Abstract
Mammalian prion diseases are fatal neurodegenerative disorders dependent on the prion protein PrP. Expansion of the oligopeptide repeats (ORE) found in PrP is associated with inherited prion diseases. Patients with ORE frequently harbor PrP aggregates, but other factors may contribute to pathology, as they often present with unexplained phenotypic variability. We created chimeric yeast-mammalian prion proteins to examine the influence of the PrP ORE on prion properties in yeast. Remarkably, all chimeric proteins maintained prion characteristics. The largest repeat expansion chimera displayed a higher propensity to maintain a self-propagating aggregated state. Strikingly, the repeat expansion conferred increased conformational flexibility, as observed by enhanced phenotypic variation. Furthermore, the repeat expansion chimera displayed an increased rate of prion conversion, but only in the presence of another aggregate, the [RNQ+] prion. We suggest that the PrP ORE increases the conformational flexibility of the prion protein, thereby enhancing the formation of multiple distinct aggregate structures and allowing more frequent prion conversion. Both of these characteristics may contribute to the phenotypic variability associated with PrP repeat expansion diseases.
The accumulation of misfolded proteins is a pathological characteristic common to neurodegenerative disorders such as Alzheimer's, Parkinson's, Huntington's, and prion diseases. Prion diseases are unique in this group by virtue of the self-propagating and transmissible nature of the misfolded prion protein (40). The prion replicates itself by converting soluble protein into the insoluble prion conformation. Only one prion protein has been identified in mammals, PrP, and its conversion into the prion conformation causes neurodegenerative diseases (40). Intriguingly, multiple proteins that behave as prions have been discovered in fungi, but these self-propagating elements do not cause disease (49). Investigation of fungal prions has provided much support for the prion hypothesis and has introduced several new tools to study prion propagation (11).
In Saccharomyces cerevisiae, the essential translation termination factor Sup35p can propagate as a prion called [PSI+] (48). Cells containing the [PSI+] prion exhibit increased nonsense suppression (48), presumably because the prion aggregates preclude the Sup35 protein from participating as efficiently in translation termination. The increase in nonsense suppression due to the [PSI+] state can be observed in yeast cells harboring premature stop codons in nutritional markers, thereby allowing the prion state to be monitored phenotypically (11).
Since the function of the mammalian prion protein is unknown, it is difficult to determine how mutations in PrP affect function and phenotype. Inherited prion diseases arise as a result of mutations in the gene encoding PrP, PRNP, some of which may favor the formation of the infectious prion conformation, PrPSc (52). Intriguingly, patients harboring disease-associated PRNP mutations often present with highly variable phenotypes. Even members of one family carrying identical PRNP mutations can present with unique symptoms, including variations in age of disease onset, disease severity, and disease duration (16, 24, 27).
Mutations that result in expansion of the PrP oligopeptide repeat (ORE) domain (ORD) are associated with dominant, inherited prion diseases (52). Insertional mutations have been identified in patients that expand the number of repeats up to 14 (27). Transgenic mice containing a 14-PrP repeat expansion, Tg PrP(PG14), develop a neurological disease similar to that of humans with inherited OREs (12, 13). The ORD is thought to be dispensable for PrP to form infectious PrPSc (41), but the PrP(PG14) protein accumulates in PrPSc-like deposits that are abundant in the brains of Tg PrP(PG14) mice (12, 13). These aggregates are not infectious (14), but protein aggregates from patients harboring other PrP OREs have been demonstrated to be transmissible to primates (7). Thus, it remains unclear how PrP repeat expansions cause disease.
Conversely, how amino acid sequence changes correlate to alterations in prion propagation has been determined for some yeast prion proteins. The sequence necessary for the conversion of Sup35p into the prion state has been defined by domain mapping and mutagenesis studies (reviewed in reference 54). The prion-forming domain (PFD) has been localized to the N terminus of Sup35p and is necessary and sufficient for prion propagation (30, 46). The PFD has a strikingly high percentage of glutamine (Q) and asparagine (N) amino acids, and mutation of certain Q or N residues eliminates [PSI+] (17). Interestingly, one region in the Sup35p PFD is strikingly similar to a region in PrP, the ORD (6). Sup35p contains 51/2 OREs, and the ORD of PrP contains five octapeptide repeats. Deletion of one or more Sup35p repeats prevents efficient propagation of [PSI+] (31, 38). Alternatively, expansion of the Sup35p ORD results in enhanced prion conversion (31), suggesting that the ORD of Sup35p influences the de novo formation of the prion state.
In order to investigate the aggregation propensity associated with the PrP repeats and the effect of disease-associated repeat expansions, we developed a novel model system in yeast. Previous research demonstrated that one PrP repeat can replace one Sup35p repeat and maintain prion competence in yeast (38), suggesting that PrP repeats may structurally mimic the Sup35p repeats. Here, we replaced the entire repeat region of Sup35p with various PrP repeat lengths and determined that the resulting chimeric proteins can behave as prions in yeast. Interestingly, we found that the PrP repeat sequence altered prion stability. The insertion of the longest repeat region (14-PrP ORE) resulted in the strongest prion phenotype and enhanced phenotypic variability. In addition, spontaneous conversion of the chimeric proteins to the prion state was dramatically increased in comparison to that of wild-type Sup35p, but only in the presence of [RNQ+]. Additional experiments demonstrated that the 14-PrP ORE could replace the N-terminal Q/N-rich region of the Sup35p PFD and successfully maintain and propagate a prion. Taken together, our results indicate that the ORE of PrP is more prion competent and enhances phenotypic variability compared to the wild-type PrP repeat length. We suggest that these properties may contribute to the variability associated with inherited prion disorders in humans.
MATERIALS AND METHODS
Strain construction.
All of the yeast strains used in this study were derivatives of 74-D694 (MATa or MATα ade1-14 trp1-289 his3Δ-200 ura3-52 leu2-3,112) (10). Yeast cells were grown and manipulated by standard techniques (22). A 74-D694 [PSI+] diploid with one copy of SUP35 replaced with a kanamycin resistance cassette (sup35::KanMX4) was transformed with a plasmid containing SUP35 (pYK810) (38). Haploid progeny cells containing pYK810 and sup35::KanMX4 were obtained (74-D694 pYK810) from the diploid. The isogenic [psi−] 74-D694 pYK810 strain was created by growth on rich medium (YPD) (22) containing 3 mM guanidine hydrochloride (GdHCl). The plasmid shuffle technique was used to create strains expressing only the Sup35p-PrP chimera (SP5, SP14, and P14MC strains). To obtain SP14/SUP35 diploid strains, [SP14+] Δsup35 cells were mated to wild-type [psi−] cells. The resulting progeny cells showed a 2:2 segregation of the sup35 deletion, and only the Δsup35 spores harbored the SP14 plasmid. The absence of the plasmid in spores containing chromosomal SUP35 is likely due to toxicity associated with the overexpression of SUP35 in strong [PSI+] strains (19). The [PSI+] Δrnq1 strain was created by PCR amplification of the antibiotic resistance marker hygromycin B with primers 5′-GAACGTACATATAGCGATACAAACGTATAGCAAAGATCTGAAATGTCGTACGCTGCAGGTCGAC-3′ and 5′-CAAATACGTAAACAAAGGATAGAAGGCGAACTGAATCATCGTTCAATCGATGAATTCGAGCTCG-3′ and subsequent transformation of the resulting product into [PSI+] 74-D694 pYK810. The plasmid shuffle approach was used to create chimeric [SP5+] Δrnq1 and [SP14+] Δrnq1 strains. To create haploid [RNQ+] strains, [sp5−] [rnq−] or [sp14−] [rnq−] strains were crossed to [psi−] [RNQ+], diploids were obtained, and tetrads were dissected. All deletion strains were verified by PCR and Western blotting.
Plasmid construction.
