Abstract
During the execution of differentiation programs, lineage-specific transcription factors are in competition with antagonistic factors that drive progenitor proliferation. Thus, the myeloid transcription factor MafB promotes macrophage differentiation of myeloid progenitors, but a constitutively active Myb transcription factor (v-Myb) can maintain proliferation and block differentiation. Little is known, however, about the regulatory mechanisms that control such competing activities. Here we report that the small ubiquitin-like protein SUMO-1 can modify MafB in vitro and in vivo on lysines 32 and 297. The absence of MafB SUMO modification increased MafB-driven transactivation and macrophage differentiation potential but inhibited cell cycle progression and myeloid progenitor growth. Furthermore, we observed that direct repression of MafB transactivation by v-Myb was strictly dependent on MafB SUMO modification. Consequently, a SUMOylation-deficient MafB K32R K297R (K32,297R) mutant could specify macrophage fate even after activation of inducible Myb alleles and resist their differentiation-inhibiting activity. Our findings suggest that SUMO modification of MafB affects the balance between myeloid progenitor expansion and terminal macrophage differentiation by controlling MafB transactivation capacity and susceptibility to Myb repression. SUMO modification of lineage-specific transcription factors may thus modulate transcription factor antagonism to control tissue homeostasis in the hematopoietic system.
The generation of cell type specificity during developmental processes depends on the action of transcription factors that drive differential gene expression programs. The hematopoietic system has served as a particularly useful model to highlight the importance of combinatorial actions of transcription factors in such differentiation processes (14, 38, 42). An example for this is the bZIP type transcription factor MafB that is up-regulated during differentiation from hematopoietic progenitors towards macrophages (2, 13, 24, 29, 43). Its ectopic expression in myeloid progenitors promotes macrophage differentiation (2, 16, 29) and inhibits alternative differentiation options towards erythroid cells (43) or dendritic cells (2), which involves antagonistic interactions with Ets-1 (43) and PU.1 (2), respectively.
To maintain homeostatic control in rapidly regenerating tissues, such as the hematopoietic system, cellular differentiation is balanced by the expansion of immature progenitors. At the molecular level, these competing programs are directed by antagonistic transcription factors. MafB appears to engage in such an antagonistic relationship with the c-Myb transcription factor in the control of myelomonocytic differentiation. MafB expression increases during differentiation (2, 13, 24, 29, 43), but c-Myb expression is high in immature cells and down-regulated as the cells differentiate towards macrophages (5, 12, 36, 44). While MafB induces differentiation (2, 16, 29), c-Myb promotes proliferation and inhibits differentiation (23, 37). This is particularly evident for the constitutively active Myb allele of E26 virus (v-Myb), which can maintain the proliferation of chicken myeloid progenitors and block their differentiation towards macrophages without immortalizing them (3, 23, 32). By temperature inactivation of the ts21 allele of v-Myb, it is possible to relieve this differentiation block and permit macrophage differentiation (2, 3, 29). Reciprocally, induction of Myb activity in macrophages results in their rapid dedifferentiation (3, 7, 36). It thus appears evident that MafB and Myb transcription factors play antagonistic roles in macrophage differentiation, but it remains unclear how these competing activities are controlled to influence the balance in one or the other direction.
Covalent posttranslational modification with small ubiquitin-like proteins, particularly SUMO-1 (28), has been recognized to play an important role in controlling various cellular functions, including transcription factor activity (15, 17-19, 21, 47). The enzymatic machinery for the covalent attachment to target proteins resembles that of ubiquitination but utilizes different components (28). A single SUMO-specific E2-conjugating enzyme (Ubc9) and a small number of SUMO-specific E3 ligases have been identified (28, 40). They mediate the attachment of SUMO to lysine residues within the consensus sequence ΨKXE, where Ψ stands for a hydrophobic amino acid and X for any amino acid (28). The process is reversible, and SUMO proteins can be removed from their targets by specific proteases (SENP1, SENP2, SENP3, SENP5, SENP6, and SENP7). In contrast to ubiquitination, SUMO modification does not directly target proteins for degradation but can have diverse effects on stability, activity, cellular localization, or protein interaction (15, 17, 28, 40, 47). The central importance of SUMO modification for the regulation of cellular, particularly nuclear, processes has been demonstrated in a number of genetic experiments. SUMO conjugation and deconjugation are required for viability in virtually all eukaryotes from Saccharomyces cerevisiae to mice (35, 48).
SUMO modification of transcription factors has generally been observed to repress transcriptional activity (15, 17, 18, 21, 47). Although in most cases these effects were observed only in transient-transactivation assays and biological consequences were not investigated or could not be detected (10), transgenic studies also implicate SUMO modification in transcriptional repression (34). Gain-of-function experiments in Caenorhabditis elegans (6) and Xenopus laevis (46) further suggest that SUMO modification can also modulate transcription factor activity in developmental processes. Generally, however, little is known about the biological significance of transcription factor SUMO modification in higher vertebrates and particularly its role in controlling the combinatorial action of transcription factors.
Here we demonstrated that MafB is subject to SUMO modification on two principal lysine residues in vitro and in vivo and showed that the absence of SUMO modification on these residues increased transcriptional activity and the abilities to induce macrophage differentiation, inhibit myeloid progenitor expansion, and repress cell cycle progression. Most interesting, however, we observed that direct repression of MafB by v-Myb is strictly dependent on MafB SUMO modification. As a consequence, SUMO modification-deficient MafB can resist v-Myb repression and promote macrophage fate in the presence of dedifferentiating v-Myb activity. Together, these results indicate that SUMO modification of MafB is a potent mechanism to balance myeloid progenitor proliferation and macrophage differentiation by controlling the Myb/MafB transcription factor antagonism. SUMO modification may thus serve an important role in controlling tissue homeostasis in the hematopoietic system.
MATERIALS AND METHODS
Cell culture, antibodies, and plasmids.
QT6 and HD11v-mybER (7) cells were grown in Dulbecco modified Eagle medium (DMEM)-8% fetal calf serum (FCS)-2% chicken serum, and HEK 293, RAW264.7, and Phoenix (ΦNX) cells were grown in DMEM-10% FCS. Both media contained 1% penicillin-streptomycin and 1% glutamine. Primary chicken myeloblast clones were cultured as described previously (29). Polyclonal antibody against MafB was generated by immunizing rabbits with a 17-amino-acid keyhole limpet hemocyanin-coupled internal peptide of mouse MafB and affinity purified by standard procedures or purchased from Calbiochem. Site-directed mutagenesis was performed by PCR on mouse MafB cDNA with overlapping mismatched primers to target the sequences: GTTCGACGTGCGGAGGAGC for lysine 32, GAGCAGCTTCGGCAGGAGGTG for lysine 281, and CAAGGTCCGGTGCGAGAAAC for lysine 297. Single, double, and triple mutants were subcloned in RC/CMV (Invitrogen) or MFG-iGFP (2) or downstream of the internal ribosome entry site (IRES) element in E26-ts21 (29). C-terminal MafB-green fluorescent protein (GFP) fusion constructs were generated by PCR-mediated in-frame cloning of wild-type or mutant MafB into pEGFP-N1 (Clontech). PMI10 was used as an expression plasmid for E26 v-Myb (34).
Virus production.
To obtain primary avian myeloid progenitor clones, hematopoietic progenitors from 2-day chicken embryos (White Leghorn chickens; Ferme Avicole HAAS, Kaltenhouse, France) were infected with E26 virus derivatives as described previously (29). MFG-iGFP retroviruses for infection of HD11v-mybER cells were produced in ecotropic Phoenix (ΦNX) packaging cells by calcium phosphate precipitation as described previously (2) and pseudotyped by cotransfection of a vesicular stomatitis virus G-protein expression construct (pCMV-VSV-G).
In vitro assays for SUMO-1 modification.
SUMO conjugation assays were performed at 37°C for 120 min in 10-μl volumes containing the following: 1 μl of wheat germ-coupled transcription-translation reaction mixture (Promega) with [35S]methionine (Amersham); 1 μg of wild-type or mutant MafB plasmid; 500 ng, 120 ng, and 150 ng of SUMO-1, SAE1/SAE2, and Ubc9 expressed in Escherichia coli (45), 50 mM Tris (pH 7.5), 5 mM MgCl2, 2 mM ATP, 10 mM creatine phosphate, 3.5 U of creatine kinase/ml, and 0.6 U inorganic pyrophosphate/ml. Reactions were terminated by the addition of sodium dodecyl sulfate (SDS) and β-mercaptoethanol (β-ME) buffer. Reaction products were separated by denaturing 10% SDS-polyacrylamide gel electrophoresis (10% SDS-PAGE) and analyzed by phosphorimaging.
