −226 to +36 is shown. The transcription start site is indicated by a bent arrow. The end points of DNA fragments used in binding assays are indicated. The regions corresponding to four TRL sites (GAGA), five ZESTE sites, and one NTF-1 site are also shown. (B) DNA binding assays. Labeled Ubx-6 fragment was incubated without (lane 1) or with 1 μl (lane 2), 2 μl (lane 3), or 3 μl (lanes 4 to 9) of purified FSH-S protein and subsequently resolved on a 3.5% native gel. For the supershift experiments, 1 μl of purified S3 (lane 5) or L3 antibody (lane 6) was included in the binding reaction. For competition experiments, 50-fold excess [comp(50×)] of unlabeled Ubx-5 (lane 7), Ubx-6 (lane 8), or a nonspecific 123-bp fragment (N, lane 9) was added to the binding reactions. Specific DNA-protein complexes are indicated by the arrow. The supershift band is indicated by an asterisk. (C) Refinement of the binding region. Labeled fragments 5a, 5b, 6a, or 6b were each incubated (in respective order) without (lanes 1, 4, 7, and 10) or with 1 μl (lanes 2, 5, 8, and 11) or 2 μl (lanes 3, 6, 9, and 12) of FSH-S protein. Specific binding was observed for 5b and 6a but not 5a and 6b.
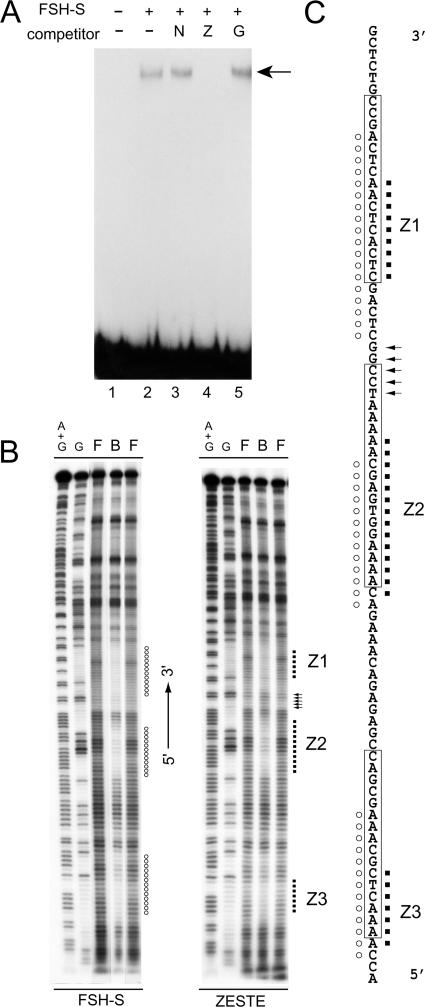
The CPR contains clusters of binding sites for ZESTE, TRL (also known as GAGA factor), and NTF-1 (Fig. 5A). To determine whether any of these sites might correspond to FRE, we performed competition assays with DNA fragments containing tandem repeats of ZESTE, TRL, or NTF-1 binding sites. Interestingly, only ZESTE repeats effectively blocked binding activity (data not shown). To exclude the possibility that fortuitous binding sites might be generated by multimerizaton of these repeats, we also tested an oligonucleotide containing one consensus ZESTE site (CGAGTG) with different flanking sequences. As shown in Fig. 6A, this oligonucleotide also blocked the binding activity of FSH-S. Thus, the ZESTE site should represent the core FRE.
FIG. 6.
FSH-S shares similar binding sites with ZESTE. (A) Identification of FSH-S binding sites. Binding assays were performed with labeled Ubx-6a in the absence (lane 2) or presence of a DNA fragment containing multiple NTF-1 sites (lane 3, N) or oligonucleotides containing the ZESTE site (lane 4, Z) or the TRL site (lane 5, G). The protein-DNA complex is indicated by the arrow. (B) In-gel footprinting assays. EMSA was carried out with either FSH-S or purified recombinant ZESTE protein, followed by OP-Cu cleavage. DNA ladders from the bound complex (B), free probe (F), or the bottom strand of Ubx-6a are shown with the promoter orientation as the reference. Protected regions are indicated by circles (FSH-S) or dots (Z). A stretch of enhanced cleavage sites by ZESTE is indicated by arrows. (C) Summary of the footprinting assay. The bottom-strand sequences from −113 to −32 are shown. The ZESTE sites (Z1 to Z3) determined by DNase I footprinting assays are boxed (4). The regions protected by FSH-S (circles) or ZESTE (dots) by the OP-Cu method are indicated.