SUP35-PrP chimeras were created by precise replacement of the repeat region of Sup35p (amino acids 40 to 96) with the octapeptide repeats of PrP. To create the chimeric Sup35-PrP plasmids, a 5′ fragment of SUP35 that added a 5′ BamHI restriction site and had the endogenous EcoRV site was cloned into an intermediate vector. The resulting plasmid was cut with HindIII and EcoRV to insert the various repeat regions of PrP containing DNA encoding 5, 8, 11, and 14 repeats. The repeat regions of PrP were PCR amplified from plasmids containing mouse PrP with 5, 8, 11, and 14 repeats (D. Harris, unpublished data) with primers A (5′-GGTTATCAAGCTTACAATGCTCAAGCCCAACCTCAGGGTGGCACCTGG-3′) and B (5′-ACCAGCTTGATATCCTTGCAAATTGTTATTGTAGTTGAAGTTTTTGTAATTTCCACGTTGGCCCCATCCACCGCC-3′). The DNA sequence of PrP was verified by DNA sequencing, and the resulting translated repeat region for wild-type PrP was as expected (PQGGTWGQ PHGGGWGQ PHGGSWGQ PHGGSWGQ PHGGGWGQ). The PrP repeat expansion amino acid sequence was PQGGTWGQ PHGGGWGQ (PHGGSWGQ PHGGSWGQ PHGGGWGQ)n, in which PrP with 8 repeats contained n = 2, PrP with 11 repeats contained n = 3, and PrP with 14 repeats contained n = 4. The chimeric fragments were removed from the intermediate vector by BamHI and EcoRV and cloned into pUCK1512 (38), which contained the rest of SUP35. The full-length chimeras were removed with XhoI (blunted) and BamHI from pUKC1512 and cloned into p413TEF (35) cut at the BamHI and SmaI sites. The resulting Sup35p-PrP chimeras with 5, 8, 11, and 14 PrP repeats were called SP5, SP8, SP11, and SP14, respectively. The positive control plasmid (pSup35) with the wild-type SUP35 sequence was created in the same manner. The P14MC plasmid was created by PCR amplification of the 14 PrP repeats with primers B and C (5′-CGGGATCCATGTCGCCTCAGGGTGGCACCTGG-3′), and the resulting fragment was cloned into pUKC1512 cut at the BamHI and EcoRV sites. The SP5-PFD-green fluorescent protein (GFP) and SP14-PFD-GFP fusion constructs were made by PCR amplification of the specific PFD region of the chimeras with primers 5′GGCGCAGGATCCATGTCGGATTCAAACCAA-3′ and 5′GCGCCGCGGATCGTTAACAACTTCGTCATCCAC-3′, and the products were digested with the BamHI and SacII restriction enzymes. The products were cloned into a SUP35-PFD-GFP plasmid (mCNMG) (39) digested with the same enzymes, replacing the PFD of wild-type SUP35. All plasmids were verified by DNA sequencing.
Phenotypic and stability assays.
To test for the prion phenotype, cells were grown to an optical density at 600 nm (OD600) of ∼1.2 in YPD, serially diluted fivefold, and spotted onto YPD, SD-ade, and YPD-3 mM GdHCl plates. To test for the mitotic stability of [PSI+], [SP5+], and [SP14+] strains, cells were resuspended and spread onto YPD plates. Cells were counted and scored as sectoring, solid red, or solid pink. To test for prion loss in [SP14+] chimera strains, cells were grown in YPD and spread onto YPD plates. The percentage of solid red colonies was determined, and the n-fold change in stability was calculated relative to the most stable strain (strong [SP14+]). The data presented represent three independent experiments in which ∼15,000 colonies were scored for each strain variant. To determine the prion conversion of [prion−] [rnq−] strains, the ura3-14 allele was used (32). Transformants containing the pLeu2-ura3-14 construct were grown overnight in SD-leu medium and plated on SD-ura-ade plates. The number of [PRION+] cells was determined by color and curability by GdHCl and compared to the total number plated. To determine the prion conversion of [prion−] [RNQ+] strains, cells were resuspended, plated on YPD, and analyzed for the number of pink colonies compared to the total number plated. All [PRION+] cells were verified by curability by GdHCl.
Protein analysis.
Sedimentation properties were analyzed as described previously with the addition of the mini EDTA-free protease inhibitor cocktail (Roche) to the ST buffer (36). Large aggregates were separated by semidenaturing detergent agarose gel electrophoresis (SDD-AGE) as previously described (1), with the following minor changes. Lysis buffer contained the mini EDTA-free protease inhibitor cocktail (Roche), and the lysates were incubated in sample buffer for 7 min at room temperature before electrophoresis. Sup35p and Sup35p-PrP chimeras were detected with a rabbit polyclonal antibody against the middle region of Sup35p that is maintained in all chimeras, anti-Sup35 (39).
PFD-GFP microscopy.
Cells were transformed with a copper-inducible plasmid containing either an SP5-PFD-GFP or an SP14-PFD-GFP fusion, and fluorescence microscopy was performed after expression of the fusion protein was induced in log-phase cells with 50 μM copper for 1 h (39).
Curing by overexpression of Hsp104p.
Hsp104p overexpression was induced from a galactose-inducible plasmid expressing Hsp104p (pYS-Gal104) by growth in minimal medium lacking uracil (liquid or plates) supplemented with 2% galactose and 1% raffinose, and final colony color was assessed on YPD (10).
Protein transformation. (i) Spheroplasting of cells.
74-D694 [prion−] cells were grown to an OD600 of ∼0.5 and harvested. The cell pellet was washed successively with 10 ml of water, 10 ml of 1 M sorbitol, and 1 ml of SCE buffer (1 M sorbitol, 10 mM EDTA, 10 mM dithiothreitol, 100 mM sodium citrate, pH 5.8). The cells were spheroplasted in 500 μl of SCE buffer with lyticase (in sodium citrate, pH 5.8) for 30 to 40 min at 30°C. After spheroplasting was complete, cells were carefully washed twice with 1 ml 1 M sorbitol and twice with 1 ml STC buffer (1 M sorbitol, 10 mM CaCl2, 10 mM Tris, pH 7.5) and then harvested at 800 × g for 1 min at 4°C. Spheroplasted cells were resuspended in 1 ml of STC buffer at 4°C.
(ii) Prion particle preparation.
[PRION+] and [prion−] cells were grown to an OD600 of ∼0.5 and harvested. Cell pellets were washed successively with 10 ml of water, 10 ml of 1 M sorbitol, and 1 ml of SCE buffer. Cells were resuspended in 600 μl of SCE buffer (containing 1 mM phenylmethylsulfonyl fluoride and protease inhibitor cocktail [Sigma Chemical Co.]) and lysed with 300 μl of sterilized glass beads by vortexing 10 times for 10 s each at 4°C. The crude lysate was centrifuged twice at 4°C for 5 min at 800 × g and 1,000 × g. The protein concentration of the supernatant was determined with the Bio-Rad protein assay reagent (Bio-Rad Laboratories). The protein lysate was sonicated three times for 10 s each time prior to transformation (Sonic Dismembrator; Fisher Scientific).
(iii) Transformation procedure.
The transformation mixture contained 5 μl of pRS316 DNA (∼300 ng/μl), ∼300 μg of sonicated protein lysate, 10 μl of carrier DNA (10 mg/ml), 150 μl of spheroplasted cells, and 5 volumes of PEG 8000 buffer (20% [wt/vol] PEG 8000, 10 mM CaCl2, 10 mM Tris, pH 7.5) and was incubated for 45 min at 25°C. The transformation mixture was harvested at 1,000 × g and resuspended in 150 μl of SOS buffer (1 M sorbitol, 7 mM CaCl2, 0.25% yeast extract, and 0.5% Bacto peptone supplemented with 0.3 mg of all of the amino acids in which the yeast strain is deficient) and incubated for 30 min at 30°C. The SOS-cell mixture was plated on SD-ura/sorbitol plates (16.4% sorbitol, 3% glucose, 2% agar, complete supplement mixture without uracil, 0.67% yeast nitrogen base without amino acids) overlaid with top agar (1.2 M sorbitol, 2.5% agar, 2% glucose, complete supplement mixture without uracil, 0.67% yeast nitrogen base without amino acids). The plates were incubated at 30°C for 5 days. Transformants were spotted onto YPD, SD-ade, and 3 mM GdHCl media to score the colonies for the prion state.
RESULTS
Chimeric Sup35-PrP proteins form prions.
To test the effects of the repeat expansion of PrP in a genetically tractable system, we expressed SUP35-PrP chimeras in yeast (Fig. 1). We replaced the ORD of Sup35p with the wild-type number of 5 PrP repeats and an expanded length of 14 PrP repeats (Fig. 1A). The chimeric molecules, termed SP5 (for Sup35-PrP 5 repeats) and SP14 (for Sup35-PrP 14 repeats), replaced the wild type by plasmid shuffle and were expressed as the only copy of SUP35. Since SUP35 is essential in yeast, this approach allowed us to evaluate the functionality of the chimeras in the absence of wild-type Sup35p. The expression of the chimeric proteins was similar to endogenous Sup35 protein levels, as determined by Western blotting (Fig. 1B). A control plasmid with wild-type SUP35 (termed pSup35) was generated and used to ensure that the episomal expression of Sup35p mimicked that of chromosomal Sup35p.
