Abstract
Two of the major histone acetyltransferases in Saccharomyces cerevisiae are NuA4 and SAGA, which acetylate histones H4 and H3, respectively. Acetylated H3 and H4 tails have been implicated in binding bromodomain proteins, including Bdf1. Bdf1 interacts with the general transcription factor TFIID, which might promote preinitiation complex (PIC) assembly. Bdf1 also interacts with the SWR complex (SWR-C). SWR-C is responsible for the deposition of the histone H2A variant H2A.Z. The placement of these interactions into a connected pathway of PIC assembly has not been fully established. Moreover, it is not known how widespread and how variable such a pathway might be on a genomic scale. Here we provide genomic evidence for S. cerevisiae that PIC assembly (TFIID occupancy) and chromatin remodeling (SWR-C and H2A.Z occupancy) are linked in large part to NuA4-directed H4 acetylation and subsequent Bdf1 binding, rather than through SAGA-directed H3 acetylation. Bdf1 and its homolog Bdf2 tend to have distinct locations in the genome. However, the deletion of BDF1 leads to the accumulation of Bdf2 at Bdf1-vacated sites. Thus, while Bdf1 and Bdf2 are at least partially redundant in function, their functions in the genome are geographically distinct.
Eukaryotic promoters are regulated in large part by the dynamic assembly and disassembly of the transcription machinery. At least in the budding yeast Saccharomyces cerevisiae, most promoters possess similar chromatin architectures with which components of the transcription machinery must contend (1, 33, 56). The interplay between chromatin and the transcription machinery is of considerable interest, and many details are now coming to light. How this plays out mechanistically on a genome-wide scale remains largely unknown.
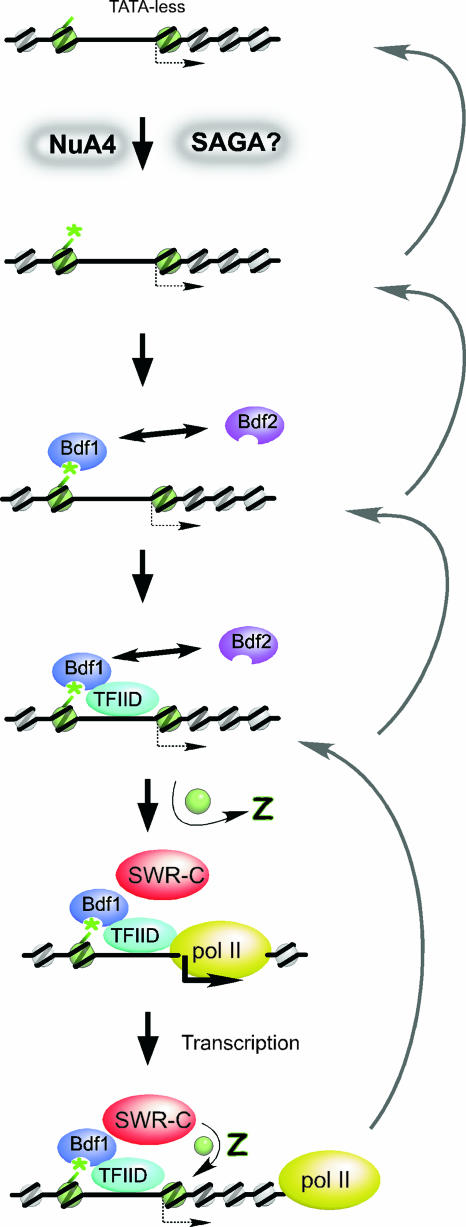
Figure 1 attempts to integrate a current view of one aspect of preinitiation complex (PIC) assembly and its linkage to chromatin. As illustrated, yeast promoters tend to possess nucleosome-free regions and are flanked by positioned nucleosomes (1, 56). These flanking nucleosomes tend to be enriched with the histone H2A variant called H2A.Z (17, 32, 44, 59). H2A.Z is largely promoter specific, affording an as yet unknown function in gene regulation. Some evidence suggests that H2A.Z might facilitate nucleosome dismantling (59), which might be important for transcription initiation (1).
FIG. 1.
Model for PIC assembly via nucleosome acetylation. Shown are a series of events starting with a typical promoter and its nucleosomal architecture. NuA4 and/or SAGA associate with promoter regions via targeting mechanisms that are not shown. These HAT complexes acetylate histone H4 and H3, respectively (and other sites as well). Bdf1 is preferentially recruited to acetylated nucleosomes, which could be replaced by Bdf2 at some promoters. It is not known to which nucleosomes Bdf1 binds. Bdf1 recruits TFIID and SWR-C, which leads to PIC assembly, the release of H2A.Z, and transcription by pol II (RNA polymerase II).