For further analyses of DNA binding properties, we compared FSH-S and recombinant ZESTE proteins by an in-gel chemical footprinting technique. The DNA-cleaving ions OP-Cu used here gain more access to unprotected sequences than DNase I and are thus capable of revealing detailed differences in binding properties (60). Similar to previous reports with DNase I (4), three sites (Z1 to Z3) were protected by ZESTE or FSH-S proteins in the Ubx-6a fragment (Fig. 6B). Despite an overall similarity, we noticed several important differences between these patterns. For example, the regions unprotected by ZESTE produced bands with intensities comparable to those from free probes. However, fainter intervening bands were produced by FSH-S, suggesting weak protection on flanking sequences. Two additional differences were found over the Z1 site. FSH-S appeared to protect more 5′ sequences than ZESTE. However, ZESTE produced several bands more intense than the control (Fig. 6B and C, arrows), suggesting DNA distortion in this region. The lack of detectable ZESTE protein and the distinct DNA binding properties exhibited by FSH-S clearly support the involvement of a novel binding factor.
If the FRE indeed corresponds to the ZESTE site, the function of the ZESTE site might be inactivated by fs(1)h mutations. Therefore, we examined the effects of fs(1)h mutation on expression of Ubx-lacZ transgenes containing two distal regulatory domains (BXD and ABX) and ∼3 kb of immediate upstream sequences in addition to a wild-type CPR (Uβ) or tandem ZESTE sites (Uβ-Z) (39) (Fig. 7A). In the wild-type background, strong lacZ signals were observed from PS5 to more posterior parts of the VNC in Uβ embryos (Fig. 7B). In addition, there was weaker misexpression in anterior parts of the VNC. As reported earlier (39), the misexpression was more pronounced in Uβ-Z embryos. More importantly, lacZ transcripts were severely reduced throughout the entire VNC in both Uβ and Uβ-Z embryos upon inactivation of fs(1)h, indicating a strict requirement of fs(1)h. Again, cad transcripts were not affected. These results strongly support the physiological relevance of FSH-S to the ZESTE site.
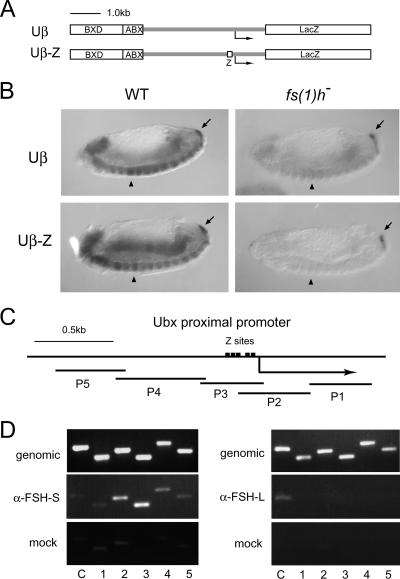
FIG. 7.