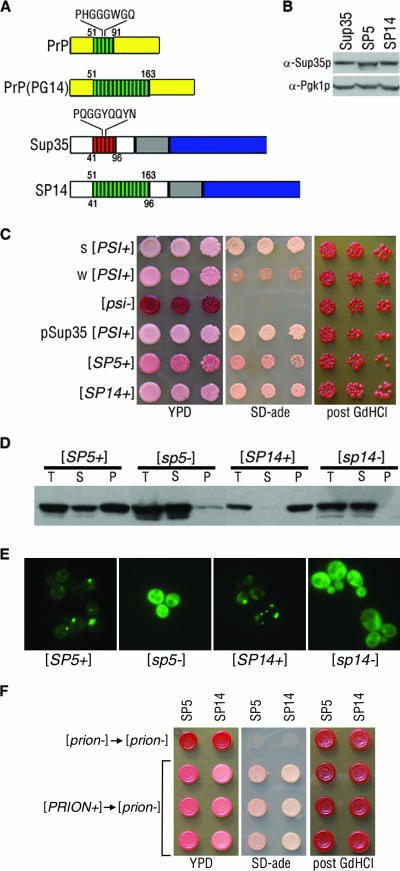
FIG. 1.
Chimeric Sup35-PrP proteins demonstrate yeast prion properties. (A) Schematic depiction of Sup35p and PrP highlighting the consensus sequence of each ORD. PrP(PG14) contains nine additional repeats. The chimeric Sup35-PrP protein (SP14) is also shown. Numbers below the diagrams represent Sup35p amino acids, and those above correspond to PrP. (B) Lysates from cells containing the Sup35p, SP5, and SP14 proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting to determine relative chimeric Sup35 protein levels. The blot was reprobed with antibodies to Pgk1p to confirm equal protein loading. (C) Strong (s) [PSI+] strain variant, weak (w) [PSI+] strain variant, [psi−], plasmid-borne SUP35-supported [PSI+] (pSup35[PSI+]), [SP5+], and [SP14+] cells were spotted onto YPD and SD-ade media. GdHCl-cured cells were subsequently spotted onto YPD (post GdHCl). (D) Cell lysates of the chimeras were subjected to ultracentrifugation, and the chimeric proteins in the total (T), supernatant (S), and pellet (P) fractions were analyzed by Western blotting. (E) [SP5+], [sp5−], [SP14+], and [sp14−] cells expressing the corresponding PFD-GFP constructs were analyzed by fluorescence microscopy. (F) Prion particles isolated from [SP5+] and [SP14+] cells can convert [prion−] cells. Protein harvested from [sp5−] or [sp14−] cells was transformed into [sp5−] or [sp14−] cells, respectively (top, [prion−] → [prion−]). Protein harvested from [SP5+] or [SP14+] cells was transformed into [sp5−] or [sp14−] cells, respectively (bottom three lines, [PRION+] → [prion−]). Individual isolates were spotted onto YPD or SD-ade, cured on YPD with 3 mM GdHCl, and subsequently spotted onto YPD (post GdHCl).
To test if the chimeric proteins could propagate the prion state, each chimera was introduced into a [PSI+] strain. The strain contains the ade1-14 allele, which harbors a premature stop codon that is read through in [PSI+] cells, thereby producing full-length, functional Ade1p. Thus, [PSI+] cells with the ade1-14 allele are adenine prototrophs and appear as light pink colonies on a rich medium (YPD). In contrast, in [psi−] cells, Sup35p is soluble and functional and therefore faithfully terminates translation. As such, [psi−] cells that harbor the ade1-14 allele are adenine auxotrophs and colonies appear red on YPD. If SP5 or SP14 protein is able to propagate the prion state, then cells containing these chimeric proteins will display a nonsense suppression phenotype in the absence of wild-type Sup35p and remain pink on YPD. Both SP5- and SP14-containing cells remained pink on YPD and grew on SD-ade medium after replacement of wild-type Sup35p in a [PSI+] strain (Fig. 1C), suggesting that these chimeric proteins maintain the prion phenotype. We refer to the prion states of SP5 and SP14 as [SP5+] and [SP14+], respectively. To determine if the [SP5+] and [SP14+] phenotypes could be eliminated (cured) in a manner similar to that of [PSI+], the cells were grown on YPD medium containing 3 mM GdHCl. The [SP5+] and [SP14+] cells became red after growth on medium containing GdHCl (Fig. 1C) and could no longer grow on SD-ade medium (data not shown). Moreover, when the chimeras were assessed as the only copy of SUP35 in a [psi−] strain, the cells remained red and could not grow on SD-ade medium (data not shown), demonstrating that both chimeras are functional in translation termination. Since the PrP repeat expansion consisting of 14 PrP repeats is pathogenic in humans, we asked if other pathogenic PrP ORD insertions could maintain prion properties in chimeras. Chimeras containing 8 and 11 PrP repeats were created and assayed. These chimeras also behaved phenotypically, biochemically, and genetically as yeast prions (data not shown). Since all of the chimeras were able to maintain the prion state, we analyzed SP5 and SP14 further to determine the effects of repeat expansions on prion properties.
The [SP14+] cells displayed more robust growth on SD-ade medium and a lighter pink colony color on YPD in comparison to [SP5+] cells (Fig. 1C). This suggests that the repeat-expanded SP14 protein maintains a stronger prion than SP5. Interestingly, cells with wild-type SUP35 can harbor strain variants of the [PSI+] prion that display heritable differences in the strength of nonsense suppression without any alteration in amino acid sequence (21). These changes in nonsense suppression can be phenotypically distinguished in yeast cells harboring the ade1-14 allele as different shades of pink on YPD and different growth rates on SD-ade medium (Fig. 1C) (50). The differences in nonsense suppression between [SP5+] and [SP14+] cells suggest that the chimeras are propagating structurally distinct prion strain variants.
The aggregation and resulting insolubility of Sup35p are defining characteristics of the [PSI+] prion state (48). To examine if the chimeric [SP5+] and [SP14+] prion phenotypes are associated with insoluble protein aggregates, we subjected lysates from [SP5+], [sp5−], [SP14+], and [sp14−] cells to ultracentrifugation to determine the fractionation pattern of the chimeric proteins. [SP5+] cells contained chimeric protein in both the supernatant and pellet fractions, while [sp5−] cells had most the protein in the supernatant (Fig. 1D). The presence of soluble protein in the [SP5+] prion cells was not unexpected, given the weak nonsense suppression phenotype observed (Fig. 1C). Lysate from [SP14+] cells, however, showed all of the chimeric protein in the pellet fraction, consistent with the stronger nonsense suppression phenotype, and lysate from [sp14−] cells displayed all of the protein in the supernatant (Fig. 1D).
To further investigate the aggregation of the chimeric prion proteins, the cellular distribution of the chimeric proteins was monitored by GFP fluorescence. An inducible PFD-GFP construct specific for each repeat region (SP5-PFD-GFP or SP14-PFD-GFP) was expressed in the corresponding [PRION+] and isogenic [prion−] cells. Cells containing [SP5+] prions displayed aggregates and diffuse background fluorescence, whereas [sp5−] cells showed only diffuse fluorescence (Fig. 1E). [SP14+] cells showed primarily a punctate fluorescence pattern, whereas [sp14−] cells displayed diffuse fluorescence. Taken together, these data demonstrate that the chimeric proteins assume an aggregated state in cells displaying a nonsense suppressor phenotype and both traits are reversed by curing.
In order to further demonstrate the prion nature of the SP5 and SP14 chimeras, we conducted protein transformations (25, 43). Protein harvested from [SP5+] and [SP14+] strains was transformed into [sp5−] and [sp14−] cells, respectively. As expected, protein obtained from [PRION+] cells, but not [prion−] cells, was able to convert [prion−] cells to the prion state (Fig. 1F). Together, these results demonstrate that the SP5 and SP14 chimeras behave as yeast prions.
Chimeric prions display enhanced instability.
To determine the mitotic stability of the chimeric prions, [SP5+], [SP14+], and [PSI+] cells were plated on YPD and the colonies were scored as solid red, pink, or sectoring. Wild-type strong (s) [PSI+] cells never displayed sectoring colonies, and less than 0.02% of the colonies spontaneously appeared red (Table 1). In contrast, approximately 85% of both [SP5+] and [SP14+] colonies sectored. This suggests that the heritable stability of the chimeric prions is much lower than that observed with [PSI+] cells. Since the ORD of PrP is known to bind copper (33), we tested whether the addition of copper to the medium influenced mitotic stability and found no change (data not shown). A striking difference between [SP5+] and [SP14+] cells was noted when the nonsectoring populations of cells were analyzed (Table 1). All nonsectoring [SP5+] colonies were red, indicating loss of the prion state. In contrast, most of the nonsectoring population of [SP14+] colonies was light pink, indicating maintenance of the prion state. Thus, although the SP5 protein propagates as a prion, the maintenance of the prion state is enhanced with the repeat expansion.