Nucleosomes in promoter regions tend to be hyperacetylated during gene activation, and the prevailing view is that acetylation at certain histone residues contributes to gene activation (5, 8, 12, 46). Although numerous histone acetyltransferases (HATs) exist in yeast (53), SAGA and NuA4 appear to be the major HAT complexes that contribute to gene activation across the genome (2, 10, 13, 16, 48, 52). SAGA prefers to acetylate histone H3 amino-terminal tails (16), whereas NuA4 prefers histone H4 tails (as well as H2A.Z tails) (2, 3, 10, 26, 39, 45, 52). Current evidence suggests that these two HAT complexes might make significant contributions to distinct PIC assembly pathways, although this is not fully known. Promoters regulated by SAGA tend to possess a TATA box core promoter element and are generally inhibited by chromatin (4, 22). SAGA might direct the TATA binding protein TBP to the TATA box in the absence of the classical TBP-containing complex TFIID (4, 22, 50). Genes that are particularly dependent upon NuA4 tend to be TATA-less and regulated by TFIID (13). Histone tails tend to play a positive role at these promoters (22).
A substantial body of evidence suggests that acetylated histone tails are binding sites for protein complexes that contain bromodomains (11, 20, 21, 23, 25, 41). Such complexes include SAGA, TFIID, RSC, and SWI/SNF, among others (7, 9, 19, 35, 37, 55). The primary bromodomain component of yeast TFIID is the dissociable Bdf1 subunit (encoded by the carboxyl-terminal domain of TAF1 in higher eukaryotes but encoded separately in yeast) (37). As shown in Fig. 1, one current view posits that H4 acetylation at promoters by NuA4 leads to Bdf1 binding (as well as the potential binding of other bromodomain factors) (19, 22, 24, 31, 35, 37, 38, 54). Bdf1 then recruits TFIID, which contributes to RNA polymerase II recruitment via other general transcription factors, to form a PIC.
The evidence linking H4 acetylation and Bdf1/TFIID binding is as follows: (i) crystallographic structures and biochemical assays show isolated Bdf1 bromodomains bound to acetylated histone H4 tails (23, 31, 38, 42); (ii) Bdf1 and TFIID colocalize throughout the yeast genome at sites that are enriched with acetylated histone H4 (13, 30); (iii) genes that are positively regulated by histone H4 tails, by the Esa1 catalytic HAT subunit of NuA4, or by Bdf1 tend to be dominated by TFIID regulation (13, 22); (iv) an Esa1 HAT mutation shows a synthetic lethal phenotype with a bdf1Δ mutation (38); (v) Bdf1 interacts directly with TFIID (37); (vi) the elimination or nucleosomal occlusion of the TATA box at the PHO5 promoter creates a transcriptional dependency on H4 tails and Bdf1 (36).
Notwithstanding the connection between Bdf1 and TFIID, a substantial fraction of Bdf1 appears to act independently of TFIID (31, 37), where it may function as a boundary preventing the spread of proteins involved in heterochromatin formation (31). Bdf1 also interacts with the SWR complex (SWR-C), which is responsible for H2A.Z deposition (Fig. 1) (27, 28, 40). SWR-C uses the energy of ATP hydrolysis through the Swr1 subunit to replace H2A with H2A.Z in nucleosomes (40, 49). Thus, Bdf1 might recruit both TFIID and SWR-C to promoter regions (44). SWR-C might catalyze the exchange of H2A.Z during transcription initiation during which nucleosomes are dismantled to allow RNA polymerase II passage and then reassembled. Consistent with the linkage of Bdf1 to H2A.Z deposition, genome-wide occupancy of the two proteins is highly correlated, with H2A.Z occupancy being dependent upon both Bdf1 and SWR-C (59). Bdf1 and H2A.Z occupancy are linked to H4 acetylation in that mutations in NuA4 subunits or in H4 tail lysines that are acetylated by NuA4 diminish Bdf1 or H2A.Z binding at specific loci (3, 30, 44).
In contrast to the apparent widespread dependency of H2A.Z deposition on Bdf1, another study saw little effect when specific loci were examined (44). The lack of effect was attributed to the functionally redundant homolog BDF2. Little is known about BDF2 except that it may be at least partially redundant with BDF1 in that BDF2 can genetically compensate for BDF1, and both have biochemical interactions with TFIID (37, 44). Open questions are whether Bdf1 is preferentially recruited over Bdf2 and whether the redundancy between Bdf1 and Bdf2 applies to all or just a subset of genomic sites.
In summary, the evidence points to a PIC assembly pathway where, in a simplified and incompletely understood way, NuA4 is recruited to promoter regions to acetylate H4 tails via Esa1. The acetylated H4 tails help recruit Bdf1, which helps recruit TFIID and SWR-C to elicit PIC assembly and H2A.Z nucleosome remodeling. In a parallel and partially redundant pathway, SAGA is recruited to acetylate H3 tails via Gcn5 and delivers TBP to TATA-containing promoters to elicit PIC assembly. The acetylated H3 tails help recruit/retain nucleosome remodeling complexes, such as SWI/SNF and RSC (18, 19, 25). Several pieces of evidence suggest that the two pathways might be interconnected to some extent. First, NuA4 (Esa1) and SAGA (Gcn5) display functional redundancy with respect to RNR3 expression (51). Second, defects in the SAGA pathway (i.e., strains lacking GCN5 or harboring nonacetylatable H3 mutants) result in defects in the NuA4 pathway (e.g., decreased H2A.Z occupancy) (44, 59). Third, H3 acetylation generally correlates with Bdf1 and H2A.Z occupancy (30, 59). One interpretation of these observations is that Gcn5-acetylated H3 tails also recruit/retain Bdf1, which recruits SWR-C and then H2A.Z. However, genetic evidence suggests that SAGA's connection to Bdf1 might not involve GCN5 (38). It remains to be determined whether H3 acetylation or Gcn5 contributes to Bdf1 occupancy at promoter regions.