Physiological relevance of FSH-S on the ZESTE site. (A and B) Requirement of fs(1)h in expression of Ubx transgenes. (A) Diagrams of transgenes containing a wild-type (Uβ) or mutated (Uβ-Z) Ubx promoter. Uβ contains ∼1.6 kb of BXD, ∼0.6 kb of ABX, and ∼4 kb of Ubx proximal sequences. Uβ-Z contains same sequences except that the region from −200 to −31 is replaced by five repeats of the ZESTE motif. (B) Effect of fs(1)h on transgene expression in embryos. Expression of transgenes and cad in wild-type (WT) or fs(1)h1 embryos was examined by whole-mount in situ hybridization as described in the legend of Fig. 1. Uβ produces strong signals from PS5 to PS13 in VNC of wild-type embryos (upper left) but very weak signals in fs(1)h1 mutants (upper right). Compared to the wild-type embryos (lower left), the expression of Uβ-Z transgene is almost completely diminished in fs(1)h1 mutants (lower right). Note that cad signals are comparable in wild-type and mutant embryos. PS5 is indicated by the arrowhead. cad signals are indicated by the arrow. (C and D) FSH-S occupancy in CPR in vivo. A diagram of the Ubx proximal promoter is shown in panel C. Five natural ZESTE sites are depicted. The PCR fragments are indicated below. They span from −1419 to +831. Panel D shows the results of immunoprecipitation of promoter-specific chromatin. Formaldehyde-fixed chromatin prepared from larval tissues of male fs(1)h17 mutants was amplified with primers spanning the Ubx promoter before (genomic) or after immunoprecipitation with S3 (α-FSH-S) or L3 (α-FSH-L) antibodies or with preimmune serum (mock). The number of each PCR pair is indicated below each lane. The promoter of caudal was used as a control (C). α, anti.
To further demonstrate that FSH-S is indeed associated with CPR of the endogenous promoter in vivo, we performed chromatin immunoprecipitation assays with formaldehyde-fixed chromatin prepared from male fs(1)h17 mutant larvae, which contain normal levels of FSH-S but diminishing amounts of FSH-L. Using five pairs of primers to cover sequences of more than 2 kb around Ubx start sites (Fig. 7C), we found that FSH-S is preferentially associated with a CPR-containing DNA fragment (Fig. 7D). Although the antibody used here could cross-react with FSH-L, the contribution of FSH-L to the binding is excluded, because only a minute amount of FSH-L was present in fs(1)h17 mutant larvae, and, more importantly, no FSH-L signal was detectable in the Ubx promoter. In addition, this association appeared to be promoter specific, since only background signal was detected in the cad promoter.
The role of FSH-S as a protein kinase.
Human RING3 protein, a FSH-S-like protein, has been shown to be a novel nuclear Ser/Thr kinase with scrambled subdomains (15). However, subsequent studies failed to show this activity in the mouse counterpart, FSRG-1, despite more than 90% sequence identity (52). To determine whether FSH-S could act as a kinase, we first examined the kinase activity in FSH-S preparations. As shown in Fig. 8A, addition of [γ-32P]ATP resulted in substantial phosphorylation of FSH-S and an additional protein of ∼56 kDa. Because this smaller protein was consistently copurified, it will be referred to as FAP56 (FSH-associated protein of 56 kDa). Phosphoamino-acid analysis of in vitro phosphorylated proteins revealed that FSH-S was phosphorylated at the serine residue, while FAP56 was phosphorylated at both serine and threonine residues (Fig. 8B). Although FSH-S phosphorylation was readily detected by radioactive labeling, no mass increase was found upon incubation with 0.1 mM ATP (Fig. 8C, lane 1). However, when treated with calf intestine phosphatase, the mass of FSH-S appeared to decrease slightly (Fig. 8C, compare lanes 3 and 4), indicating a limited phosphorylation of FSH-S.
The kinase activity of RING3 kinase could be restored by renaturation on nitrocellular filter after SDS-PAGE (15). Using this procedure, we failed to detect FSH-S phosphorylation in parallel experiments (data not shown). However, we reasoned that if FSH-S is a kinase, it must be able to bind ATP. An ATP analog, FSBA, has been used for affinity labeling of ATP binding proteins including kinases (72). Therefore, we examined the reactivity of FSH-S toward FSBA. Using an FSBA-specific antibody, we found that FSH-S could indeed be covalently linked to FSBA (Fig. 8D, lane 1). More importantly, the degree of cross-linking was substantially reduced by excessive ATP (lane 2), indicating that FSH-S can bind ATP specifically.