TABLE 1.
Mitotic stability of the [PRION+] state in the chimeras
| Cell type | Avg % of sectoring colonies ± SD | Avg % of nonsectoring colonies ± SD
|
|
|---|---|---|---|
| Red | Pink | ||
| s [PSI+] | 0 | <0.02 | >99.98 |
| [SP5+] | 85.7 ± 3.8 | 14.2 ± 3.8 | 0 |
| [SP14+] | 84.1 ± 4.8 | 1.8 ± 3.1 | 14.1 ± 2.6 |
[SP14+] prions display epigenetic inheritance and strain variants.
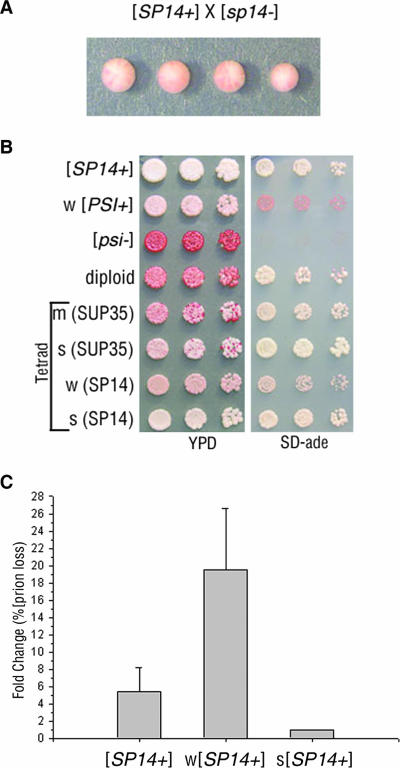
A hallmark of yeast prions is dominant, epigenetic transmission of the prion phenotype (48). In a cross between [psi−] and [PSI+] cells, the [PSI+] state is dominant in the diploid and when sporulated, all four meiotic progeny display the prion phenotype. To determine if the [SP14+] prion phenotype could be inherited in an epigenetic manner, [SP14+] cells were crossed to [sp14−] cells. The resulting diploid displayed the nonsense suppression phenotype, as did all meiotic progeny (Fig. 2A). Moreover, [SP14+] cells were also able to convert [sp5−] cells to [SP5+] in a cross between heterologous strains (data not shown). These results demonstrate that [SP14+] does indeed behave as a prion genetically. To determine if [SP14+] could transmit the prion phenotype to wild-type Sup35p, [SP14+] cells were crossed to [psi−] cells. The resulting diploid and meiotic progeny were analyzed for the prion phenotype. [SP14+] cells, [psi−] cells and SP14/SUP35 diploid cells were spotted onto YPD and SD-ade media to assess nonsense suppression (Fig. 2B). The SP14/SUP35 [PRION+] diploid grew on SD-ade medium but displayed both pink and red colonies on YPD, suggesting inefficient inheritance of the nonsense suppression phenotype. Tetrads from the SP14/SUP35 [PRION+] diploid were spotted onto YPD and SD-ade media to assess inheritance of the prion phenotype in the progeny. The tetrads were also spotted onto selective medium to follow the 2:2 segregation of the sup35 deletion and the presence of SP14. Spores expressing SP14 protein displayed phenotypic variability, as shown in the example tetrad (Fig. 2B): one light pink colony and one dark pink colony (YPD). The light pink SP14 colony grew better on SD-ade medium than the dark pink colony. Spores containing wild-type SUP35 also inherited the prion phenotype but showed a high degree of instability, as both pink and red colonies were seen on YPD. The loss of nonsense suppression in the progeny expressing wild-type Sup35p suggests inefficient templating of the [SP14+] prion structure onto the Sup35 protein. However, once a single pink colony containing wild-type Sup35p was purified by restreaking, a stable [PSI+]-dependent nonsense suppression phenotype was established (data not shown). Different strengths of nonsense suppression in [PSI+] cells are observed in prion strain variants (Fig. 1C, top two rows). The SP14-containing progeny displayed differential nonsense suppression phenotypes, suggesting that the SP14 protein propagates distinct strain variants in [SP14+] cells.
FIG. 2.
[SP14+] prions display non-Mendelian inheritance and mitotic instability. (A) A representative tetrad from a cross of [SP14+] to [sp14−] illustrated epigenetic inheritance of the prion phenotype. (B) [SP14+], weak (w) [PSI+], [psi−], an SP14/SUP35 [PRION+] diploid (containing one copy of SP14 and one copy of SUP35), and a tetrad from the SP14/SUP35 [PRION+] diploid were spotted onto YPD and SD-ade media. The relative strength of nonsense suppression in the progeny is described as strong (s), medium (m), or weak (w). The SUP35 allele expressed in the progeny is indicated parenthetically. (C) The original isolate of [SP14+] and phenotypically weak and strong [SP14+] variants were plated on YPD and analyzed for prion loss (red colonies). Prion loss was calculated as the change compared to the most stable [SP14+] variant.
Strain variants of [SP14+] show a distinct change in mitotic stability.
Weak [PSI+] strain variants are mitotically unstable in comparison to strong [PSI+] strain variants (21, 26). To explore the possibility that [SP14+] meiotic progeny shared this characteristic of prion strain variants, we examined the mitotic stability of the chimeric prions in the SP14 progeny. SP14-containing progeny displaying a strong and a weak nonsense suppression phenotype, in addition to the original [SP14+] isolate, were analyzed for prion loss (the appearance of red colonies). Cells were plated on YPD, and the number of solid red colonies was scored as a percentage of the total. The weak [SP14+] strain variant had almost a 20-fold higher prion loss compared to the strong strain variant (Fig. 2C). These data show that the weak [SP14+] strain variant displaying less nonsense suppression is mitotically less stable than the strong [SP14+] strain variant.
Since strain variants of [PSI+] and PrPSc are due to structurally distinct aggregates (15, 28, 44), we hypothesized that weak and strong yeast prion strain variants might show differential transmission of their respective structures onto soluble proteins with different amino acid sequences. Therefore, we tested whether each isolated strain variant could transmit its prion state to soluble wild-type Sup35p or soluble SP14 protein. Strong and weak [PSI+] cells were mated to [psi−] and [sp14−] cells. Diploids were obtained, and tetrads were dissected. As expected, analysis of the tetrads showed that both strong (s) and weak (w) [PSI+] could transmit to [psi−] and convert Sup35p to [PSI+] (Table 2). However, only strong [PSI+] could transmit prion properties to [sp14−] to generate [SP14+] cells. The capacity of strong [PSI+], but not weak [PSI+], to propagate the prion structure in SP14-containing cells provides additional genetic evidence for the structural difference between wild-type [PSI+] strain variants. Similarly, to examine the transmission of the [SP14+] variants, strong (s) and weak (w) variants were crossed to [psi−] and [sp14−] cells. Analysis of the tetrads demonstrated that two independent strong [SP14+] variants could efficiently transmit the prion structure to either [psi−] or [sp14−] cells (Table 2). In contrast, two independent weak [SP14+] strain variants could transmit the prion state to [sp14−] cells but could not transmit the prion state to wild-type [psi−] cells. Thus, weak variants of either [PSI+] or [SP14+] cells were unable to transmit the prion structure to proteins containing amino acid sequence differences; however, both were able to transmit the prion structure to proteins with identical sequences. These results suggest that the aggregate structures of strong and weak [SP14+] prion variants are different from one another.
TABLE 2.
Meiotic transmission of [SP14+] strain variants
| Cell type | Growth on SD-adeb | Transmission to [psi−] | Transmission to [sp14−] |
|---|---|---|---|
| s [PSI+] | ++++ | Yes | Yesa |
| w [PSI+] | + | Yes | Noa |
| s [SP14+] v1 | ++++ | Yes | Yes |
| s [SP14+] v2 | ++++ | Yes | Yes |
| w [SP14+] v1 | ++ | No | Yes |
| w [SP14+] v2 | ++ | Rare | Yes |
Weak [PSI+] and strong [PSI+] also show this difference in transmission to [sp5−].
++++, very good; ++, good; +, fair.
[SP5+] and [SP14+] cells contain large protein aggregates.