Assays for in vivo SUMO-1 modification, protein interaction, and Western blotting.
For in vivo assays of SUMO-1 modification, QT6 cells were transfected with 200 ng/cm2 of wild-type or mutant MafB expression plasmid, lysed in 5 mM imidazole-containing G buffer (6 M guanidinium-Cl in PT buffer [100 mM Na2HPO4/NaH2PO4, 20 mM Tris-Cl {pH 8.0}, 10 mM β-ME, 1× protease inhibitor mix]). Lysates were ultrasound treated, incubated with Ni-nitrilotriacetic acid-agarose (QIAGEN) for 2 h and washed for 30 min each with G buffer, buffer W (pH 8.0) (8 M urea in PT buffer), twice with buffer W (pH 6.3)-0.2% Triton X-100 and buffer W (pH 6.3)-0.3% Triton X-100. Matrix-bound protein was eluted with 200 mM imidazole, 5% SDS, 150 mM Tris-Cl (pH 6.7), 30% glycerol, 720 mM β-ME, and 1× protease inhibitor mix and separated by 10% SDS-PAGE and revealed by Western blotting. Immunoblotting was performed with anti-MafB (1/500), antihemagglutinin tag (anti-HA tag) (1/1,000; Roche), anti-Ubc9 (1/500; Calbiochem), antitubulin (1/3,000; Sigma), and secondary anti-rabbit (Santa Cruz, CA) or anti-rat antibodies conjugated to horseradish peroxidase (Jackson, MN) using ECL detection kit (Amersham) as described previously (29). Glutathione S-transferase (GST) pull-down assays were performed as described previously (29).
Reporter gene assays.
QT6 cells were plated at 20,000 cells/cm2, transfected after 24 h by CaPO4 precipitation as indicated, and harvested 48 h later in 40 mM Tris-Cl (pH 7.5)-1 mM EDTA-150 mM NaCl (TEN). The 3xMARE (29) and 3xMRE (36) reporter plasmids have been described previously. Cell lysates were assayed for firefly luciferase activity and normalized to β-galactosidase activity from a cotransfected RSV-lacZ plasmid as described previously (29).
Phagocytosis assays.
Phagocytosis assays were essentially performed as described previously (2, 29) with 250,000 cells in 250 μl medium incubated with 4 μl suspension of 1 μm phycoerythrin (PE)-coated latex beads (F-8851; Molecular Probes) for 2 h. Cells were harvested in TEN, washed three times with medium at 4°C, and analyzed by fluorescence-activated cell sorting (FACS). Mean fluorescence and fluorescence-positive cells were quantified using FloWJo.
Growth rate and cell cycle analysis.
A total of 100,000 cells were seeded, and after the indicated time, 10 μl of 5-mg/ml 3-(4,5-dimethylthiazol-2-yl)2,5-diphenyltetrazolium bromide (MTT; Sigma) was added for 5 h at 37°C. Water-insoluble formazan blue formed in viable cells was dissolved in HCl-isopropanol. Optical density was read at 492 nm, and cell number was determined using a standard curve. For cell cycle analysis, QT6 cells were plated at 20,000 cells/cm2, transfected with 200 ng/cm2 GFP fusion plasmids and after 12 h shifted to low serum for 36 h (DMEM-0.5% FCS), trypsinized, washed in phosphate-buffered saline, fixed overnight in 70% ethanol-30%glycerol, and incubated with 1-mg/ml RNase A (Q-Biogen) and 50-ng/ml propidium iodide (Sigma) for 30 min at 37°C. Cell cycle analysis on GFP-positive cells was performed by selecting fluorescence area versus width discrimination on the propidium iodide signal and using the range method (partec) in CellQuest.
siRNA inactivation.
Subconfluent 12-well cultures of HEK 293 cells were incubated for 5 h with 130 μl OPTIMEM 1 (Gibco BRL) containing a mix of 6 μl oligofectamine reagent (Invitrogen) and 6 μl of 20 μM small interfering RNA (siRNA) (DHARMACON) that were mixed and preincubated for 20 min according to the supplier's instructions. After overnight addition of 1 ml serum-free DMEM, the cells were again transfected for 5 h with a mixture containing 8 μl oligofectamine, 6 μl of 20 μM siRNA, 400 ng v-Myb expression plasmid, 10 ng MafB or MafB K32,297R expression plasmid, and 200 ng MARE reporter in 130 μl OPTIMEM 1 after appropriate mixing and preincubation, incubated for 24 h in 1 ml serum-free DMEM, and assayed 6 h later for luciferase activity. siRNA duplexes against ubc9 have been described previously (20). Stealth siRNA negative-control duplex (Invitrogen) served as a negative control.
RESULTS
MafB is SUMO-1 modified in vitro.
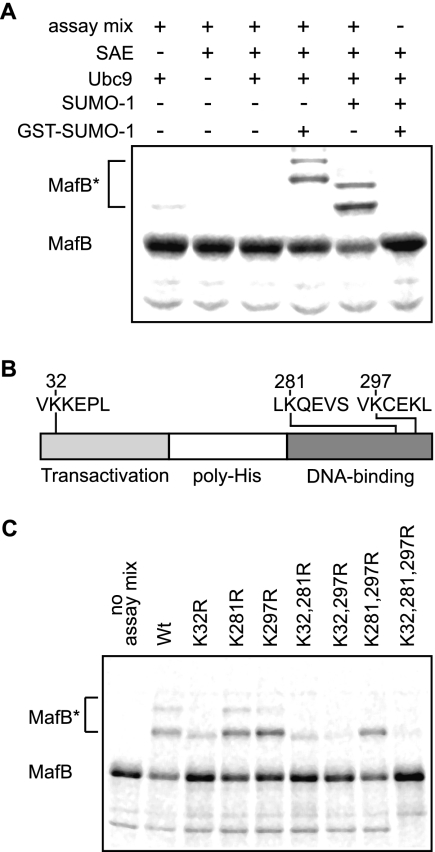
To determine whether MafB was a substrate for posttranslational modification with SUMO-1, we subjected MafB to an in vitro assay with 35S-labeled, in vitro-translated MafB, recombinant SUMO E1-activating and E2-conjugating enzymes and SUMO-1 or GST fused to SUMO-1 (GST-SUMO-1) (45). As shown in Fig. 1A, higher-molecular-weight forms of MafB (MafB*) were detected only in the presence of all assay components and SUMO-1 or GST-SUMO-1, with GST-SUMO-1 resulting in slower-migrating bands than SUMO-1, reflecting the additional molecular weight of the GST moiety. In both cases, two prominent high-molecular-weight bands indicated that MafB was modified with at least two SUMO-1 molecules as targets.
FIG. 1.
MafB is SUMO-1 modified in vitro. (A) Incubation of in vitro-translated, 35S-labeled MafB with recombinant SAE and Ubc9 SUMO modification enzymes, SUMO-1 or GST-SUMO-1, and an ATP-generating system, as indicated, results in the appearance of high-molecular-weight species representing SUMO-modified MafB (MafB*). (B) SUMO modification consensus sites ψKXE (ψ for an aliphatic amino acid) are found three times in the primary sequence of MafB at lysine residues 32, 281, and 297. (C) Single, double, or triple lysine-to-arginine MafB mutants of ψKXE consensus sites were subjected to in vitro modification for SUMO-1 as in panel A. Loss of high-molecular-weight MafB species (MafB*) identified two principal SUMO modification sites at lysines 32 and 297 with loss of SUMO-1 modification observed for the K32,297R mutant. Wt, wild type.