Addition of FSH-S to cell extracts in which endogenous kinases were heat inactivated resulted in phosphorylation of many proteins (data not shown), suggesting the presence of many kinase substrates. The clustering of multiple binding sites for FSH-S (or ZESTE) and TRL in the CPR suggests that they might be spatially juxtaposed upon binding to the CPR, raising the possibility that TRL might be a potential kinase substrate. Using in vitro kinase assays, we found that addition of FSH-S to purified recombinant TRL indeed resulted in its phosphorylation (Fig. 8E).
DISCUSSION
In this report, we provide several lines of evidence to support a direct role of FSH-S in HOX gene activation. Unlike other trxG proteins, FSH-S acts directly on the CPR of the Ubx promoter. The revelation of several interesting properties of FSH-S offers important mechanistic insights into the Ubx regulatory circuitry.
Lack of functional fs(1)h is known to cause complex developmental defects including homeotic transformation and early embryonic lethality. The contribution of two different fs(1)h products to these effects had not been determined. Based on the following observations, we suggest that FSH-S is primarily involved in HOX regulation. First, we show that FSH-S, but not FSH-L, can effectively activate homeotic promoters in imaginal discs and cultured cells. Second, FSH-S is a nuclear protein, while FSH-L is mainly found in centrosomes and is involved in organization of mitotic spindles in early embryos (unpublished data). Third, FSH-S can bind and function both in vitro and in vivo through a specific motif in the CPR. Lastly, no homeotic phenotype has been observed in an fs(1)h17 mutant lacking FSH-L (17; also data not shown). Thus, FSH-S is directly responsible for the homeotic effect of fs(1)h. Although FSH-L contains the entire sequence of FSH-S, our results clearly indicate that it does not play any significant role in the homeotic effect. The complex developmental functions of fs(1)h are very likely to be divided between different isoforms.
The abilities of FSH-S to bind a specific motif and to affect promoter activity through the CPR indicate that FSH-S plays an important role at the CPR for activation of the Ubx promoter. Among 18 trxG genes we have examined, fs(1)h, trx, and ash1 form a small but interesting subgroup that is most critical for Ubx activation and is known to act through specific regulatory sequences. Previous studies have shown that TRX and ASH1 act primarily through distal sequences that are essential for domain-specific Ubx expression (9, 49, 56, 65). Recently, they have also been implicated in transcriptional elongation by their association with promoter and transcribed sequences (50, 51). FSH-S is the only factor that functions primarily, if not entirely, on the CPR. Given the critical role of the CPR in promoter activity, FSH-S is very likely to play a key role in integration of activating signals from distal elements and factors. The strong synergistic effects reported here for fs(1)h, trx, and ash1 mutations indicate that they are involved in a critical step of Ubx promoter activation and that intimate functional relationships probably exist between these factors. Although they appear to exist in distinct protein complexes, it is highly likely that they interact directly or through associated factors. Such interactions may facilitate the action of TRX and ASH1 in the promoter and more downstream regions. An alternative (but not mutually exclusive) possibility is that FSH-S might be involved in attenuation of the repressing activity of PcG proteins. Since the distal response elements for PcG proteins and TRX/ASH1 are largely overlapping and their histone modification activities are functionally antagonistic, destabilization of PcG complexes could result in more efficient occupancy and/or more potent chromatin modification by TRX and ASH1. In either case, the activities of these distal factors might also be modulated by the kinase activity associated with FSH-S (see discussion below).
The stimulatory effects of FSH-S on the Ubx basal promoter also suggest that FSH-S may directly affect the basal transcription machinery. A closely related Saccharomyces cerevisiae protein, BDF1, has been shown to be a TFIID-associated factor, acting potentially as a functional substitute for TAF1 in higher organisms (43). Mouse BRD4 stimulates transcription by binding to positive transcription elongation factor b (31, 71). Although it is unclear whether FSH-S possesses similar activities, the presence of structurally similar domains suggests that it may interact with these basal transcription factors. Therefore, we speculate that FSH-S provides a dual interface for interactions with distal factors and basal transcriptional machinery for optimal Ubx transcription.