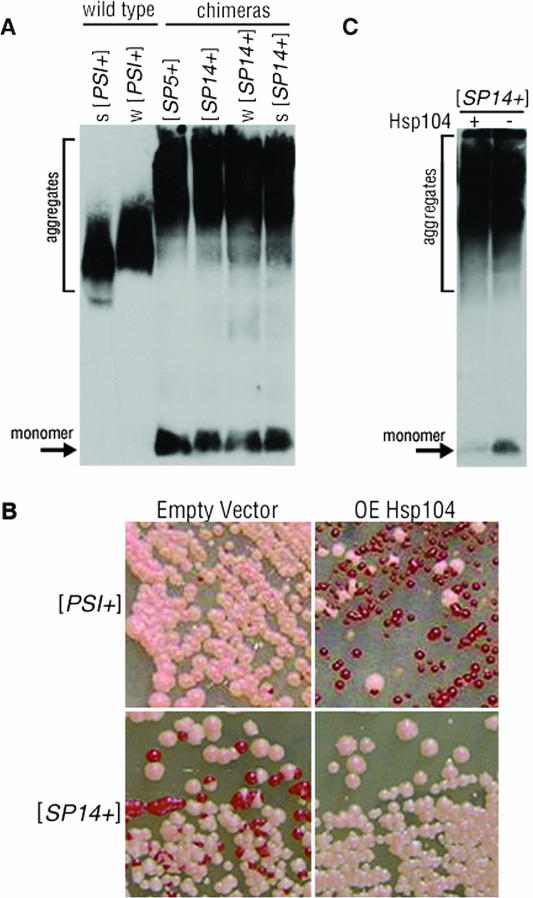
Changes in mitotic stability of [PSI+] have been previously correlated to increased aggregate size (4, 5, 29). To investigate if the decreased mitotic stability of the chimeric prions might result from different aggregate sizes, we performed SDD-AGE and detected Sup35p by Western blotting. The aggregated Sup35p from [PSI+] cells migrates as a large smear (Fig. 3A), whereas Sup35p from [psi−] cells is monomeric and appears as a faster-migrating band (29). Aggregates of Sup35p from weak [PSI+] strain variants typically display a larger smear than aggregates isolated from strong [PSI+] variants (Fig. 3A) (29). The chimeric protein from [SP5+] and [SP14+] cells showed much larger smears of protein and a broader range of sizes in comparison to Sup35p from [PSI+] strain variants (Fig. 3A). Interestingly, protein from weak and strong [SP14+] strain variants showed the same large aggregates as seen in [SP5+] and [SP14+] cells. These data suggest that the prion protein aggregates contained in [SP5+] and [SP14+] cells differ from those in weak or strong [PSI+] cells and the large aggregates may contribute to the decreased mitotic stability of the chimeras.
FIG. 3.
[SP14+] contains large aggregates which are unaffected by prion stabilization. (A) Protein aggregates and monomeric Sup35p from wild-type [PSI+] (strong [s] and weak [w]) and [SP5+] and [SP14+] (original isolate, weak and strong) strains were separated by SDD-AGE and detected by Western blotting. (B) Wild-type [PSI+] and [SP14+] cells with and without the overexpression (OE) of Hsp104p plated on YPD. (C) Protein aggregates and soluble protein (monomer) from lysates of [SP14+] cells with (+) or without (−) Hsp104p overexpression were separated by SDD-AGE and detected by Western blotting.
Hsp104p overexpression stabilizes the [SP14+] prion phenotype.
Hsp104p is required for the propagation of the [PSI+] prion (10) and is hypothesized to break up Sup35p aggregates in [PSI+] cells to facilitate the propagation and inheritance of the prion into daughter cells through mitosis (reviewed in reference 47). If the endogenous level of Hsp104p is insufficient to perform this function on the large aggregates of the chimeric proteins, then this could contribute to the observed mitotic instability (4, 5, 29). Deletion of HSP104 in the chimeras cured the prion phenotype (data not shown), indicating that Hsp104p is required for chimeric prion maintenance. Therefore, we investigated whether the overexpression of Hsp104p would affect their propagation. As expected, overexpression of Hsp104p had a curing effect on wild-type [PSI+] cells (Fig. 3B, upper right) (10). However, the overexpression of Hsp104p in [SP14+] cells produced primarily pink colonies (Fig. 3B, lower right). The phenotypic instability in [SP14+] cells remained in the empty-vector control, as evident by the frequent appearance of red colonies (Fig. 3B, lower left). Hence, in contrast to its curing effect on wild-type [PSI+] cells, Hsp104p overexpression stabilized the [SP14+] prion phenotype. Therefore, we asked if the [SP14+] aggregates were dramatically altered by the overexpression of Hsp104p. Protein from [SP14+] cells subjected to overexpression of Hsp104p was analyzed by SDD-AGE and Western blotting (Fig. 3C). Neither the size nor the range of the aggregates from [SP14+] cells changed following the overexpression of Hsp104p. However, the amount of monomeric protein was decreased in the presence of excess Hsp104p. Taken together, these data suggest that the overexpression of Hsp104p stabilizes the [SP14+] phenotype without changing the size of the observable protein aggregates. Given the broad range of aggregate sizes and the ability of SP14 to maintain multiple independent structures in a cell, we hypothesize that the stabilization of the [SP14+] prion could occur because Hsp104p is disaggregating the more unstable aggregates in the [SP14+] prion population (47). Indeed, it remains unclear how these aggregates observed by SDD-AGE relate to the prion phenotype.
[SP14+] strain variants interconvert readily.
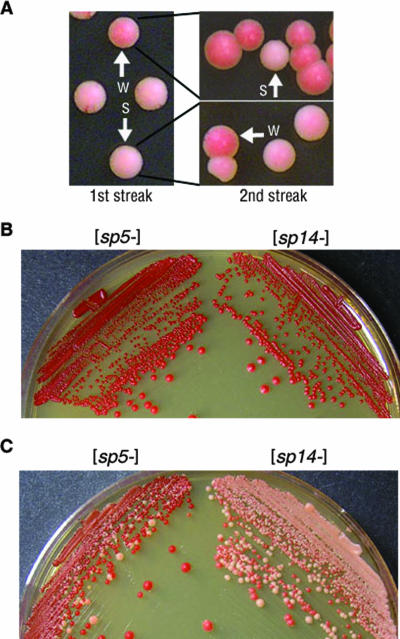
Our genetic analysis suggests that strain variants of the chimeric [SP14+] prions behave similarly to wild-type [PSI+] strain variants. Prion strain variants that occur with wild-type Sup35p rarely interconvert (26). However, we observed that [SP14+] strain variants frequently interconvert between strong and weak nonsense suppression phenotypes (Fig. 4A). Interconversion was detected by plating a [SP14+] culture on YPD and subsequently restreaking isolated colonies. Individual colonies were determined to be strong or weak phenotypically on the basis of colony color (Fig. 4A, first streak). An individual weak or strong [SP14+] colony was selected, resuspended in liquid medium, and plated on YPD. Colonies that started as one phenotype did not maintain the same phenotype in all mitotic progeny (Fig. 4A, second streak). This suggests that not only does the SP14 protein display multiple prion strain variants, but an individual colony potentially contains more than one self-propagating structure.
FIG. 4.
[SP14+] variants show frequent interconversion and higher spontaneous conversion in a [RNQ+]-dependent manner. (A) [SP14+] cells were plated on YPD (1st streak). A single strong [SP14+] variant and a single weak variant were isolated (as indicated by the arrows) and replated on YPD (2nd streak) to analyze variant stability. (B) Red colonies expressing SP5 and SP14 in [rnq−] cells remained red on YPD and were stable as [prion−]. (C) Red colonies expressing SP5 and SP14 in [RNQ+] cells gave rise to pink colonies and frequently converted to [PRION+].
Alternatively, the interconversion of the [SP14+] variants could suggest that the prion conformation of SP14 protein may be lost and then reacquired frequently. To determine if this was the case, we tested if the spontaneous conversion of [sp14−] to the prion state was enhanced in comparison to the conversion of [sp5−] or [psi−]. The spontaneous conversion of both chimeras and [psi−] to their respective [PRION+] states occurred at about the same rate (∼1 in 105 cells). Thus, if there is increased conformational flexibility in the SP14 protein due to the repeat expansion, it does not enhance the de novo appearance of the prion state. Intriguingly, this result differed from that of the wild-type Sup35p repeat expansion in which a dramatic increase in frequency of prion conversion was observed (31).