The model of NuA4- or SAGA-directed PIC assembly via acetylation and Bdf1 binding is far from established and thus would benefit from additional experimental investigation. Here we examine tenets of the model on a genome-wide scale. We explore the consequence of the loss of acetylation by NuA4 for the recruitment of Bdf1, TFIID, SWR-C, and H2A.Z at the majority of yeast promoter regions. We also examine the effect of the loss of Bdf1 on the recruitment of these factors and whether the loss of Bdf1 results in a physical relocation of Bdf2 to Bdf1 binding sites. Requirements for SAGA (Gcn5) are also examined. The results presented here provide multiple lines of evidence for a general model of PIC assembly and H2A.Z nucleosome remodeling derived largely from the direction of NuA4 rather than that of SAGA. The genome-wide analysis further reveals that the disruption of factor binding at preferred sites results in the accumulation of factors at less-preferred sites which we speculate to be default repositories for such factors.
MATERIALS AND METHODS
Yeast strains.
A list of the strains used in this study is provided in Table 1. Deletions were created by replacing the entire open reading frame with the kanamycin resistance gene. Proteins were tagged by generating PCR products of the tandem affinity purification (TAP) tag from the Open Biosystems TAP-tagged yeast collection strains and incorporating the PCR product into the wild-type or mutant yeast strains by homologous recombination.
TABLE 1.
Characteristics of strains used in this studya
| Strain | Chromosomal deletion(s) | Plasmid 1 | Plasmid 2 | Tagged protein | Source |
|---|---|---|---|---|---|
| yMD52 | taf1Δ esa1Δ | pJI12 (WT TAF1) | pSAPE1 (WT ESA1) | BDF1-TAP | This paper |
| yMD55 | taf1Δ esa1Δ | pJI12 (WT TAF1) | pSAPE1 (WT ESA1) | SWR1-TAP | This paper |
| yMD58 | taf1Δ esa1Δ | pJI12 (WT TAF1) | pSAPE1 (WT ESA1) | HTZ1-TAP | This paper |
| yMD59 | taf1Δ esa1Δ | pJI11 (taf1-ts2 mutant) | pSAPE1 (WT ESA1) | BDF1-TAP | This paper |
| yMD67 | taf1Δ esa1Δ | pJI12 (WT TAF1) | pSAPE2 (esa1-414 mutant) | BDF1-TAP | This paper |
| yMD69 | taf1Δ esa1Δ | yCP1 (WT TAF1) | pSAPE2 (esa1-414 mutant) | TAF1-TAP | This paper |
| yMD70 | taf1Δ esa1Δ | pJI12 (WT TAF1) | pSAPE2 (esa1-414 mutant) | SWR1-TAP | This paper |
| yMD73 | taf1Δ esa1Δ | pJI12 (WT TAF1) | pSAPE2 (esa1-414 mutant) | HTZ1-TAP | This paper |
| yMD75 | bdf1Δ | TAF1-TAP | This paper | ||
| yMD76 | bdf1Δ | SWR1-TAP | This paper | ||
| yMD79 | bdf1Δ | HTZ1-TAP | This paper | ||
| yMD87 | bdf1Δ | BDF2-TAP | This paper | ||
| BDF1-TAP | BDF1-TAP | Open Biosystems | |||
| BDF2-TAP | BDF2-TAP | Open Biosystems | |||
| HTZ1-TAP | HTZ1-TAP | Open Biosystems | |||
| SWR1-TAP | SWR1-TAP | Open Biosystems | |||
| TAF1-TAP | TAF1-TAP | Open Biosystems |
WT, wild type.
Chromatin immunoprecipitation and microarray analysis (ChIP-chip).
Cultures were grown in standard yeast peptone dextrose medium at 25°C to an optical density at 600 nm of 0.8 and then rapidly shifted to 37°C for 45 min. Cells were cross-linked with 1% formaldehyde and simultaneously cooled to 25°C; strains containing TAP-tagged histones were cross-linked for 15 min, and TAP tag transcription factors were cross-linked for 2 h. Parental strains lacking the TAP tag served as negative “null” controls. Cross-linking reactions were quenched with 125 mM glycine for 5 min, and cells were harvested. Cells were lysed with zirconia beads, and the chromatin was washed prior to sonication. Sonication was performed using the Diagenode BioRuptor. The BioRuptor was used at the “high” setting, with alternating sessions of 30 s of sonication and 30 s of resting. Samples that were cross-linked for 15 min were sonicated four at a time for a total of 6 min, and 2-h-cross-linked samples were sonicated two at a time for a total of 13 min. The final DNA fragment size for all samples was between 300 and 500 base pairs. The sonicated DNA was amplified by 20 (for histones) or 25 (for transcription factors) cycles of PCR and subsequently hybridized to spotted microarrays containing >6,000 PCR-generated probes spanning each intergenic region as previously described (58). Data were filtered and analyzed as previously described (57). Negative controls showed very low levels of DNA recovery, as expected.