The sharing of the same target sequences between FSH-S and ZESTE may help clarify a long-standing enigma about the role of ZESTE in Ubx regulation. The function of zeste was revealed by a pairing-dependent phenomenon called transvection in which Ubx alleles with defective promoters can partially complement alleles with impaired regulatory sequences (18, 35). Thus, zeste can facilitate the transutilization of the regulatory sequences on one chromosome by the Ubx promoter on a paired homologous chromosome. However, zeste is not required for expression of an intact endogenous Ubx gene or expression of a Ubx transgene containing more complete regulatory sequences (i.e., 35-kb sequences in 35UZ transgene) (38, 39), despite the fact that ZESTE binds to the CPR and is required for expression of Ubx transgenes containing partial regulatory sequences. Moreover, zeste is dispensable for viability (22). These findings indicate that zeste is not essential for Ubx expression under normal genetic contexts. In contrast, FSH-S is indispensable for Ubx regulation and for development. The ability of FSH-S to bind the same target sequences indicates that FSH-S represents a critical component of a regulatory circuitry that utilizes regulatory signals present on the same chromosome to insure proper transcription of intact Ubx promoter. It is interesting that zeste-independent transvection has also been found that appears to employ the mechanisms that normally operate between the distal elements and the proximal promoter (42). We speculate that FSH-S is also very likely to play a role in zeste-independent transvection.
Our finding of DNA binding activity in FSH-S is surprising, since the double-bromodomain and the C-terminal ET domain, two prominent domains required for the function of FSH-S, are not known for DNA binding activity. It is possible that FSH-S may possess a novel DNA binding domain. Alternatively, the binding activity might be contributed by a factor that is associated with FSH-S, since several proteins were copurified with FSH-S and since recombinant FSH-S did not show the same activity (data not shown). Further characterization of FSH-S and associated factors is necessary to resolve this question.
Another interesting feature of FSH-S is the kinase activity. The structural similarities to the RING3 nuclear kinase (15), the detection of a similar kinase activity, and the ATP binding activity are consistent with the notion that FSH-S contains a Ser/Thr kinase activity. However, the lack of kinase activity in bacterially expressed FSH-S suggests that posttranslational modification or an additional factor(s) is required for such an activity. It is interesting that, in addition to FSH-S and FAP56, many other proteins including TRL can be phosphorylated by FSH-S in vitro, suggesting a broad substrate specificity. Thus, it seems plausible that FSH-S may modulate the activities of factors that are brought into its proximity. Once it occupies the CPR, we speculate that FSH-S may affect multiple factors that are in close contact with CPR by either short- or long-range interactions.
Acknowledgments
We are very grateful to the following people for providing reagents and flies: M. Biggin, D. Brower, I. Dawid, S. Haynes, D. Hogness, M. Koelle, M. Krasnow, J. Laney, P. Macdonald, J. Manley, V. Pirrotta, F. Sauer, M. Scott, A. Shearn, and the Bloomington Drosophila Stock Center. We acknowledge that FSH antibodies were generated in I. Dawid's laboratory at the National Institutes of Health.