Given this difference, we set out to explore extragenic factors that may play a role in conversion frequency. It is now known that another epigenetic element, [RNQ+], influences the induction of Sup35p into the prion state (18, 20, 37) but is not necessary for continued propagation of [PSI+] (20). We determined that the [RNQ+] prion was not required for continued propagation of the chimeric prions [SP5+] and [SP14+] by expressing the chimeras in a Δrnq1 strain (data not shown). Since we initially assessed spontaneous conversion in cells that were [rnq−] (Fig. 4B), we examined the impact of the [RNQ+] prion on the spontaneous conversion of the chimeric proteins. Unlike the induction of [PSI+], which is enhanced by [RNQ+] (20), we found that the spontaneous conversion of [psi−] to [PSI+] is not affected by [RNQ+] (∼1 in 105 cells in both [rnq−] and [RNQ+] cells). However, the spontaneous conversion of [sp5−] [RNQ+] and [sp14−] [RNQ+] cells to the [PRION+] state was markedly enhanced (Fig. 4C). Strikingly, cells containing the SP14 protein converted to [SP14+] more frequently (∼3 × 10−1) than cells containing the SP5 protein converted to [SP5+] (∼6 × 10−2). This suggests that the repeat expansion does influence the spontaneous prion conversion of the chimeric protein, but only in the presence of another aggregate, [RNQ+].
Expanded PrP repeats can replace the Sup35p PFD.
We observed that the expanded repeat domain in SP14 enables the protein to initiate and propagate many structural conformations, thereby suggesting that the repeat expansion confers enhanced structural flexibility to PrP. Both the PFD of Sup35p and the N terminus of PrP, including the ORD, are highly unstructured regions (42, 51). The Q/N-rich character of the extreme N terminus of Sup35p has been shown to be critical for the maintenance of [PSI+] (17). Our initial chimeras characterized retained the Q/N-rich region of the N terminus of Sup35p. Since the SP14 results suggested enhanced conformational flexibility, we asked if the 14-PrP repeats could also replace the essential N-terminal portion of the Sup35p PFD. Therefore, we created a new chimera termed P14MC (PrP 14 repeats fused to the middle [M] and carboxy-terminal [C] domains of Sup35p) and expressed it in yeast as the sole copy of Sup35p by the plasmid shuffle approach. The expression of P14MC protein was similar to endogenous Sup35p levels (data not shown). Cells expressing the P14MC protein were analyzed for the maintenance and propagation of the prion state. Strong [PSI+], weak [PSI+] and [psi−] cells, as well as cells expressing P14MC protein in the prion state, [P14MC+], and nonprion state, [p14MC−], were grown on YPD, SD-ade, and medium containing GdHCl to assess nonsense suppression and curing (Fig. 5A). [P14MC+] cells were pink on YPD and grew well on SD-ade medium, suggesting that the protein maintains the nonsense suppression phenotype in the prion state. Conversely, [p14MC−] cells were red on YPD and could not grow on SD-ade medium, suggesting that the protein is functional in the nonprion state.
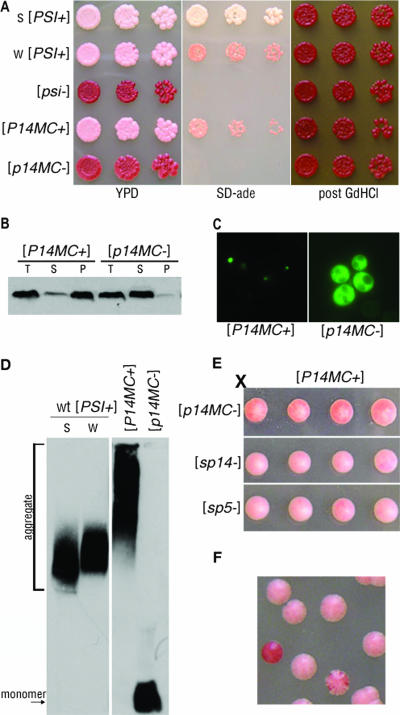
FIG. 5.
PrP repeat expansion maintains prion competence in the absence of the Q/N region of Sup35p. (A) Strong (s) [PSI+] and weak (w) [PSI+], [psi−], and P14MC in [P14MC+] and [p14MC−] cells were spotted onto YPD and SD-ade media. GdHCl-cured cells were subsequently spotted onto YPD (post GdHCl). (B) Lysates from [P14MC+] and [p14MC−] cells were subjected to ultracentrifugation, and the total (T), supernatant (S), and pellet (P) fractions were analyzed by Western blotting. (C) Fluorescence microscopy of SP14-PFD-GFP expressed in [P14MC+] and [p14MC−] cells. (D) Cell lysates from wild-type (wt) strong [PSI+] and weak [PSI+] and strains with a P14MC chimera in either the [P14MC+] or the [p14MC−] state were analyzed by SDD-AGE and Western blotting. (E) A [P14MC+] strain was crossed to [p14MC−], [sp14−], and [sp5−] strains. Tetrads dissected from the resulting diploids are shown. (F) [P14MC+] cells struck onto YPD generated colonies that maintained stable [P14MC+] (pink), colonies that completely lost the prion (red), and colonies that frequently lost the prion (sectored).
We next analyzed the biochemical properties of the P14MC protein. Cell lysates were subjected to ultracentrifugation to determine the aggregation state of the protein in [P14MC+] and [p14MC−] cells. Most of the protein from [P14MC+] cells was present in the pellet fraction, whereas that from [p14MC−] cells was in the supernatant (Fig. 5B). Thus, the nonsense suppression phenotype observed in the [P14MC+] cells correlates with the aggregation of the P14MC protein. In addition, curing [P14MC+] cells concomitantly resolubilized the protein and reversed the nonsense suppression phenotype (Fig. 5A and B). In agreement with the sedimentation results, expression of SP14-PFD-GFP showed a distinct punctate fluorescence pattern in [P14MC+] cells and diffuse fluorescence in [p14MC−] cells (Fig. 5C). To further examine the aggregates, chimeric Sup35p from [P14MC+] lysate was analyzed by SDD-AGE and Western blotting. Protein aggregates from [P14MC+] cells were larger than those from wild-type strong or weak [PSI+] cells (Fig. 5D) and resembled aggregates from [SP5+] and [SP14+] cells (Fig. 3A). Furthermore, this assay demonstrated that [p14MC−] cells contain only monomeric P14MC protein. These results suggest that the P14MC protein biochemically behaves as a prion.
Next, we determined if the [P14MC+] prion aggregates could be inherited in an epigenetic manner, as is characteristic of yeast prions. We mated [P14MC+] cells to [psi−] (data not shown), [p14MC−], [sp14−], or [sp5−] cells, obtained diploids, and dissected tetrads. The meiotic progeny all showed a 4:0 nonsense suppression phenotype, indicating that the [P14MC+] prion was transmitted in an epigenetic fashion to the chimeras (Fig. 5E) but displayed a slightly weaker nonsense suppression phenotype than [SP14+] (Fig. 5E; also compare SD-ade panels in Fig. 1C and 5A). In addition, protein obtained from [P14MC+] cells converted [p14MC−] cells to [P14MC+] after protein transformation (data not shown). We also examined if the lack of the Q/N region in the P14MC protein affected the stability of the [P14MC+] prion in comparison to [SP14+] cells (Fig. 5F). [P14MC+] cells plated on YPD displayed both red and sectoring colonies, suggesting that the change in the context of the 14-PrP repeats did not alter the stability of the chimeric prion, as [SP14+] cells also showed red and sectoring colonies (Table 1). Taken together, these results indicate that 14-PrP repeats can replace both the N-terminal Q/N region and the ORD of Sup35p and still maintain prion competence. While the PFD of wild-type Sup35p has a striking 44% Q/N content, the prion-competent P14MC PFD contains only 18% Q/N. This suggests that the structural requirements for a yeast prion protein are not limited by a critical percentage of Q/N residues and that there may be other ways to achieve the structural flexibility required for yeast prion propagation.
DISCUSSION
Here, we describe a novel model system using a chimeric yeast-mammalian prion to evaluate the functionality of the PrP repeats in the context of the yeast prion protein Sup35p. Precise replacement of the entire repeat domain of Sup35p with the PrP ORD resulted in a tractable system to evaluate the effect of the PrP repeat expansion. The wild-type number of PrP repeats and all ORD expansions in the Sup35p-PrP chimeras behaved phenotypically, biochemically, and genetically as prions, although the PrP repeats did confer unique characteristics on the resulting prions. SP14 displayed a stronger prion phenotype in comparison to SP5, suggesting that the addition of octapeptide repeats allowed for a more stable self-replicating structure. This resembles what is seen in human patients with PrP repeat expansions, considering that the repeat expansion is associated with aggregation and spontaneous development of prion disease (16). The 14-PrP repeats also facilitated the establishment and propagation of prion strain variants, as indicated by differences in phenotype, mitotic stability, and prion transmission. Furthermore, in the presence of [RNQ+], SP14 converted into the prion form more readily than SP5. Strikingly, the 14-PrP repeats replaced the Q/N-rich region and the repeat region of Sup35p and maintained prion properties. Our data suggest that the PrP repeat expansion can influence prion conversion and enhance the formation of multiple aggregate structures.