Examination of the primary sequence of MafB identified lysine residues 32, 281, and 297 as lying within the consensus sequence ΨKXE for SUMO modification (Fig. 1B). To test which of these sites were preferentially targeted for SUMO modification, we generated single, double, and triple point mutations of lysine to arginine and subjected them to the in vitro SUMO modification assay. As shown in Fig. 1C, mutation K281R had no effect, mutation K297R had a weak effect, and mutation K32R had the strongest effect. Consistent with this, the double mutant K32,297R showed a dramatic loss of SUMO modification similar to the triple mutant K32,281,297R, whereas the K32,281R mutant was still weakly modified and the K281,297R double mutant was still strongly modified. Together these results indicated that MafB is a substrate for modification with more than one SUMO-1 molecule via lysine 32 as the primary target and lysine 297 as a secondary attachment site. All further experiments were performed with MafB K32,297R as the minimal mutation showing loss of SUMO modification.
SUMO-1 modification of MafB in vivo.
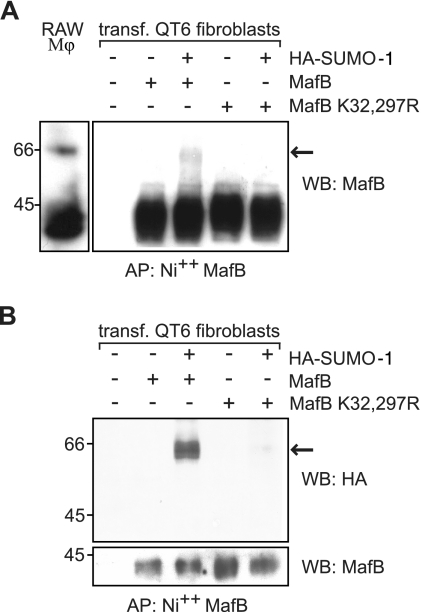
To determine whether SUMO-1 modification of MafB occurred in vivo, we analyzed protein extracts of RAW264.7 cells, a macrophage line expressing endogenous MafB, by Western blotting with anti-MafB antibody. As shown in the left panel of Fig. 2A, we could indeed detect a more slowly migrating form of MafB that is consistent with SUMO modification. To verify that this more slowly migrating form of MafB was a result of SUMO modification, we cotransfected QT6 fibroblasts with MafB or the SUMO modification-deficient MafB K32,297R mutant with or without HA-tagged SUMO-1 (HA-SUMO-1). MafB proteins were enriched by affinity purification from cell lysates on nickel agarose taking advantage of the intrinsic MafB polyhistidine domains, which confer high binding affinity to chelated nickel matrix, and subsequently immunoblotted with an antibody specific for MafB. As shown in the right panel of Fig. 2A, we could detect the more slowly migrating species only under conditions where both wild-type MafB and HA-SUMO-1 had been cotransfected but not in the absence of HA-SUMO-1 or with the SUMO modification-deficient mutant MafB K32,297R. The low proportion of modified protein observed is consistent with the previously reported small percentage of highly expressed target proteins existing as the SUMO-modified form at steady state, which is likely a reflection of the dynamic nature and potential long-term effects of transient SUMO modification (25). To further confirm that this high-molecular-weight form of MafB corresponded to SUMO-modified MafB, we purified MafB on Ni-agarose and probed a Western blot with antibodies against the HA tag of the cotransfected SUMO-1. This staining revealed again a more slowly migrating species only in the presence of HA-SUMO-1 for wild-type MafB but not mutant MafB K32,297R and of the same size as in the anti-MafB blot (Fig. 2B). Together these results confirmed that SUMO-1 modification via lysine residues 32 and 297 of MafB also occurred in vivo.
FIG. 2.
MafB is SUMO-1 modified in vivo. (A) Western blotting with an antibody directed against MafB on protein extracts prepared from RAW264.7 macrophages (RAW Mφ) (left) or transfected (transf.) QT6 fibroblasts (right). QT6 cells were transfected (+) as indicated with expression constructs for wild-type MafB or MafB K32,297R mutant and HA-tagged SUMO-1, affinity purified on a nickel matrix for polyhistidine stretches contained in the MafB sequences (AP: Ni++ MafB), and analyzed by Western blotting (WB) with anti-MafB antibody. The arrow points to a more slowly migrating form of MafB that is consistent with SUMO modification. (B) Protein extracts from transfected QT6 cells were prepared and treated as in panel A and analyzed by Western blotting with an anti-HA antibody (top blot). The blot was reprobed with an antibody against MafB and revealed at low exposure to control for equal expression and loading (bottom blot). The positions of molecular size markers in kilodaltons are indicated to the left of the blots.
SUMO modification represses MafB transcriptional activity.
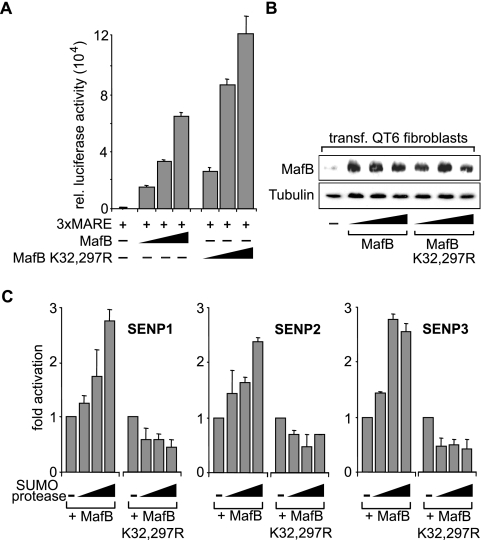
To investigate the functional consequences of MafB SUMO modification, we tested its influence on transcriptional activity. We used a synthetic Maf-responsive reporter containing three Maf consensus binding sites (3xMARE) upstream of a minimal promoter that we have shown previously to be activated by MafB (29). As shown in Fig. 3A, cotransfection of this reporter construct with wild-type MafB resulted in the expected dose-dependent transactivation. Interestingly, the SUMO modification-deficient MafB K32,297R mutant showed increased transactivation activity. This effect was not due to increased expression of MafB K32,297R as verified by immunoblotting (Fig. 3B). The previously established SUMO site preference (Fig. 1) corresponded to the strength of SUMO-mediated transcriptional repression. A single point mutation of K297 had no effect on transcriptional activity, and a single mutation of K32 had a less pronounced effect than the double mutation did (data not shown).
FIG. 3.
SUMO modification represses transactivation capacity of MafB. (A) QT6 cells were transfected (+) with a luciferase reporter containing three Maf-responsive MARE sites (3xMARE) and increasing amounts of expression constructs for wild-type MafB or MafB K32,297R mutant as indicated. rel., relative. (B) Protein extracts of QT6 cells transfected (transf.) with MafB or MafB K32,297R expression constructs from the assay shown in panel A were Western blotted and probed with anti-MafB antibody (top) and antitubulin antibody (bottom) as a control. (C) QT6 cells were transfected with 3xMARE reporter, MafB or MafB K32,297R mutant expression construct and increasing amounts of expression constructs for SUMO-specific protease SENP1, SENP2, or SENP3 as indicated. Results are expressed as changes in transactivation. All transfection assays were filled up with empty vector to a constant amount of expression plasmid and performed in duplicate, and luciferase activity was normalized to β-galactosidase activity from a cotransfected control plasmid. Error bars indicate standard errors of the means, and data are representative of at least two independent experiments.
To confirm that the observed effect of the MafB mutants on transactivation activity was indeed based on SUMO modification, we interfered with it enzymatically. SUMO modification is a dynamic process (25), and SUMO-specific proteases that remove SUMO tags from target proteins have been identified (25, 33). Therefore, we tested the SUMO-specific proteases SENP1, SENP2, and SENP3 for their effect on MafB activity. As shown in Fig. 3C, all of these enzymes increased the transcriptional activity of MafB in a dose-dependent manner severalfold but failed to activate the transcriptional activity of the SUMO modification-deficient MafB K32,297R mutant. This indicated that the activating effect of SENP1, SENP2, and SENP3 on MafB transactivation was due to removal of SUMO from residues K32 and K297 on MafB.
Together these results showed that decreased SUMO modification increased MafB transactivation activity and that the MafB K32,297R mutant mimics the activity of unmodified protein.
Absence of MafB SUMO modification increases its potential to drive macrophage differentiation and inhibit myeloid progenitor growth.