This work was supported by grants to D.-H.H. from Academia Sinica and National Science Council, Republic of China (grants NSC 83-0412-B-001-069, 84-2311-B-001-045, and 85-2311-B-001-010) and in part by the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Ashburner, M. 1989. Drosophila: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 2.Beisel, C., A. Imhof, J. Greene, E. Kremmer, and F. Sauer. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419:857. [DOI] [PubMed] [Google Scholar]
- 3.Biggin, M. D., S. Bickel, M. Benson, V. Pirrotta, and R. Tjian. 1988. Zeste encodes a sequence-specific transcription factor that activates the Ultrabithorax promoter in vitro. Cell 53:713-722. [DOI] [PubMed] [Google Scholar]
- 4.Biggin, M. D., and R. Tjian. 1988. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell 53:699-711. [DOI] [PubMed] [Google Scholar]
- 5.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 6.Byrd, K. N., and A. Shearn. 2003. ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc. Natl. Acad. Sci. USA 100:11535-11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y.-L., Y.-H. Peng, I. C. Pan, D.-S. Sun, B. King, and D.-H. Huang. 2001. Essential role of Drosophila Hdac1 in homeotic gene silencing. Proc. Natl. Acad. Sci. USA 98:9730-9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y.-L., B. O. King, M. O'Connor, A. Mazo, and D. H. Huang. 1995. Functional reconstruction of trans regulation of the Ultrabithorax promoter by the products of two antagonistic genes, trithorax and Polycomb. Mol. Cell. Biol. 15:6601-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, R. T., T. Furukawa, N. Tanese, and J. E. Treisman. 1999. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 18:7029-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Condie, J. M., J. A. Mustard, and D. L. Brower. 1991. Generation of anti-Antennapedia monoclonal antibodies and Antennapedia protein expression in imaginal discs. Drosophila Info. Serv. 70:52-54. [Google Scholar]
- 12.Crosby, M. A., C. Miller, T. Alon, K. L. Watson, C. P. Verrijzer, R. Goldman-Levi, and N. B. Zak. 1999. The trithorax group gene moira encodes a Brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 19:1159-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 14.Debec, A., A. Szollosi, and D. Szollosi. 1982. A Drosophila melanogaster cell line lacking centriole. Biol. Cell. 44:133-138. [Google Scholar]
- 15.Denis, G. V., and M. R. Green. 1996. A novel, mitogen-activated nuclear kinase is related to a Drosophila developmental regulator. Genes Dev. 10:261-271. [DOI] [PubMed] [Google Scholar]
- 16.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, M.-M. Zhou, and M.-M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 17.Digan, M. E., S. R. Haynes, B. A. Mozer, I. B. Dawid, F. Forquignon, and M. Gans. 1986. Genetic and molecular analysis of fs(1)h, a maternal effect homeotic gene in Drosophila. Dev. Biol. 114:161-169. [DOI] [PubMed] [Google Scholar]
- 18.Duncan, I. W. 2002. Transvection effects in Drosophila. Annu. Rev. Genetics 36:521-556. [DOI] [PubMed] [Google Scholar]
- 19.Forquignon, F. 1981. A maternal effect mutation leading to deficiencies of organs and homeotic transformations in the adults of Drosophila. Wilhelm Roux's Arch. 190:132-138. [DOI] [PubMed] [Google Scholar]
- 20.Fritsch, C., J. L. Brown, J. A. Kassis, and J. Muller. 1999. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 126:3905-3913. [DOI] [PubMed] [Google Scholar]
- 21.Gans, M., F. Forquignon, and M. Masson. 1980. The role of dosage of the region 7D1-7D5-6 of the X chromosome in the production of homeotic transformation in Drosophila melanogaster. Genetics 96:887-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg, M. L., R. A. Colvin, and A. F. Mellin. 1989. The Drosophila zeste locus is nonessential. Genetics 123:145-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Haynes, S. R., C. Dollard, F. Winston, S. Beck, J. Trowsdale, and I. B. Dawid. 1992. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 20:2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haynes, S. R., B. A. Mozer, N. Bhatia-Dey, and I. B. Dawid. 1989. The Drosophila fsh locus, a maternal effect homeotic gene, encodes apparent membrane proteins. Dev. Biol. 134:246-257. [DOI] [PubMed] [Google Scholar]