Strain variants of both yeast prions and PrPSc are believed to be composed of structurally unique self-propagating aggregates (15, 25, 28, 43). Although conformationally distinct variants have been associated with PrPSc strains (2, 3), they have not been associated with inherited PrP mutations. The phenotypic variability associated with some pathogenic point mutations in PrP has been attributed to changes in glycoform ratio, but this did not correlate with the repeat expansion mutations (23). We suggest that the phenotypic variability observed in inherited repeat expansion prion diseases could result, in part, from differences in the structures acquired by the mutant protein.
Our data show that the repeat-expanded chimera displays phenotypic variation reminiscent of prion strains. The phenotypically distinct [SP14+] strain variants exhibited different mitotic stabilities (Fig. 2C) and differential transmission to imperfect prion protein sequences (Table 2). Taken together, these data suggest that the prions within these variants possess unique characteristics that might be structural, although the nature of the structural differences could not be elucidated. To date, the precise nature of the structural differences in mammalian and yeast prion strain variants also has not been described. A unique feature of the [SP14+] strain variants was their enhanced ability to interconvert. Strong variants could spontaneously give rise to weak variants, and vice versa (Fig. 4A). This observation suggests that the SP14 protein is not entirely committed to one particular prion conformation. There are two possibilities of how the interconversion could arise. First, the [SP14+] cells could maintain multiple structures and distinct phenotypes emerge when they separate and multiply (upon restreaking). The phenotype observed in one colony need not indicate structural homogeneity of the chimeric protein, but rather the colony might only show the phenotype of the dominant structure. Second, the SP14 protein may spontaneously create new prion aggregates when the prion state is lost, and these need not be the same strain variant. The influence of [RNQ+] could then contribute to interconversion via reacquisition. In the absence of the [RNQ+] prion, the [sp5−] and the [sp14−] cells did not spontaneously convert to the [PRION+] state more readily than [psi−] converts to [PSI+] (Fig. 4B). However, in the presence of [RNQ+], the spontaneous conversion of the chimeric proteins was greatly enhanced (Fig. 4C). This result is similar to that obtained with the Sup35p repeat expansion, which also had a higher frequency of spontaneous conversion in comparison to wild-type Sup35p (31). We have found that the previously described Sup35p repeat expansion (31) also converted more frequently to [PSI+] only in the presence of the [RNQ+] prion (E. M. H. Tank and H. L. True, unpublished data).
In vitro studies suggest that the repeat expansion in PrP decreases the lag phase of amyloid formation (34) and increases the accessibility of the N terminus of the mutant protein (53), thereby allowing a quicker conformational change into a pathogenic state. Our in vivo model indicates that the spontaneous conversion of the repeat expansion protein to an aggregated state need not be the sole mechanism whereby these mutations affect structure and, ultimately, disease. Instead, the repertoire of conformations achieved by the 14-PrP repeat chimeric protein appeared to be increased. However, in the presence of an additional aggregate, the conversion rate of SP14 was considerably enhanced. Therefore, other misfolded proteins may act as a potential source for initiating a conformational change in the repeat-expanded PrP mutants expressed in the brain. The ORD expansion proteins may interact with other preexisting aggregates to foster a change in conformation or create a high local concentration of the mutant protein which then aggregates.
Our model provides a possible explanation for the behavior of PrP repeat expansions and their propensity not only to cause disease but to contribute to the large degree of phenotypic variability observed in patients. Our data suggest that additional repeats in PrP may allow the formation of multiple unique aggregate conformations that could propagate in different tissues at different rates. In addition, if multiple aggregate structures are present in expanded PrP repeat diseases, then this could offer an explanation as to why infectious aggregate structures may also be produced in some cases (7) but not others (8, 14, 45). When we investigated the interaction between SP5 and SP14 by analyzing strains expressing both chimeras, indeed, we did observe a wide range of phenotypes (data not shown). Previous analyses of repeat expansion mutants isolated from human brain tissue demonstrated coaggregation with wild-type PrP, suggesting that phenotypic variability might be explained by the extent of this association (9). We suggest that differential association of mutant and wild-type PrP might be a consequence of different structures of the repeat expansion-containing PrP. Thus, while other genetic modifiers may play a role in disease variability, at least some of the phenotypic variability observed with PrP repeat expansion diseases may be based on intragenic features of the mutant.
Acknowledgments
We thank members of the True lab for helpful discussions and J. Weber and S. Stewart for comments on the manuscript. We thank K. Blumer and J. Cooper for the use of equipment and A. Li for advice. We thank M. Tuite, S. Liebman, and S. Lindquist for plasmids, strains, and antibodies. We thank S. Liebman for assistance with the transformation protocol.
This research was supported by National Institutes of Health grants F31 NS054513 (E.M.H.T.) and GM072228 (H.L.T.), the Ellison Medical Foundation (H.L.T.), and the Edward Mallinckrodt, Jr., Foundation (H.L.T.).
Footnotes
Published ahead of print on 4 June 2007.
REFERENCES
- 1.Bagriantsev, S., and S. W. Liebman. 2004. Specificity of prion assembly in vivo. J. Biol. Chem. 279:51042-51048. [DOI] [PubMed] [Google Scholar]
- 2.Bessen, R. A., D. A. Kocisko, G. J. Raymond, S. Nandan, P. T. Lansbury, and B. Caughey. 1995. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature 375:698-700. [DOI] [PubMed] [Google Scholar]
- 3.Bessen, R. A., and R. F. Marsh. 1992. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J. Gen. Virol. 73(Pt. 2):329-334. [DOI] [PubMed] [Google Scholar]
- 4.Borchsenius, A. S., S. Muller, G. P. Newnam, S. G. Inge-Vechtomov, and Y. O. Chernoff. 2006. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr. Genet. 49:21-29. [DOI] [PubMed] [Google Scholar]
- 5.Borchsenius, A. S., R. D. Wegrzyn, G. P. Newnam, S. G. Inge-Vechtomov, and Y. O. Chernoff. 2001. Yeast prion protein derivative defective in aggregate shearing and production of new ‘seeds’. EMBO J. 20:6683-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bousset, L., and R. Melki. 2002. Similar and divergent features in mammalian and yeast prions. Microbes Infect. 4:461-469. [DOI] [PubMed] [Google Scholar]
- 7.Brown, P., C. J. Gibbs, Jr., P. Rodgers-Johnson, D. M. Asher, M. P. Sulima, A. Bacote, L. G. Goldfarb, and D. C. Gajdusek. 1994. Human spongiform encephalopathy: the National Institutes of Health series of 300 cases of experimentally transmitted disease. Ann. Neurol. 35:513-529. [DOI] [PubMed] [Google Scholar]
- 8.Castilla, J., A. Gutierrez-Adan, A. Brun, B. Pintado, F. J. Salguero, B. Parra, F. D. Segundo, M. A. Ramirez, A. Rabano, M. J. Cano, and J. M. Torres. 2005. Transgenic mice expressing bovine PrP with a four extra repeat octapeptide insert mutation show a spontaneous, non-transmissible, neurodegenerative disease and an expedited course of BSE infection. FEBS Lett. 579:6237-6246. [DOI] [PubMed] [Google Scholar]
- 9.Chen, S. G., P. Parchi, P. Brown, S. Capellari, W. Zou, E. J. Cochran, C. L. Vnencak-Jones, J. Julien, C. Vital, J. Mikol, E. Lugaresi, L. Autilio-Gambetti, and P. Gambetti. 1997. Allelic origin of the abnormal prion protein isoform in familial prion diseases. Nat. Med. 3:1009-1015. [DOI] [PubMed] [Google Scholar]
- 10.Chernoff, Y. O., S. L. Lindquist, B. Ono, S. G. Inge-Vechtomov, and S. W. Liebman. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268:880-884. [DOI] [PubMed] [Google Scholar]
- 11.Chernoff, Y. O., S. M. Uptain, and S. L. Lindquist. 2002. Analysis of prion factors in yeast. Methods Enzymol. 351:499-538. [DOI] [PubMed] [Google Scholar]
- 12.Chiesa, R., B. Drisaldi, E. Quaglio, A. Migheli, P. Piccardo, B. Ghetti, and D. A. Harris. 2000. Accumulation of protease-resistant prion protein (PrP) and apoptosis of cerebellar granule cells in transgenic mice expressing a PrP insertional mutation. Proc. Natl. Acad. Sci. USA 97:5574-5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiesa, R., P. Piccardo, B. Ghetti, and D. A. Harris. 1998. Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron 21:1339-1351. [DOI] [PubMed] [Google Scholar]
- 14.Chiesa, R., P. Piccardo, E. Quaglio, B. Drisaldi, S. L. Si-Hoe, M. Takao, B. Ghetti, and D. A. Harris. 2003. Molecular distinction between pathogenic and infectious properties of the prion protein. J. Virol. 77:7611-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collinge, J. 2001. Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci. 24:519-550. [DOI] [PubMed] [Google Scholar]
- 16.Croes, E. A., J. Theuns, J. J. Houwing-Duistermaat, B. Dermaut, K. Sleegers, G. Roks, M. Van den Broeck, B. van Harten, J. C. van Swieten, M. Cruts, C. Van Broeckhoven, and C. M. van Duijn. 2004. Octapeptide repeat insertions in the prion protein gene and early onset dementia. J. Neurol. Neurosurg. Psychiatry 75:1166-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DePace, A. H., A. Santoso, P. Hillner, and J. S. Weissman. 1998. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 93:1241-1252. [DOI] [PubMed] [Google Scholar]
- 18.Derkatch, I. L., M. E. Bradley, J. Y. Hong, and S. W. Liebman. 2001. Prions affect the appearance of other prions: the story of [PIN+]. Cell 106:171-182. [DOI] [PubMed] [Google Scholar]
- 19.Derkatch, I. L., M. E. Bradley, and S. W. Liebman. 1998. Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc. Natl. Acad. Sci. USA 95:2400-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derkatch, I. L., M. E. Bradley, S. V. Masse, S. P. Zadorsky, G. V. Polozkov, S. G. Inge-Vechtomov, and S. W. Liebman. 2000. Dependence and independence of [PSI+] and [PIN+]: a two-prion system in yeast? EMBO J. 19:1942-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derkatch, I. L., Y. O. Chernoff, V. V. Kushnirov, S. G. Inge-Vechtomov, and S. W. Liebman. 1996. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144:1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guthrie, C., and G. Fink. 2004. Guide to yeast genetics and molecular and cell biology, vol. 194. Elsevier Academic Press, San Diego, CA.