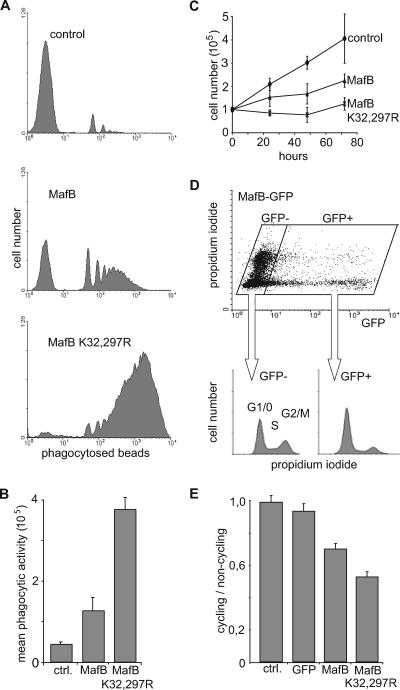
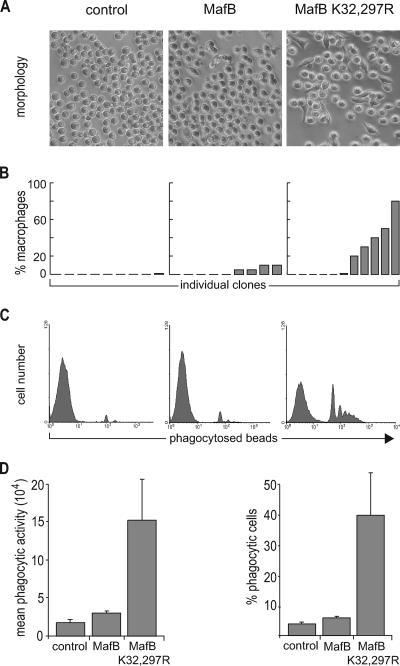
We and others have shown previously that expression of MafB in myeloid progenitors promoted macrophage differentiation (2, 16, 29). To investigate whether MafB SUMO modification was important for this biological activity, we used primary transformed avian myeloblasts infected with bicistronic retroviral vectors expressing either MafB or MafBK32,297R mutant from an E26-ts21 retrovirus (29). We measured phagocytic activity in these cells as an indicator of functional macrophage differentiation. We cultured individual myeloid clones for 24 h at 42°C to relieve a differentiation block imposed by the temperature-sensitive ts21 v-Myb allele of E26-ts21 (3) and quantified the ability of the cells to phagocytose fluorescent beads by FACS. As shown in representative FACS profiles of Fig. 4A, MafB expression resulted in macrophage differentiation and conferred significant phagocytic activity to the cells. Expression of MafB K32,297R, however, caused a more dramatic increase in phagocytic activity, both in terms of the number of beads ingested per cell and in the percentage of phagocytic cells (Fig. 4A). Quantitative analysis of several clones confirmed this observation and revealed an approximately threefold increase in the mean phagocytic activity of MafB K32,297R clones over MafB clones (Fig. 4B). Together these data indicated that deficiency for SUMO modification strongly increased the ability of MafB to induce functional macrophage differentiation in myeloid progenitors.
FIG. 4.
Increased functional macrophage differentiation and growth inhibition of myeloid progenitors in the absence of MafB SUMO modification. (A) Representative FACS profiles of E26-ts21 myeloid clones expressing no transgene (control), MafB, or MafB K32,297R mutant after 24 h at 42°C to relieve a E26-ts21-imposed differentiation block and 2 h of incubation with fluorescent PE-conjugated latex beads to monitor phagocytic activity as a measure of functional macrophage differentiation. (B) Quantification of mean phagocytic activity indicating the mean fluorescence intensity as a measure of the number of phagocytosed beads per cell, calculated from FACS profiles as shown in panel A. The average of three independent clones from each genotype is shown. ctrl., control. (C) Growth rate of E26-ts21 myeloid progenitor clones expressing no transgene (control), MafB, or MafB K32,297R mutant. Cell numbers were quantified every 24 h using an MTT assay. (D) QT6 cells were transfected with an expression construct for a MafB-GFP fusion protein and stained with propidium iodide for DNA contents. The cell cycle profiles of GFP-positive (GFP+) and GFP-negative (GFP−) cells were analyzed by propidium iodide staining on fixed cells and plotted separately. (E) The same analysis as in panel D was performed for nontransfected cells (control [ctrl.]) and cells transfected with GFP, MafB-GFP, or MafB K32,297R-GFP. Results are expressed as ratio of cycling (S and G2/M) to noncycling (G1/G0) cells. Error bars in all panels indicate standard errors of the means, and data are representative of at least two independent experiments.
Upon terminal macrophage differentiation, cells exit the cell cycle. Therefore, we wanted to determine whether the increased ability of SUMO-deficient MafB to promote macrophage differentiation might also result in growth inhibition of myeloid progenitors. To test this, we measured the growth rates of control ts21-E26 myeloblast clones and ts21-E26 myeloblast clones expressing MafB or MafB K32,297R by determining cell numbers at daily intervals. As shown in Fig. 4C, control clones grew rapidly with a doubling time of 1 to 1.5 days, whereas MafB-expressing clones doubled approximately only every 3 days. MafB K32,297R-expressing clones, however, hardly grew during the 3-day observation period and had an extrapolated doubling time of more than 7 days. To further analyze whether this might involve an effect of MafB SUMOylation on cell cycle progression, we generated fusion constructs of MafB and MafB K32,297R with GFP for the analysis of the cell cycle profile in GFP-positive, fusion protein-expressing cells. GFP fusion did not impede biological activity, since MafB-GFP constructs were still able to promote macrophage differentiation (data not shown). We transfected QT6 cells with these fusion constructs or GFP-only controls and analyzed the cell cycle profile of transfected cells by FACS after propidium iodide staining for DNA content. As shown in Fig. 4D, the profile of MafB-GFP-transfected cells revealed that GFP-positive, MafB-expressing cells showed a significantly reduced proportion of cells in G2/M or S phase compared to nontransfected GFP-negative cells. As shown in Fig. 4E, the ratio of cycling cells in G2/M or S to noncycling cells was about 1 for nontransfected control cells and cells expressing only GFP but 0.7 in cells expressing MafB-GFP, reflecting the reduced amount of cycling cells. This ratio was further reduced to 0.5 in MafB K32,297R-GFP-expressing cells, indicating that the absence of SUMO modification also increased the ability of MafB to inhibit cell cycle progression.
Together these results suggested that SUMO modification increased the ability of MafB to induce macrophage differentiation and inhibit proliferation in myeloid progenitors.
The absence of SUMO modification enables MafB to overcome a v-Myb-imposed differentiation block.
Promotion of macrophage differentiation by MafB in E26-transformed myeloid progenitors requires relief of a v-Myb-imposed differentiation block by temperature inactivation of the v-Myb ts21 allele (2, 29). Surprisingly, cells infected with the SUMO modification-deficient MafB K32,297R mutant frequently acquired a typical macrophage morphology even without a temperature shift, and thus in the presence of active v-Myb, whereas clones expressing control or wild-type MafB did not (Fig. 5A). As shown in Fig. 5B, quantitative analysis of individual clones revealed that about half of MafB K32,297R-expressing clones had 20 to 80% of cells with macrophage morphology after 2 weeks in liquid culture. By comparison, MafB-expressing clones only occasionally showed a small percentage of cells with macrophage morphology. Similarly, all control virus-infected clones had a round and nonadherent myeloid progenitor phenotype with <0.1% macrophage-type cells. Wright-Giemsa staining also revealed cells with typical macrophage morphology in MafB K32,297R-expressing clones but not in MafB-expressing clones or control clones (data not shown). These morphological differences correlated with functional criteria of increased macrophage differentiation. As shown in representative histograms of Fig. 5C, only clones expressing MafB K32,297R but not clones expressing wild-type MafB or control clones showed significant phagocytic activity. This observation was confirmed in a quantification of the mean phagocytic activity and the percentage of phagocytic cells for three independent clones of each genotype (Fig. 5D). Western blotting did not reveal increased MafB protein expression in MafB K32,297R-expressing clones (data not shown). Together these results indicated that the absence of SUMO modification enabled MafB to overcome a v-Myb-induced differentiation block and to induce macrophage differentiation in myeloid progenitor cells.
FIG. 5.