Microarray data accession number.
Raw data are available at GEO under accession number GSE6707. Processed microarray data are available in Table S1 in the supplemental material.
RESULTS
The model in Fig. 1 places NuA4-directed (and/or SAGA-directed) acetylation as an early step in PIC assembly, followed by Bdf1 binding. We therefore focused on the downstream events that are potentially dependent upon these upstream events. NuA4-directed acetylation is catalyzed by Esa1 (2). Since Esa1 is essential for cell growth, it was necessary to employ the temperature-sensitive allele esa1-414 (10). Bdf1 was eliminated using a bdf1Δ strain. Chromatin immunoprecipitations of TAP-tagged proteins were performed on a genome-wide scale (by ChIP-chip) (58) in which the binding patterns of Bdf1, Bdf2, TFIID (via Taf1), SWR-C (via Swr1), and H2A.Z nucleosomes (via Htz1) were examined in mutant and wild-type strains. All formaldehyde cross-linking was performed under the same conditions in which cells were grown exponentially in rich medium at 25°C, followed by an abrupt shift to 37°C for 45 min (58), which eliminates esa1-414 when present (10, 13).
Occupancy of Bdf1, TFIID (Taf1), SWR-C (Swr1), and H2A.Z (Htz1) at many of the same promoter regions depends upon NuA4 (Esa1).
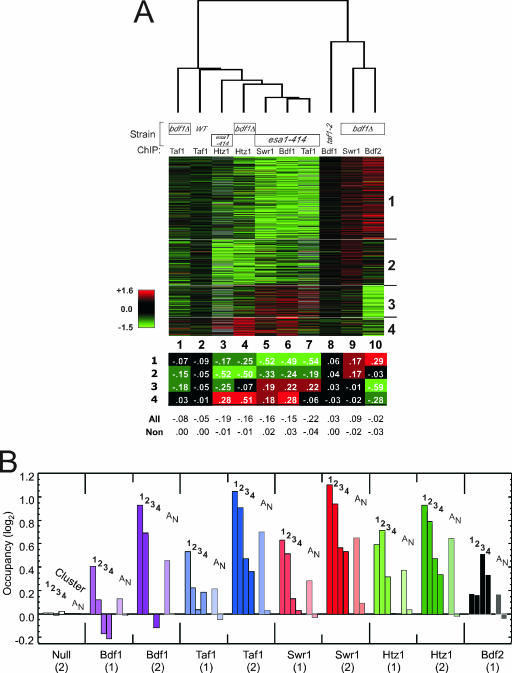
Log2-transformed ratios of factor occupancy in a mutant relative to that in a wild-type strain are presented as a cluster plot in Fig. 2A. Each row corresponds to an intergenic promoter region. Only those intergenic regions that met a specified cutoff (Fig. 2 legend) for changes in occupancy in at least one set of experiments are shown. The remainder may represent genes whose expression is increasingly rate limited by factors other than the ones studied here and which thus display fewer changes. These genes are nevertheless likely to be regulated in part by the mechanisms described here, particularly under conditions where they might be more highly expressed. The ratios were centered (log2 = 0) to the median value of all nonpromoter intergenic regions (i.e., regions between two convergently transcribed genes). The data were clustered by k means into four clusters, representing the maximum number of visually nonredundant clusters. None of these clusters were enriched with genes belonging to specific Gene Ontology terms (data not shown), which may simply reflect the fact that NuA4 contributes to the expression of most genes, regardless of their biological function or process.
FIG. 2.
Cluster plot of genome-wide changes in factor occupancy in mutant versus wild-type strains. (A) Changes in factor occupancy in the indicated mutants. ChIP-chip was performed on the indicated factors in the indicated strains. Assays were run in parallel with mutant and wild-type (WT) strains, which were labeled separately with Cy3 and Cy5 fluorescent dyes and cohybridized to spotted microarrays containing ∼6,000 intergenic-region-length PCR-generated probes. Data are presented as a cluster diagram (14) in which each row is an intergenic promoter region and each column is an average of two dye-swapped replicates. Red, green, and black denote increases, decreases, and no change in binding, respectively. Data were filtered to include only those intergenic regions that contained a single promoter region and had the largest change in occupancy (i.e., <10th or >90th percentile in at least one column). Six hundred eleven intergenic regions (∼10% of the analyzed genome) met these criteria. These intergenic regions are intended to be the strongest representatives of the genome so as to generate the strongest patterns. Such patterns are nevertheless likely to be applicable, with lower intensity, to most other intergenic promoter regions. Rows were grouped according to k means into four clusters (with 283, 158, 108, and 62 intergenic regions) (14). Columns were clustered hierarchically (14). The table below the cluster plot provides the average log2 ratio in each cluster for each experiment. “All” denotes all promoters, and “Non” denotes nonpromoter intergenic regions (i.e., regions between two convergently transcribed genes). (B) Average occupancy of the indicated factors for the promoter regions of clusters 1 to 4. “A” denotes all single-promoter intergenic regions (>3,000), and “N” denotes nonpromoter intergenic regions (>1,000). Data for sets designated “(1)” are the log2 ratios of the wild-type reference data set presented in panel A (binding after a shift to 37°C for 45 min) divided by the ChIP result for a “Null” untagged control (58). Data for sets designated “(2)” are the log2 ratios of the wild-type data set from reference 58 (binding after a shift to 37°C for 15 min) divided by a “Null” result. The two data sets were collected from two different studies which differed only in the time at 37°C (15 versus 45 min) before binding was measured. Their trends were similar, thereby providing additional confidence in the conclusions drawn from the data. Log2 values are relative to those for nonpromoter regions.