- 23.Hill, A. F., S. Joiner, J. A. Beck, T. A. Campbell, A. Dickinson, M. Poulter, J. D. Wadsworth, and J. Collinge. 2006. Distinct glycoform ratios of protease resistant prion protein associated with PRNP point mutations. Brain 129:676-685. [DOI] [PubMed] [Google Scholar]
- 24.King, A., L. Doey, M. Rossor, S. Mead, J. Collinge, and P. Lantos. 2003. Phenotypic variability in the brains of a family with a prion disease characterized by a 144-base pair insertion in the prion protein gene. Neuropathol. Appl. Neurobiol. 29:98-105. [DOI] [PubMed] [Google Scholar]
- 25.King, C. Y., and R. Diaz-Avalos. 2004. Protein-only transmission of three yeast prion strains. Nature 428:319-323. [DOI] [PubMed] [Google Scholar]
- 26.Kochneva-Pervukhova, N. V., M. B. Chechenova, I. A. Valouev, V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 2001. [Psi+] prion generation in yeast: characterization of the ‘strain’ difference. Yeast 18:489-497. [DOI] [PubMed] [Google Scholar]
- 27.Kovács, G. G., G. Trabattoni, J. A. Hainfellner, J. W. Ironside, R. S. Knight, and H. Budka. 2002. Mutations of the prion protein gene phenotypic spectrum. J. Neurol. 249:1567-1582. [DOI] [PubMed] [Google Scholar]
- 28.Krishnan, R., and S. L. Lindquist. 2005. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature 435:765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kryndushkin, D. S., I. M. Alexandrov, M. D. Ter-Avanesyan, and V. V. Kushnirov. 2003. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278:49636-49643. [DOI] [PubMed] [Google Scholar]
- 30.Li, L., and S. Lindquist. 2000. Creating a protein-based element of inheritance. Science 287:661-664. [DOI] [PubMed] [Google Scholar]
- 31.Liu, J. J., and S. Lindquist. 1999. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature 400:573-576. [DOI] [PubMed] [Google Scholar]
- 32.Manogaran, A. L., K. T. Kirkland, and S. W. Liebman. 2006. An engineered nonsense URA3 allele provides a versatile system to detect the presence, absence and appearance of the [PSI+] prion in Saccharomyces cerevisiae. Yeast 23:141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millhauser, G. L. 2007. Copper and the prion protein: methods, structures, function, and disease. Annu. Rev. Phys. Chem. 58:299-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore, R. A., C. Herzog, J. Errett, D. A. Kocisko, K. M. Arnold, S. F. Hayes, and S. A. Priola. 2006. Octapeptide repeat insertions increase the rate of protease-resistant prion protein formation. Protein Sci. 15:609-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 36.Ness, F., P. Ferreira, B. S. Cox, and M. F. Tuite. 2002. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 22:5593-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osherovich, L. Z., and J. S. Weissman. 2001. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell 106:183-194. [DOI] [PubMed] [Google Scholar]
- 38.Parham, S. N., C. G. Resende, and M. F. Tuite. 2001. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J. 20:2111-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patino, M. M., J. J. Liu, J. R. Glover, and S. Lindquist. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273:622-626. [DOI] [PubMed] [Google Scholar]
- 40.Prusiner, S. B. 1998. Prions. Proc. Natl. Acad. Sci. USA 95:13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogers, M., F. Yehiely, M. Scott, and S. B. Prusiner. 1993. Conversion of truncated and elongated prion proteins into the scrapie isoform in cultured cells. Proc. Natl. Acad. Sci. USA 90:3182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheibel, T., and S. L. Lindquist. 2001. The role of conformational flexibility in prion propagation and maintenance for Sup35p. Nat. Struct. Biol. 8:958-962. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka, M., P. Chien, N. Naber, R. Cooke, and J. S. Weissman. 2004. Conformational variations in an infectious protein determine prion strain differences. Nature 428:323-328. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka, M., P. Chien, K. Yonekura, and J. S. Weissman. 2005. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell 121:49-62. [DOI] [PubMed] [Google Scholar]
- 45.Tateishi, J., T. Kitamoto, M. Z. Hoque, and H. Furukawa. 1996. Experimental transmission of Creutzfeldt-Jakob disease and related diseases to rodents. Neurology 46:532-537. [DOI] [PubMed] [Google Scholar]
- 46.Ter-Avanesyan, M. D., A. R. Dagkesamanskaya, V. V. Kushnirov, and V. N. Smirnov. 1994. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 137:671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.True, H. L. 2006. The battle of the fold: chaperones take on prions. Trends Genet. 22:110-117. [DOI] [PubMed] [Google Scholar]
- 48.Tuite, M. F., and B. S. Cox. 2003. Propagation of yeast prions. Nat. Rev. Mol. Cell Biol. 4:878-890. [DOI] [PubMed] [Google Scholar]
- 49.Uptain, S. M., and S. Lindquist. 2002. Prions as protein-based genetic elements. Annu. Rev. Microbiol. 56:703-741. [DOI] [PubMed] [Google Scholar]
- 50.Uptain, S. M., G. J. Sawicki, B. Caughey, and S. Lindquist. 2001. Strains of [PSI+] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 20:6236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Viles, J. H., D. Donne, G. Kroon, S. B. Prusiner, F. E. Cohen, H. J. Dyson, and P. E. Wright. 2001. Local structural plasticity of the prion protein. Analysis of NMR relaxation dynamics. Biochemistry 40:2743-2753. [DOI] [PubMed] [Google Scholar]
- 52.Wadsworth, J. D., A. F. Hill, J. A. Beck, and J. Collinge. 2003. Molecular and clinical classification of human prion disease. Br. Med. Bull. 66:241-254. [DOI] [PubMed] [Google Scholar]
- 53.Yin, S., S. Yu, C. Li, P. Wong, B. Chang, F. Xiao, S. C. Kang, H. Yan, G. Xiao, J. Grassi, P. Tien, and M. S. Sy. 2006. Prion proteins with insertion mutations have altered N-terminal conformation and increased ligand binding activity and are more susceptible to oxidative attack. J. Biol. Chem. 281:10698-10705. [DOI] [PubMed] [Google Scholar]
- 54.Zhouravleva, G., V. V. Alenin, S. Inge-Vechtomov, and Y. O. Chernoff. 2002. To stick or not to stick: prion domains from yeast to mammals. Recent Res. Dev. Mol. Cell. Biol. 3:185-218. [Google Scholar]