Absence of MafB SUMO modification enhances spontaneous macrophage differentiation. (A) Representative phase-contrast micrographs of E26-ts21 myeloid progenitor clones expressing no transgene (control), MafB, or MafB K32,297R mutant 2 days after transfer of colonies from methocel into liquid culture. (B) Quantification of cells with macrophage morphology in individual clones expressing no transgene (control), MafB, or MafB K32,297R mutant after 2 weeks of liquid culture. (C) Representative FACS profiles of individual clones expressing either no transgene (control), MafB, or MafB K32,297R mutant that were incubated for 2 h with fluorescent PE-conjugated latex beads to measure phagocytic activity as a indication of functional macrophage differentiation. (D) Quantification of mean phagocytic activity indicating the mean fluorescence intensity as a measure of the number of phagocytosed beads per cell (left) and of the percentage of fluorescence-positive cells as a measure of phagocytosing cells (right), calculated from FACS profiles as shown in panel C. The average of three independent clones of each genotype derived from the same experiment is shown. Error bars indicate standard errors of the means, and data are representative of at least two independent experiments.
v-Myb inhibits MafB transactivation.
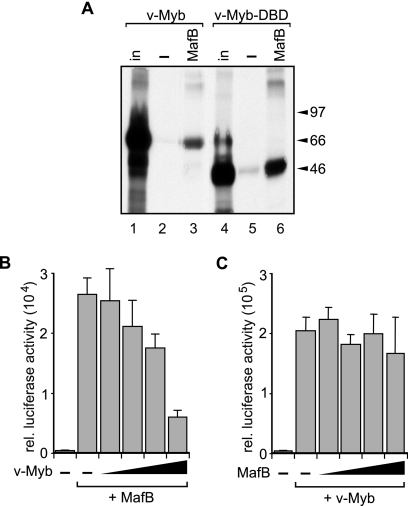
These observations prompted us to investigate whether the v-Myb-induced macrophage differentiation block involved direct MafB inhibition. MafB and Myb transcription factors act functionally in an antagonistic way during myelomonocytic differentiation inducing differentiation and proliferation, respectively (2, 16, 23, 29, 37). Therefore, we wondered whether the ability of v-Myb to block MafB macrophage-inducing activity in E26 myeloid progenitors might be due to direct physical interaction and repression. As shown in Fig. 6A, v-Myb bound via its DNA binding domain to MafB in a GST pull-down assay. To further test whether this correlated with a direct repression of MafB, we tested the effect of v-Myb on MafB transactivation activity. As shown in Fig. 6B, v-Myb repressed cotransfected MafB on a MARE reporter in a dose-dependent manner severalfold. By contrast, in the reciprocal experiment shown in Fig. 6C, increasing amounts of MafB could not repress v-Myb activity on a synthetic Myb reporter containing three Myb response elements (3xMRE) from the Myb target gene mim-1 (36). Together these results indicated that direct repression of MafB by v-Myb may explain the inability of MafB to overcome the v-Myb-imposed differentiation block in E26-transformed myeloid progenitors.
FIG. 6.
v-Myb binds and represses MafB. (A) MafB binds to v-Myb DNA binding domain (v-Myb-DBD). In vitro-translated, [35S]methionine-labeled E26 v-Myb (lanes 1 to 3) or E26 v-Myb DNA binding domain (lanes 4 and 5) was incubated with gluthathione bead-immobilized GST protein (lanes 2 and 5) or a GST-MafB fusion protein (lanes 3 and 6). Amount of input protein (in) is shown as a reference (lanes 1 and 3). The positions of molecular size markers are indicated to the right of the blot in kilodaltons. (B) v-Myb represses MafB transactivation. QT6 cells were transfected with 3xMARE reporter, constant amounts of expression constructs for MafB, and increasing amounts of v-Myb as indicated. rel., relative. (C) MafB does not repress v-Myb transactivation. QT6 cells were transfected with 3× MRE reporter, constant amounts of expression constructs for v-Myb, and increasing amounts of MafB as indicated. Transient-transactivation assays were filled up with empty vector to a constant amount of expression plasmid and performed in duplicate, and luciferase activity was normalized to β-galactosidase activity from a cotransfected control plasmid. Error bars indicate standard errors of the means, and data are representative of at least two independent experiments.
v-Myb repression of MafB is SUMO dependent.
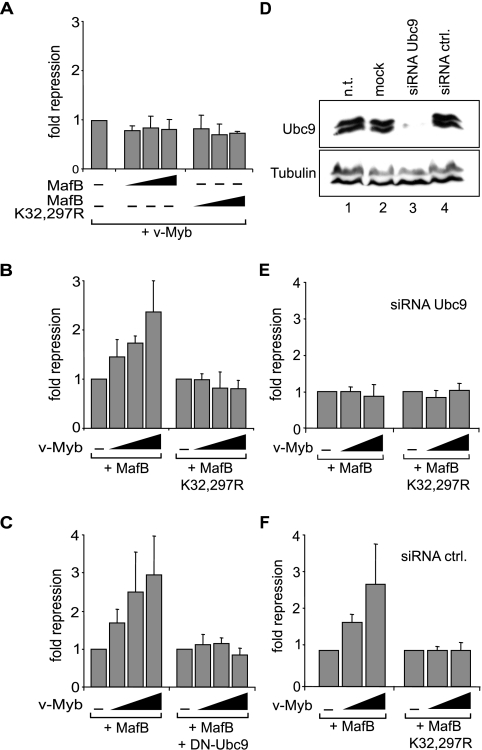
Since we observed that the SUMO-deficient mutant MafB K32,297R could escape a v-Myb-imposed differentiation block in E26-transformed myeloid progenitors (Fig. 5), we tested whether the MafB K32,297R mutant might have gained the ability to repress v-Myb transactivation activity. However, as shown in Fig. 7A, this mutant was still incapable of repressing v-Myb. To test whether reciprocally the inhibition of MafB by v-Myb might be dependent on SUMO modification, we analyzed whether the MafB K32,297R mutant could still be repressed by v-Myb. However, v-Myb could not inhibit MafB when SUMO modification of MafB was prevented by mutation of SUMO acceptor sites (Fig. 7B). To confirm that this effect was due to the loss of SUMO modification, we cotransfected a catalytically inactive C93S mutant of Ubc9 that acts as a dominant-negative version (20) of this essential E2-conjugating enzyme in the SUMO modification pathway (35, 40). In the presence of Ubc9 C93S, v-Myb could not inhibit MafB (Fig. 7C). To further corroborate that MafB repression was dependent on its SUMO modification, we inhibited endogenous Ubc9 expression using an siRNA approach. The siRNA efficiency was verified by Western blotting and revealed inhibition of Ubc9 expression levels of over 95% (Fig. 7D). When we cotransfected this siRNA together with v-Myb and MafB, it completely abolished the ability of v-Myb to repress MafB but had no influence on repression of MafB K32,297R (Fig. 7E). By contrast, a control siRNA that did not affect Ubc9 expression (Fig. 7D) had no effect on v-Myb repression of MafB transactivation (Fig. 7F). Together these results strongly indicated that the repression of MafB by v-Myb is dependent on SUMO modification of MafB K32 and K297 acceptor sites.
FIG. 7.
v-Myb repression of MafB requires MafB SUMOylation. (A) MafB SUMO target site mutation does not affect v-Myb transactivation activity. QT6 cells were transfected with 3xMRE reporter, constant amounts of expression constructs for v-Myb, and increasing amounts of MafB or MafB K32,297R as indicated. (B) MafB SUMO target site mutation abolishes v-Myb repression. QT6 cells were transfected with 3xMARE reporter and expression constructs for MafB or MafB K32,297R mutant and increasing amounts of v-Myb as indicated. (C) v-Myb repression of MafB is sensitive to inhibition of SUMO E2-conjugating enzyme Ubc9. QT6 cells were transfected with 3xMARE reporter and expression constructs for MafB and increasing amounts of v-Myb in the absence (left) or presence (right) of dominant-negative Ubc9 expression constructs as indicated. (D) Ubc9 siRNA strongly reduces Ubc9 expression. Protein extracts from nontransfected (n.t.) cells (lane 1), mock-transfected cells (lane 2), or HEK293 cells transfected with Ubc9 siRNA (lane 3) or control (ctrl.) siRNA (lane 4) were Western blotted with antibody against Ubc9 (top) or tubulin as a control (bottom). (E and F) Ubc9 knockdown abolishes SUMO site-dependent v-Myb repression of MafB. HEK293 cells were transfected with 3xMARE reporter and expression constructs for MafB or MafB K32,297R mutant and increasing amounts of v-Myb as indicated in the presence of Ubc9-specific siRNA (E) or control (ctrl.) siRNA (F). All transactivation assays were filled up with empty vector to a constant amount of expression plasmid and performed in duplicate, and luciferase activity was normalized to β-galactosidase activity from a cotransfected control plasmid. Error bars indicate standard errors of the means, and data are representative of at least two independent experiments.