As revealed by the four basic patterns in Fig. 2A, the loss of NuA4 (Esa1) resulted in a coordinated loss of Bdf1, TFIID (Taf1), SWR-C (Swr1), and H2A.Z (Htz1) at promoter regions in clusters 1 and 2. As expected, clusters 1 and 2 had the highest levels of these factors in wild-type cells (Fig. 2B). We interpret the strong linkage of factor loss to mean that the NuA4-directed acetylation of nucleosomes plays an important role in the recruitment or retention of these proteins by a significant subset of all yeast promoters in accord with the model presented in Fig. 1. Most other genes may not be sufficiently expressed to reliably evaluate factor occupancy. Formally, we cannot exclude the possibility that NuA4 might have additional unknown targets that contribute to factor recruitment/retention. We also do not exclude the possibility of a more complex scenario whereby H4 acetylation by NuA4 promotes H3 acetylation via SAGA, with the latter possibility or some combination of both contributing to factor recruitment. The possibility of a contribution from SAGA is addressed further below.
Promoter regions that are dependent upon NuA4 for factor recruitment are highly acetylated.
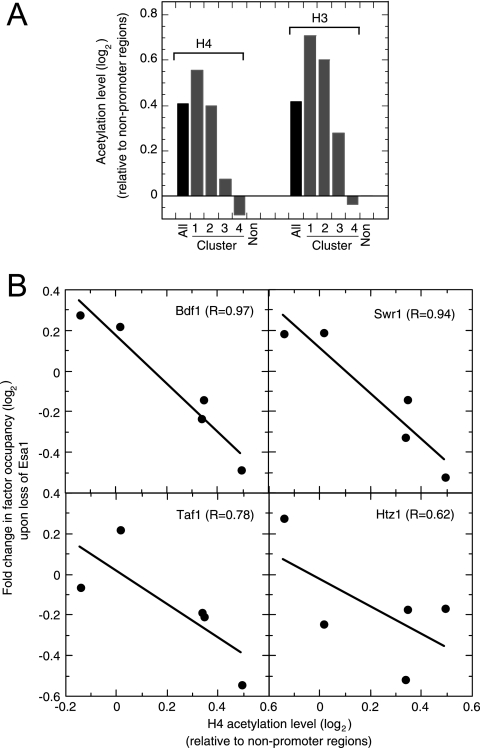
Based upon the model shown in Fig. 1, we expect that the promoter regions that are most susceptible to losing factors in the NuA4 pathway would be those that are highly acetylated at H4. We addressed this by examining the average H4 acetylation status of genes in all four clusters and throughout the rest of the genome by using existing H4 acetylation data (6). As shown in Fig. 3A (left side), the cluster 1 promoter regions had a relatively high H4 acetylation status, followed by that of the cluster 2 regions. Compared to the rest of the genome, clusters 3 and 4 had comparatively low H4 acetylation levels which were similar to levels found in nonpromoter regions. A similar pattern was also observed for H3 acetylation (Fig. 3A, right side), leaving open the possibility of involvement by the H3-specific SAGA (Gcn5) acetyltransferase. Figure 3B quantifies factor loss as a function of acetylation levels, showing that higher initial levels of H4 acetylation strongly correlate with greater losses of Bdf1, TFIID (Taf1), SWR-C (Swr1), and H2A.Z (Htz1) when NuA4 is eliminated. The further up the pathway (shown in Fig. 1) that a factor resided, the stronger the correlation. Together, these findings place Bdf1, TFIID (Taf1), SWR-C (Swr1), and H2A.Z (Htz1) recruitment in the same pathway and downstream of H4 acetylation by NuA4 at genes represented by clusters 1 and 2.
FIG. 3.
Intrinsic H4 acetylation levels at genes from clusters 1 to 4. (A) Relative occupancy level of acetylated H4 (K5,8,12,16) and H3 (K9,14) at clusters 1 to 4. Acetylation levels were obtained from reference 6 and were normalized to the levels for nonpromoter intergenic regions. Data were transformed into log2 ratios, and the average for each cluster was determined. “Non” denotes the average of >2,000; “All” denotes all promoter regions. (B) Changes (n-fold) in factor occupancy upon the loss of Esa1 correlate with the initial H4 acetylation levels. The H4 acetylation levels are from panel A. Changes in factor occupancy levels represent averages from Fig. 2A.