- 26.Huang, D.-H., and Y.-L. Chang. 2004. Isolation and characterization of CHRASCH, a Polycomb-containing silencing complex. Methods Enzymol. 377:267-282. [DOI] [PubMed] [Google Scholar]
- 27.Huang, D.-H., Y.-L. Chang, C.-C. Yang, I. C. Pan, and B. King. 2002. pipsqueak encodes a factor essential for sequence-specific targeting of a Polycomb group protein complex. Mol. Cell. Biol. 22:6261-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, D.-H., and I. B. Dawid. 1990. The maternal-effect gene fsh is essential for the specification of the central region of the Drosophila embryo. New Biol. 2:163-170. [PubMed] [Google Scholar]
- 29.Hur, M.-W., J. D. Laney, S.-H. Jeon, J. Ali, and M. D. Biggin. 2002. Zeste maintains repression of Ubx transgenes: support for a new model of Polycomb repression. Development 129:1339-1343. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 31.Jang, M. K., K. Mochizuki, M. Zhou, H.-S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523-534. [DOI] [PubMed] [Google Scholar]
- 32.Jiang, Y. W., P. Veschambre, H. Erdjument-Bromage, P. Tempst, J. W. Conaway, R. C. Conaway, and R. D. Kornberg. 1998. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc. Natl. Acad. Sci. USA 95:8538-8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jorgensen, E. M., and R. L. Garber. 1987. Function and misfunction of the two promoters of the Drosophila Antennapedia gene. Genes Dev. 1:544-555. [DOI] [PubMed] [Google Scholar]
- 34.Kennison, J. A. 1995. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29:289-303. [DOI] [PubMed] [Google Scholar]
- 35.Kennison, J. A., and J. W. Southworth. 2002. Transvection in Drosophila. Adv. Genet. 46:399-420. [DOI] [PubMed] [Google Scholar]
- 36.Kennison, J. A., and J. W. Tamkun. 1988. Dosage-dependent modifiers of Polycomb and Antennapedia mutations in Drosophila. Proc. Natl. Acad. Sci. USA 85:8136-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krasnow, M. A., E. E. Saffman, K. Kornfeld, and D. S. Hogness. 1989. Transcriptional activation and repression by Ultrabithorax proteins in cultured Drosophila cells. Cell 57:1031-1043. [DOI] [PubMed] [Google Scholar]
- 38.Laney, J. D., and M. D. Biggin. 1996. Redundant control of Ultrabithorax by zeste involves functional levels of zeste protein binding at the Ultrabithorax promoter. Development 122:2303-2311. [DOI] [PubMed] [Google Scholar]
- 39.Laney, J. D., and M. D. Biggin. 1992. zeste, a nonessential gene, potently activates Ultrabithorax transcription in the Drosophila embryo. Genes Dev. 6:1531-1541. [DOI] [PubMed] [Google Scholar]
- 40.Lygerou, Z., C. Conesa, P. Lesage, R. N. Swanson, A. Ruet, M. Carlson, A. Sentenac, and B. Séraphin. 1994. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 22:5332-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mansukhani, A., P. H. Gunaratne, P. W. Sherwood, B. J. Sneath, and M. Goldberg. 1988. Nucleotide sequence and structural analysis of the zeste locus of Drosophila melanogaster. Mol. Gen. Genet. 211:121-128. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Laborda, A., A. Gonzalez-Reyes, and G. Morata. 1992. Trans regulation in the Ultrabithorax gene of Drosophila: alterations in the promoter enhance transvection. EMBO J. 11:3645-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 44.Mazo, A. M., D. Huang, B. A. Mozer, and I. B. Dawid. 1990. The trithorax gene, a trans-acting regulator of the Bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc. Natl. Acad. Sci. USA 87:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGinnis, W., and R. Krumlauf. 1992. Homeobox genes and axial patterning. Cell 68:283-302. [DOI] [PubMed] [Google Scholar]
- 46.Moreno, E., and G. Morata. 1999. Caudal is the Hox gene that specifies the most posterior Drosophila segment. Nature 400:873. [DOI] [PubMed] [Google Scholar]