SUMO-deficient MafB resists v-Myb-induced macrophage dedifferentiation.
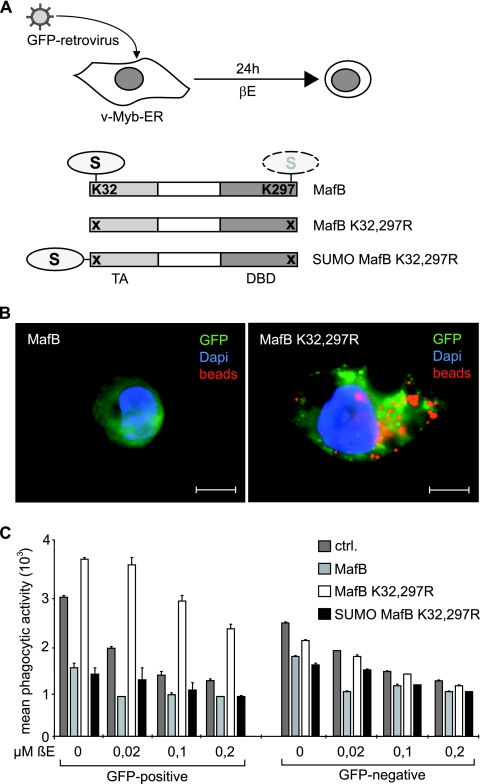
It has been shown previously that macrophages stably expressing an inducible version of v-Myb fused to the estrogen receptor hormone binding domain (v-Myb-ER) can be induced to dedifferentiate and lose macrophage attributes in the presence of β-estradiol (β-E) (7). We hypothesized that if the resistance of SUMO-deficient MafB to v-Myb repression was responsible for its ability to escape a v-Myb-imposed differentiation block, it should also resist v-Myb-induced dedifferentiation of macrophages. To address this question, we infected v-Myb-ER-expressing HD11 macrophages (7) with MFG-iGFP retroviruses (2) expressing no transgene or different versions of MafB together with an IRES element-driven GFP and analyzed macrophage phenotype and phagocytic activity of these cells after β-E stimulation (Fig. 8A). As shown in Fig. 8B, control or MafB virus-infected, GFP-positive cells behaved like uninfected cells and acquired a round myeloblast-like appearance 24 h after v-Myb induction, whereas SUMO-deficient MafB K32,297R mutant-infected cells maintained a typical macrophage morphology. Consistent with maintained macrophage function, MafB K32,297R virus-infected cells were also still capable of phagocytosing large numbers of fluorescent latex beads, whereas MafB or control virus-infected cells had lost this capacity (Fig. 8B). To quantify these observations, we measured phagocytic activity by FACS analysis of bead fluorescence in GFP-positive and GFP-negative cells. As shown in Fig. 8C, control infected cells rapidly lost macrophage phenotype in a β-E concentration-dependent fashion, whereas MafB K32,297R but not MafB virus-infected cells resisted this stimulus. To further confirm that this effect of the MafB K32,297R mutant was dependent on the loss of SUMO modification and not to some other fortuitous effect of the mutations, we reestablished the SUMOylated state on this mutant by covalently fusing SUMO-1 to its N terminus (Fig. 8A). As shown in Fig. 8C, this mutant behaved again like wild-type protein and could not resist v-Myb-induced dedifferentiation of macrophages. Significantly, uninfected, GFP-negative cells from all cultures behaved similarly, indicating that the observed effects were dependent on the expression of MafB proteins. Together these results indicate that in the absence of SUMO modification, MafB-driven macrophage differentiation becomes resistant to v-Myb inhibition. They also suggest that a v-Myb-induced block of macrophage differentiation critically involves SUMO-dependent repression of MafB.
FIG. 8.
SUMOylation-deficient MafB confers resistance to v-Myb-induced macrophage dedifferentiation. (A) Experimental setup. HD11 macrophages stably expressing v-MybER were infected with MFG-iGFP virus expressing an IRES-GFP cassette together with either no transgene or the indicated constructs, MafB, MafB K32,297R mutant, or SUMO-1 fused to MafB K32,297R. Infected cultures were induced for 24 h with β-estradiol (βE) to activate v-Myb. DBD, DNA binding domain. (B) Fluorescence micrographs of HD11v-MybER cells 24 h after v-Myb induction by 0.2 μM β-E and after 2 h of incubation with PE-conjugated fluorescent latex beads. Cells were infected with an MFG-iGFP virus expressing wild-type MafB or MafB K32,297R mutant as indicated. Dapi, 4′,6′-diamidino-2-phenylindole. Bars, 10 μm. (C) Control (ctrl.) HD11v-MybER cultures or HD11v-MybER cultures infected with virus expressing MafB, MafB K32,297R, or SUMO-MafB K32,297R mutant were treated for 24 h with the indicated concentrations of β-E, incubated for 2 h with PE-conjugated fluorescent beads, and analyzed by FACS for phagocytosed beads in infected, GFP-positive cells and in uninfected, GFP-negative cells. Quantification of phagocytic activity was expressed as their mean fluorescence intensity as a measure of the number of phagocytosed beads per cell. Error bars indicate standard errors of the means from three independent samples, and data are representative of two independent experiments.
DISCUSSION
We have shown that SUMO modification of the myeloid transcription factor MafB controls its transcriptional activity and ability to specify macrophage fate. We have identified two principal SUMO acceptor sites, whose modification inhibits transcriptional activity and renders MafB susceptible to repression by constitutively active Myb transcription factor. As a consequence, SUMOylation-deficient MafB has a strongly increased ability to limit myeloid progenitor proliferation and promote macrophage differentiation. In addition, the loss of MafB SUMOylation confers resistance to v-Myb inhibition of macrophage differentiation. Our results suggest that MafB SUMO modification can control the balance of myeloid progenitor expansion and macrophage differentiation by shifting the equilibrium between antagonistic Myb and MafB activities. Modulating the relative strengths of antagonistic transcription factor pairs by SUMO modification may thus serve as a potent mechanism for homeostatic control in the hematopoietic system.
Lysine residues of proteins can be subject to multiple posttranslational modifications (15). However, several lines of evidence indicate that the observations reported in this study are based on SUMO modification of MafB. Besides a perfect match of the target lysines to the SUMO modification consensus sequence, the effects of their mutation were fully reproduced by interfering in other ways with SUMO modification, such as SUMO removal by SUMO-specific proteases or by inhibiting SUMO conjugation using dominant-negative and siRNA inhibition of Ubc9, an essential enzyme of the SUMO modification pathway. Furthermore, these experimental manipulations had no influence on the MafB K32,297R SUMO target site mutant, indicating that it faithfully reflected the full activity of non-SUMOylated MafB. In addition, reimposing a SUMO-modified state on the K32,297R mutant protein by covalent fusion of SUMO-1 reestablished sensitivity to v-Myb-induced macrophage dedifferentiation.
SUMO modification has been shown to affect transcription factors, histones, polycomb group genes, and other nuclear proteins that influence transcription (15, 17, 18, 21, 41, 47, 49). Although SUMO modification can affect DNA binding (21), protein stability, and nuclear localization (19), in the majority of the cases, it appears to repress transcriptional activity (18, 21, 47). The detailed mechanisms of repression by SUMOylation have yet to be determined, but it has been proposed to involve changes in subnuclear localization (15, 21) or recruitment of histone deacetylases (15, 18, 21) and other corepressors (9, 31). Our new results indicate that beyond global transcriptional repression, SUMO modification may selectively regulate transcription factor cross inhibition.
Reciprocally inhibitory interactions between two different lineage-specific transcription factors have been invoked to maintain a balance between alternative differentiation programs (8, 22, 30). The work presented here and other studies (39) indicate that transcription factor cross inhibition may also be important for the homeostatic control between progenitor proliferation and differentiation into postmitotic mature cells. Regulatory mechanisms, however, that shift the equilibrium towards one or the other direction have not been identified. Our observations on the Myb/MafB antagonism in macrophage differentiation provide support for the hypothesis that SUMOylation may be such a mechanism.