Occupancy of Bdf1 in promoter regions is not promoted by SAGA (Gcn5) or TFIID (Taf1).
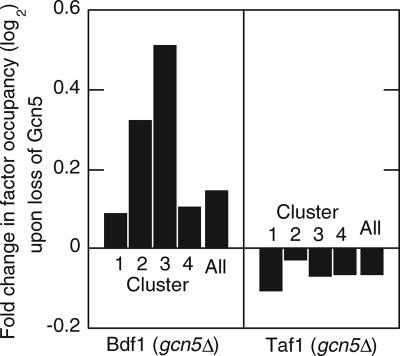
We next examined the potential involvement of SAGA in factor occupancy since H3 acetylation and SAGA have been linked to Bdf1 and Htz1 occupancy (30, 59). To address the possibility that SAGA-directed acetylation contributed to Bdf1 occupancy, we performed ChIP-chip on Bdf1 and Taf1 in gcn5Δ and wild-type strains. As shown in Fig. 4 (left panel), the promoter regions of genes in all four clusters actually gained Bdf1 when Gcn5 was missing, suggesting that SAGA or H3 acetylation is not contributing positively to Bdf1 recruitment/retention at promoters. Since both Bdf1 and Gcn5 possess bromodomains, the findings are more consistent with the possibility that the two bromodomain proteins compete for occupancy of a common target, such as acetylated H4 tails. That possibility remains to be tested. Taken together, these findings are consistent with the involvement of the NuA4 pathway in recruitment/retention of Bdf1 at promoter regions. Since H3 acetylation levels correlate with H4 acetylation levels (6, 30, 34, 43), this might account for previous observations that the acetylation of certain H3 residues correlates with Bdf1 occupancy (30).
FIG. 4.
Changes in Bdf1 and Taf1 occupancy when Gcn5 is eliminated. ChIP-chip assays were performed with Bdf1-TAP and Taf1-TAP as described in the legend to Fig. 2A; the test and reference samples were derived from gcn5Δ and GCN5 strains, respectively. The average n-fold change in occupancy between the mutant and wild-type strains was determined for the set of genes in clusters 1 to 4 (Fig. 2A), log2 transformed, and plotted. The data were centered such that there was, on average, no change in occupancy in nonpromoter intergenic regions. “All” denotes the value for all intergenic promoter regions for which there were valid data.
In contrast to our observations with Bdf1, we observed somewhat less TFIID (Taf1) promoter occupancy in the absence of Gcn5 (Fig. 4, right panel), which is consistent with the notion put forward by other studies that SAGA and H3 acetylation make significant, albeit modest, global contributions to transcription (16, 29, 43, 47). Our finding that the changes in Taf1 occupancy did not mirror those in Bdf1 when GCN5 was deleted suggests that the additional Bdf1 occupancy at promoter regions is not sufficient to promote PIC formation. The idea that Bdf1 is maintained at promoter regions independent of its interactions with TFIID is demonstrated in Fig. 2A (column 8), where loss of TFIID in a taf1-2 temperature-sensitive strain did not alter Bdf1 occupancy.
Bdf1 and Bdf2 are geographically separate but interchangeable at certain genes.
The NuA4-mediated pathway outlined in Fig. 1 indicates that the binding of Bdf1 to acetylated nucleosomes contributes to the subsequent recruitment of SWR-C and TFIID. Thus, we examined the impact of Bdf1 loss on factor recruitment, which, as shown in Fig. 2A, had a wider range of effects than the loss of NuA4 (Esa1) did. The loss of Bdf1 resulted in the depletion of H2A.Z (Htz1) at promoter regions in a manner akin to what was observed upon the loss of NuA4 (Esa1) (compare clusters 1 and 2 in columns 3 and 4 in Fig. 2A). This is consistent with the model in Fig. 1 and elsewhere (59) in which Bdf1 recruits/retains SWR-C which deposits H2A.Z (Htz1).
Despite the promoter regions in cluster 1 having H2A.Z (Htz1) occupancy similar to that of cluster 2 (Fig. 2B), cluster 2 experienced more of a loss of H2A.Z than cluster 1, when either Bdf1 or NuA4 (Esa1) was eliminated (Fig. 2A, columns 3 and 4; compare clusters 1 and 2). If Bdf1 and Bdf2 are functionally redundant, as previously proposed (37, 44), and if we consider the additional constraint that this redundancy is promoter specific, then perhaps Bdf2 is able to replace Bdf1 at cluster 1 but not at cluster 2. We were unable to test this by mutating both BDF1 and BDF2 since the deletion of both genes is lethal and a deletion of BDF1 in the context of a number of nonlethal BDF2 point mutations reverted rapidly in our hands.