- 47.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 48.Naar, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 49.Orlando, V., E. P. Jane, V. Chinwalla, P. J. Harte, and R. Paro. 1998. Binding of Trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 17:5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papp, B., and J. Muller. 2006. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 20:2041-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petruk, S., Y. Sedkov, K. M. Riley, J. Hodgson, F. Schweisguth, S. Hirose, J. B. Jaynes, H. W. Brock, and A. Mazo. 2006. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127:1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhee, K., M. Brunori, V. Besset, R. Trousdale, and D. J. Wolgemuth. 1998. Expression and potential role of Fsrg1, a murine bromodomain-containing homologue of the Drosophila gene female sterile homeotic. J. Cell Sci. 111:3541-3550. [DOI] [PubMed] [Google Scholar]
- 53.Ringrose, L., and R. Paro. 2004. Epigenetic regulation of cellular memory by the Polycomb and trithorax group proteins. Annu. Rev. Genet. 38:413-443. [DOI] [PubMed] [Google Scholar]
- 54.Rubin, G. M., and A. C. Spradling. 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218:348-353. [DOI] [PubMed] [Google Scholar]
- 55.Ruther, U., and B. Muller-Hill. 1983. Easy identification of cDNA clones. EMBO J. 2:1791-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanchez-Elsner, T., D. Gou, E. Kremmer, and F. Sauer. 2006. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science 311:1118-1123. [DOI] [PubMed] [Google Scholar]
- 57.Schneuwly, S., R. Klemenz, and W. J. Gehring. 1987. Redesigning the body plan of Drosophila by ectopic expression of the homeotic gene Antennapedia. Nature 325:816-818. [DOI] [PubMed] [Google Scholar]
- 58.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C.-T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 59.Shearn, A. 1989. The ash-1, ash-2 and trithorax genes of Drosophila melanogaster are functionally related. Genetics 121:517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sigman, D. S., M. D. Kuwabara, C. H. Chen, and T. W. Bruice. 1991. Nuclease activity of 1,10-phenanthroline-copper in study of protein-DNA interactions. Methods Enzymol. 208:414-433. [DOI] [PubMed] [Google Scholar]
- 61.Smale, S. T., and J. T. Kadonaga. 2003. The RNA polymerase II core promoter. Annu. Rev. Biochem. 72:449-479. [DOI] [PubMed] [Google Scholar]
- 62.Smith, C. L., and C. L. Peterson. 2005. ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65:115-148. [DOI] [PubMed] [Google Scholar]
- 63.Tamkun, J. W., R. Deuring, M. P. Scott, M. Kissinger, A. M. Pattatucci, T. C. Kaufman, and J. A. Kennison. 1992. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68:561-572. [DOI] [PubMed] [Google Scholar]
- 64.Tie, F., T. Furuyama, J. Prasad-Sinha, E. Jane, and P. J. Harte. 2001. The Drosophila Polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding p55 and the histone deacetylase RPD3. Development 128:275-286. [DOI] [PubMed] [Google Scholar]
- 65.Tillib, S., S. Petruk, Y. Sedkov, A. Kuzin, M. Fujioka, T. Goto, and A. Mazo. 1999. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Mol. Cell. Biol. 19:5189-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Treisman, J. 2001. Drosophila homologues of the transcriptional coactivation complex subunits TRAP240 and TRAP230 are required for identical processes in eye-antennal disc development. Development 128:603-615. [DOI] [PubMed] [Google Scholar]
- 67.Tripoulas, N., D. LaJeunesse, J. Gildea, and A. Shearn. 1996. The Drosophila ash1 gene product, which is localized at specific sites on polytene chromosomes, contains a SET domain and a PHD finger. Genetics 143:913-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vazquez, M., L. Moore, and J. A. Kennison. 1999. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the Brahma chromatin-remodeling factor to regulate transcription. Development 126:733-742. [DOI] [PubMed] [Google Scholar]
- 69.White, R. A., and M. Wilcox. 1984. Protein products of the bithorax complex in Drosophila. Cell 39:163-171. [DOI] [PubMed] [Google Scholar]
- 70.Winston, F., and C. D. Allis. 1999. The bromodomain: a chromatin-targeting module? Nat. Struct. Mol. Biol. 6:601-604. [DOI] [PubMed] [Google Scholar]
- 71.Yang, Z., J. H. N. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19:535-545. [DOI] [PubMed] [Google Scholar]
- 72.Zoller, M. J., and S. S. Taylor. 1979. Affinity labeling of the nucleotide binding site of the catalytic subunit of cAMP-dependent protein kinase using p-fluorosulfonyl-[14C]benzoyl 5′-adenosine. Identification of a modified lysine residue. J. Biol. Chem. 254:8363-8368. [PubMed] [Google Scholar]