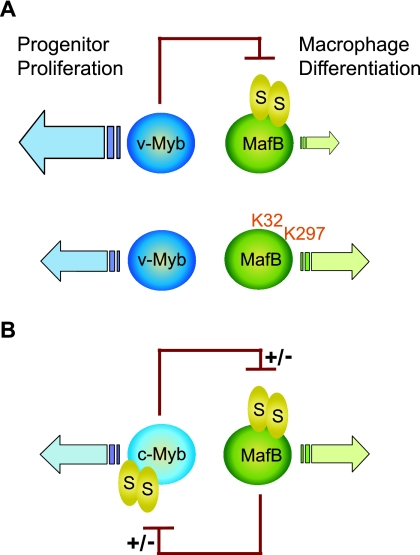
As mentioned above, v-Myb inhibits macrophage differentiation and promotes extended self renewal of myeloid progenitors (3, 23), whereas MafB enhances differentiation and inhibits their proliferation (2, 29, 43). Our results suggest that this functional antagonism between Myb and MafB activities may depend on a direct inhibitory interaction between v-Myb and MafB. We observed that while v-Myb could repress MafB transactivation, MafB could not inhibit v-Myb activity. This observation could provide a molecular explanation for the ability of v-Myb to arrest myeloid progenitors in a proliferative state and the incapacity of MafB to drive macrophage differentiation in the presence of active v-Myb (shown schematically in Fig. 9A, top row). We could further show that in the absence of SUMO modification, MafB could resist v-Myb repression and overcome the v-Myb-imposed differentiation block, thus reestablishing an equilibrium between myeloid progenitor proliferation and macrophage differentiation (shown schematically in Fig. 9A, bottom row). While it is clear that SUMO modification of MafB is required for the ability of v-Myb to repress MafB-dependent transcription, this is not simply a consequence of SUMO modification facilitating binding of v-Myb, as unmodified MafB still interacts strongly with v-Myb (Fig. 6A). In summary, the observation that repression of MafB by v-Myb is strictly dependent on MafB SUMO modification indicates that SUMOylation may provide a mechanism to regulate transcription factor antagonism in the control of progenitor proliferation and differentiation.
FIG. 9.
Model of SUMO-dependent Myb/MafB antagonism in macrophage differentiation. (A) v-Myb inhibits MafB transactivation but reciprocally is not inhibited by MafB, resulting in v-Myb-activated progenitor proliferation and a block of MafB-driven macrophage differentiation (top row). The absence of SUMO modification in the MafB K32,297R mutant frees MafB from v-Myb repression and enables macrophage differentiation but has no effect on v-Myb activity (bottom row), as SUMO modification determines the susceptibility to repression but not the ability to repress. (B) Hypothetical model of SUMO-dependent c-Myb/MafB cross inhibition in myelomonocytic progenitors. In contrast to v-Myb, c-Myb contains SUMO modification sites and is susceptible to MafB repression, suggesting mutual SUMO-dependent inhibition of MafB and c-Myb and a balance maintained between progenitor proliferation and macrophage differentiation. Selective alterations in SUMO modification status on either protein may control susceptibility to repression and modulate the balance between progenitor proliferation and differentiation according to physiological needs.
The constitutive activity of v-Myb appears to conserve a transient activity of c-Myb in the dynamic equilibrium between progenitor proliferation and differentiation, since c-Myb and v-Myb have similar effects on progenitor proliferation (23, 37) and c-Myb can also repress MafB transactivation (unpublished observations). Reciprocally, and in contrast to v-Myb, c-Myb can also be inhibited by MafB and the closely related family member c-Maf (26), which like MafB can induce monocyte differentiation (27) and appears to compensate for MafB function during the differentiation of MafB-deficient macrophages (1). The reason for the differential susceptibility of Myb proteins to MafB repression is currently unknown, but it is tempting to speculate that the deletion of c-Myb SUMO acceptor sites in the truncated v-Myb protein (4, 11) renders it resistant to MafB repression, analogous to the resistance of the MafB K32,296R mutant to Myb repression. Interestingly, SUMOylation-deficient MafB could still repress c-Myb (unpublished observations), similar to the ability of unmodified v-Myb to repress MafB. Together these observations suggest that SUMO modification of MafB and c-Myb selectively affects their susceptibility to repression but not their ability to repress. It thus appears that Maf and Myb proteins reciprocally inhibit each other during myelomonocytic differentiation and mutually keep their antagonistic activities in check in a SUMOylation-dependent manner (shown schematically in Fig. 9B). Selective removal of SUMO modification from one of the transcription factor partners would thus permit escape from repression by its antagonist while maintaining its own repressive capacities and thus shift the equilibrium towards progenitor proliferation or differentiation as required by the physiological conditions.
In summary, our results show that SUMO modification of a lineage-specific transcription factor can control the dynamic equilibrium of progenitor expansion and differentiation and establish SUMO modification as a mechanism to regulate tissue homeostasis in the hematopoietic system. It will be interesting to determine whether SUMO modification more generally regulates the activity of antagonistic transcription factor pairs in homeostasis and lineage choice.
Acknowledgments
S.T. was supported by the Böhringer Ingelheim Fonds (BIF) and the Societé Francaise d'Hématologie (SFH), C.O. and L.V. by the French Ministère de l'Éducation Nationale de l'Enseignement Supérieur et de la Recherche, C.O. by La Ligue Nationale contre le Cancer, Y.B. by the Centre National de la Recherche Scientifique (CNRS) and the Fondation pour la Recherche Médicale (FRM), and S.S. by the Association pour la Recherche sur le Cancer (ARC), SFH, and the Fondation de France (FdF). The work was supported by the ATIPE program of the CNRS; an installation grant from FRM; grant 2004004150 from FdF; grants 5387, 8453, and 3422 from ARC and grant 05-79 from AICR to M.H.S.; a Convention Franco-Marrocain CNRS/CNRST to M.H.S. and Y.B.; and a grant from CRUK to R.T.H.
We thank K.-H. Klempnauer for providing HD11v-MybER cells.
Footnotes
Published ahead of print on 4 June 2007.