The possibility that Bdf2 might replace Bdf1 in a promoter-specific manner was instead suggested by the binding pattern of Bdf2 in a bdf1Δ strain. Consistent with that notion, there was a gain of Bdf2 at cluster 1 (Fig. 2A, column 10) and no change at cluster 2. This result fits the predicted behavior of Bdf2 if it were substituting for Bdf1 in a promoter-specific manner (i.e., at cluster 1 but not at cluster 2) when Bdf1 is absent. At the promoter regions of both clusters 1 and 2, however, Bdf1 was preferentially recruited over Bdf2 when both were present (Fig. 2B).
The deletion of BDF1 resulted in only a modest loss of TFIID (Taf1) at cluster 1 (Fig. 2A, column 1) compared to the loss experienced by the esa1-414 mutant. This finding is consistent with the Bdf1/Bdf2 functional redundancy model, although the data do not exclude the possibility of other recruitment mechanisms for TFIID, such as through activators (15). At cluster 2 genes, the losses of TFIID (Taf1) were of similar magnitudes in the bdf1Δ and esa1-414 strains (columns 1 versus 7 in cluster 2), which is also consistent with the lack of Bdf2 involvement at those genes.
The loss of Bdf1 resulted in a small but consistent gain of SWR-C (Swr1) at the promoter regions of clusters 1 and 2 (Fig. 2A, column 9). These promoter regions normally had relatively high levels of SWR-C (Swr1) in wild-type strains (Fig. 2B). At cluster 1, the maintenance/augmentation of SWR-C (Swr1) in the absence of Bdf1 could be from a redundant action of Bdf2, but this possibility would seem to require NuA4-mediated acetylation (Fig. 2A, column 5). Acetylation and Bdf2 have no established connection.
Coordinate movement of factors throughout the genome.
Compared to those in clusters 1 and 2, the promoter regions in clusters 3 and 4 had low H4 acetylation levels (Fig. 3A) and low occupancy levels of Bdf1, TFIID (Taf1), SWR-C (Swr1), and H2A.Z (Htz1) but high levels of Bdf2 (Fig. 2B). These findings suggest that Bdf2 is normally directed to a different class of promoters than are these other factors. These Bdf2-enriched promoters have relatively low levels of H4 acetylation (Fig. 3A), which is consistent with in vitro studies demonstrating that Bdf2 interacts with unacetylated histone tails (37). Acetylation might confer preferential occupancy for Bdf1 over Bdf2 at the promoter regions in cluster 1, but this remains to be tested.
A consistent pattern shown in the cluster plot in Fig. 2A is that clusters 3 and 4 tended to respond oppositely to the way clusters 1 and 2 respond. We interpret this to mean that when the specific targeting of factors is disrupted, these factors are coordinately redistributed to lower-affinity sites. The coordinated redistribution of factors residing within the same pathway further illustrates their functional interconnectivity.
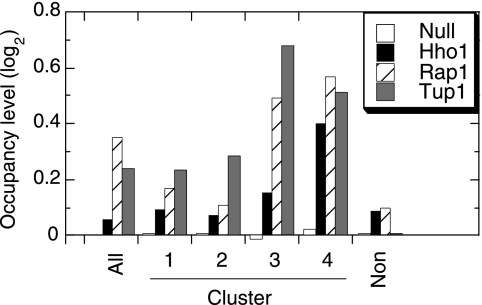
To further examine the composition of these opposing classes of promoters, we looked at the average occupancy levels of a variety of other regulatory factors in each cluster using existing ChIP-chip data. As shown in Fig. 5, a number of factors are more enriched in clusters 3 and 4 than in clusters 1 and 2, including proteins associated with repression, such as histone H1 (Hho1), Tup1, and Rap1. Thus, the findings in Fig. 2 suggest that on a genomic scale, PIC assembly components are directed to specific promoters, as expected, but when this delivery breaks down, these factors are redistributed to less-occupied regions of the genome that tend to be enriched with repressor proteins.
FIG. 5.
Regions that are depleted of NuA4-assembled transcription components tend to be enriched with repressor proteins. Average occupancy data (log2 scale) for the promoter regions in the indicated clusters were obtained from reference 58, as described in the legend to Fig. 2B.
DISCUSSION
H4 acetylation by NuA4 directs PIC assembly and chromatin remodeling via Bdf1.
Histone acetylation is likely to play multiple roles in the regulation of gene expression and other nuclear processes. It remains uncertain as to what mechanistic role individual acetylated lysines play in gene control. A significant advance came with the finding that bromodomains bind to acetylated lysines, potentially linking bromodomain-containing components of the transcription regulatory machinery directly to nucleosome acetylation.