REFERENCES
- 1.Aziz, A., L. Vanhille, P. Mohideen, L. M. Kelly, C. Otto, Y. Bakri, N. Mossadegh, S. Sarrazin, and M. H. Sieweke. 2006. Development of macrophages with altered actin organization in the absence of MafB. Mol. Cell. Biol. 26:6808-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakri, Y., S. Sarrazin, U. P. Mayer, S. Tillmanns, C. Nerlov, A. Boned, and M. H. Sieweke. 2005. Balance of MafB and PU.1 specifies alternative macrophage or dendritic cell fate. Blood 105:2707-2716. [DOI] [PubMed] [Google Scholar]
- 3.Beug, H., P. Blundell, and T. Graf. 1987. Reversibility of differentiation and proliferative capacity in avian myelomonocytic cells transformed by tsE26 leukemia virus. Genes Dev. 1:277-286. [DOI] [PubMed] [Google Scholar]
- 4.Bies, J., J. Markus, and L. Wolff. 2002. Covalent attachment of the SUMO-1 protein to the negative regulatory domain of the c-Myb transcription factor modifies its stability and transactivation capacity. J. Biol. Chem. 277:8999-9009. [DOI] [PubMed] [Google Scholar]
- 5.Bjerregaard, M. D., J. Jurlander, P. Klausen, N. Borregaard, and J. B. Cowland. 2003. The in vivo profile of transcription factors during neutrophil differentiation in human bone marrow. Blood 101:4322-4332. [DOI] [PubMed] [Google Scholar]
- 6.Broday, L., I. Kolotuev, C. Didier, A. Bhoumik, B. P. Gupta, P. W. Sternberg, B. Podbilewicz, and Z. Ronai. 2004. The small ubiquitin-like modifier (SUMO) is required for gonadal and uterine-vulval morphogenesis in Caenorhabditis elegans. Genes Dev. 18:2380-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burk, O., and K. H. Klempnauer. 1991. Estrogen-dependent alterations in differentiation state of myeloid cells caused by a v-myb/estrogen receptor fusion protein. EMBO J. 10:3713-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantor, A. B., and S. H. Orkin. 2001. Hematopoietic development: a balancing act. Curr. Opin. Genet. Dev. 11:513-519. [DOI] [PubMed] [Google Scholar]
- 9.Chang, C. C., D. Y. Lin, H. I. Fang, R. H. Chen, and H. M. Shih. 2005. Daxx mediates the small ubiquitin-like modifier-dependent transcriptional repression of Smad4. J. Biol. Chem. 280:10164-10173. [DOI] [PubMed] [Google Scholar]
- 10.Collavin, L., M. Gostissa, F. Avolio, P. Secco, A. Ronchi, C. Santoro, and G. Del Sal. 2004. Modification of the erythroid transcription factor GATA-1 by SUMO-1. Proc. Natl. Acad. Sci. USA 101:8870-8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahle, O., T. O. Andersen, O. Nordgard, V. Matre, G. Del Sal, and O. S. Gabrielsen. 2003. Transactivation properties of c-Myb are critically dependent on two SUMO-1 acceptor sites that are conjugated in a PIASy enhanced manner. Eur. J. Biochem. 270:1338-1348. [DOI] [PubMed] [Google Scholar]
- 12.Duprey, S. P., and D. Boettiger. 1985. Developmental regulation of c-myb in normal myeloid progenitor cells. Proc. Natl. Acad. Sci. USA 82:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eichmann, A., A. Grapin-Botton, L. Kelly, T. Graf, N. M. Le Douarin, and M. Sieweke. 1997. The expression pattern of the mafB/kr gene in birds and mice reveals that the kreisler phenotype does not represent a null mutant. Mech. Dev. 65:111-122. [DOI] [PubMed] [Google Scholar]
- 14.Fisher, A. G. 2002. Decision making in the immune system: cellular identity and lineage choice. Nat. Rev. Immunol. 2:977-982. [DOI] [PubMed] [Google Scholar]
- 15.Freiman, R. N., and R. Tjian. 2003. Regulating the regulators: lysine modifications make their mark. Cell 112:11-17. [DOI] [PubMed] [Google Scholar]
- 16.Gemelli, C., M. Montanari, E. Tenedini, T. Zanocco Marani, T. Vignudelli, M. Siena, R. Zini, S. Salati, E. Tagliafico, R. Manfredini, A. Grande, and S. Ferrari. 2006. Virally mediated MafB transduction induces the monocyte commitment of human CD34+ hematopoietic stem/progenitor cells. Cell Death Differ. 13:1686-1696. [DOI] [PubMed] [Google Scholar]
- 17.Gill, G. 2003. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 13:108-113. [DOI] [PubMed] [Google Scholar]
- 18.Gill, G. 2005. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15:536-541. [DOI] [PubMed] [Google Scholar]
- 19.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18:2046-2059. [DOI] [PubMed] [Google Scholar]
- 20.Girdwood, D., D. Bumpass, O. A. Vaughan, A. Thain, L. A. Anderson, A. W. Snowden, E. Garcia-Wilson, N. D. Perkins, and R. T. Hay. 2003. p300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11:1043-1054. [DOI] [PubMed] [Google Scholar]
- 21.Girdwood, D. W., M. H. Tatham, and R. T. Hay. 2004. SUMO and transcriptional regulation. Semin. Cell Dev. Biol. 15:201-210. [DOI] [PubMed] [Google Scholar]
- 22.Graf, T. 2002. Differentiation plasticity of hematopoietic cells. Blood 99:3089-3101. [DOI] [PubMed] [Google Scholar]
- 23.Graf, T. 1992. Myb: a transcriptional activator linking proliferation and differentiation in hematopoietic cells. Curr. Opin. Genet. Dev. 2:249-255. [DOI] [PubMed] [Google Scholar]
- 24.Hamada, M., T. Moriguchi, T. Yokomizo, N. Morito, C. Zhang, and S. Takahashi. 2003. The mouse mafB 5′-upstream fragment directs gene expression in myelomonocytic cells, differentiated macrophages and the ventral spinal cord in transgenic mice. J. Biochem. (Tokyo) 134:203-210. [DOI] [PubMed] [Google Scholar]
- 25.Hay, R. T. 2005. SUMO: a history of modification. Mol. Cell 18:1-12. [DOI] [PubMed] [Google Scholar]
- 26.Hegde, S. P., A. Kumar, C. Kurschner, and L. H. Shapiro. 1998. c-Maf interacts with c-Myb to regulate transcription of an early myeloid gene during differentiation. Mol. Cell. Biol. 18:2729-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hegde, S. P., J. Zhao, R. A. Ashmun, and L. H. Shapiro. 1999. c-Maf induces monocytic differentiation and apoptosis in bipotent myeloid progenitors. Blood 94:1578-1589. [PubMed] [Google Scholar]
- 28.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355-382. [DOI] [PubMed] [Google Scholar]
- 29.Kelly, L. M., U. Englmeier, I. Lafon, M. H. Sieweke, and T. Graf. 2000. MafB is an inducer of monocytic differentiation. EMBO J. 19:1987-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laiosa, C. V., M. Stadtfeld, and T. Graf. 2006. Determinants of lymphoid-myeloid lineage diversification. Annu. Rev. Immunol. 24:705-738. [DOI] [PubMed] [Google Scholar]
- 31.Lin, D. Y., H. I. Fang, A. H. Ma, Y. S. Huang, Y. S. Pu, G. Jenster, H. J. Kung, and H. M. Shih. 2004. Negative modulation of androgen receptor transcriptional activity by Daxx. Mol. Cell. Biol. 24:10529-10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McNagny, K. M., and T. Graf. 1996. Acute avian leukemia viruses as tools to study hematopoietic cell differentiation. Curr. Top. Microbiol. Immunol. 212:143-162. [DOI] [PubMed] [Google Scholar]
- 33.Melchior, F., M. Schergaut, and A. Pichler. 2003. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28:612-618. [DOI] [PubMed] [Google Scholar]
- 34.Motohashi, H., F. Katsuoka, C. Miyoshi, Y. Uchimura, H. Saitoh, C. Francastel, J. D. Engel, and M. Yamamoto. 2006. MafG sumoylation is required for active transcriptional repression. Mol. Cell. Biol. 26:4652-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nacerddine, K., F. Lehembre, M. Bhaumik, J. Artus, M. Cohen-Tannoudji, C. Babinet, P. P. Pandolfi, and A. Dejean. 2005. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell 9:769-779. [DOI] [PubMed] [Google Scholar]
- 36.Ness, S. A., Å. Marknell, and T. Graf. 1989. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell 59:1115-1125. [DOI] [PubMed] [Google Scholar]
- 37.Oh, I. H., and E. P. Reddy. 1999. The myb gene family in cell growth, differentiation and apoptosis. Oncogene 18:3017-3033. [DOI] [PubMed] [Google Scholar]
- 38.Orkin, S. H. 2000. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 1:57-64. [DOI] [PubMed] [Google Scholar]
- 39.Porse, B. T., T. A. Pedersen, X. Xu, B. Lindberg, U. M. Wewer, L. Friis-Hansen, and C. Nerlov. 2001. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell 107:247-258. [DOI] [PubMed] [Google Scholar]
- 40.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 41.Shiio, Y., and R. N. Eisenman. 2003. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100:13225-13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sieweke, M. H., and T. Graf. 1998. A transcription factor party during blood cell differentiation. Curr. Opin. Genet. Dev. 8:545-551. [DOI] [PubMed] [Google Scholar]
- 43.Sieweke, M. H., H. Tekotte, J. Frampton, and T. Graf. 1996. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell 85:49-60. [DOI] [PubMed] [Google Scholar]
- 44.Studzinski, G. P., and Z. S. Brelvi. 1987. Changes in proto-oncogene expression associated with reversal of macrophage-like differentiation of HL 60 cells. J. Natl. Cancer Inst. 79:67-76. [PubMed] [Google Scholar]
- 45.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 46.Taylor, K. M., and C. Labonne. 2005. SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev. Cell 9:593-603. [DOI] [PubMed] [Google Scholar]
- 47.Verger, A., J. Perdomo, and M. Crossley. 2003. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 4:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaguchi, T., P. Sharma, M. Athanasiou, A. Kumar, S. Yamada, and M. R. Kuehn. 2005. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol. Cell. Biol. 25:5171-5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, H., G. A. Smolen, R. Palmer, A. Christoforou, S. van den Heuvel, and D. A. Haber. 2004. SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat. Genet. 36:507-511. [DOI] [PubMed] [Google Scholar]