As a bromodomain protein that interacts with both TFIID and SWR-C, Bdf1 has the ability to link nucleosome acetylation to PIC assembly and chromatin remodeling. To better understand this process from a genome regulation perspective, we sought to identify which acetylation activities are responsible for recruiting/retaining Bdf1 at promoter regions. Cells have a multitude of HATs, of which the NuA4 and SAGA complexes appear to play the largest gene regulatory role. Both are implicated in Bdf1 occupancy at promoters in that Bdf1 occupancy correlates with H4 and H3 acetylation across the yeast genome (13, 30, 59). Indeed, H4 and H3 acetylation also correlates across the yeast genome (6, 30, 34, 43). Our findings demonstrate a strong connection between Bdf1 and NuA4 in that the loss of NuA4 (Esa1) function resulted in a loss of Bdf1 binding to many promoter regions. We examined an alternative possibility that H4 acetylation might instead be helping to recruit or retain the bromodomain complex SAGA, which would employ its Gcn5 subunit to acetylate histone H3. The acetylated H3 might then recruit/retain Bdf1. Despite the fact that SAGA and NuA4 tend to occupy and acetylate similar sets of genes (47), we found that the loss of Gcn5 did not lead to a selective loss of Bdf1 at promoter regions, thereby ruling out a SAGA-directed recruitment of Bdf1. Rather, there was a significant tendency for Bdf1 occupancy to increase upon the loss of Gcn5. Conceivably, SAGA (Gcn5) and Bdf1 might compete for occupancy at the same NuA4 acetylated sites. Indeed, SAGA binds to nucleosomal arrays acetylated at H4 by NuA4 in vitro (19), and Bdf1 binds to acetylated H4 tails (23, 31, 38, 42). The activation of SAGA-regulated genes is linked to the dissociation of Bdf1 from their promoter regions (57), which is consistent with binding competition between SAGA and Bdf1. Nevertheless, this possibility remains to be tested further.
Bdf1 and Bdf2 are partially redundant but are geographically distinct.
Genetic evidence suggests that Bdf1 and Bdf2 are functionally redundant (37). It is striking that the clustering patterns of ChIP-chip genomic data in Fig. 2 reveal instances of functional redundancy between these two factors but paradoxically also suggest geographically distinct functions. As inferred from Fig. 2, in a bdf1Δ strain, the promoter regions of clusters 2 and 3 are depleted of both Bdf1 and Bdf2, the latter as a result of the redistribution of Bdf2 from cluster 3 to cluster 1. With both Bdf1 and Bdf2 missing, these two clusters displayed the greatest loss of TFIID (Taf1) compared to other clusters. Cluster 1, on the other hand, which gained Bdf2 in the bdf1Δ strain, lost very little TFIID. These findings are consistent with the notion of functional redundancy.
Bdf1 and Bdf2 occupy distinct locations in the genome, with Bdf1 occupying regions of high H4 acetylation and Bdf2 occupying regions of low H4 acetylation. This acetylation difference may account for the proteins' distinct genomic distribution in that Bdf1, and not Bdf2, preferentially binds acetylated histone H4 tails (38). The observation that Bdf2 redistributes itself to sites vacated by Bdf1 in a bdf1Δ strain suggests that Bdf1 normally occludes Bdf2 binding at those sites. Interestingly, when H4 acetylation is lost upon the inactivation of NuA4, Bdf1 relocates from its bound genomic sites to sites where Bdf2 normally resides. Both of these observations suggest a degree of interchangeability between Bdf1 and Bdf2. If so, then some promoter regions have mechanisms that favor Bdf1 while mechanisms of other regions favor Bdf2. Regions bound by Bdf1 versus Bdf2 are occupied by distinct repertoires of transcriptional regulators, which could contribute to the differential occupancy of Bdf1 and Bdf2. How promoter specificity is achieved for the Bdf proteins remains to be determined.
Coordinated redistribution of transcription regulators.
The findings presented here provide genome-wide support for the model of PIC assembly presented in Fig. 1. This model appears to be general rather than selective for genes involved in a specific physiological process or function. The genomic data also reveal a substantial tailoring of this model in which factors such as Bdf1 and Bdf2 have differing influences, depending upon the promoter. Most yeast genes are expressed at low levels. These genes also have low levels of acetylation and generally lack assembled PICs, but they might nevertheless be permissive for PIC assembly. The loss of assembly factors elsewhere in the genome, based on mutagenesis experiments, appears to result in adventitious assembly at such transcriptionally quiescent locations. The redistribution appears to be highly coordinated with other assembly factors. Taken together, all of these findings clearly indicate that assembly and disassembly events are coordinated at individual loci as well as across the entire genome.
Supplementary Material
Acknowledgments
We thank J. Reese for many helpful suggestions and S. Zanton for technical assistance.
This work was supported by NIH grant GM059055.
Footnotes
Published ahead of print on 25 May 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albert, I., T. N. Mavrich, L. P. Tomsho, J. Qi, S. J. Zanton, S. C. Schuster, and B. F. Pugh. 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446:572-576. [DOI] [PubMed] [Google Scholar]
- 2.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babiarz, J. E., J. E. Halley, and J. Rine. 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20:700-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basehoar, A. D., S. J. Zanton, and B. F. Pugh. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699-709. [DOI] [PubMed] [Google Scholar]
- 5.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, and S. L. Schreiber. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99:8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 8.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 9.Chandy, M., J. L. Gutiérrez, P. Prochasson, and J. L. Workman. 2006. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot. Cell 5:1738-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke, A. S., J. E. Lowell, S. J. Jacobson, and L. Pillus. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19:2515-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 12.Dion, M. F., S. J. Altschuler, L. F. Wu, and O. J. Rando. 2005. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA 102:5501-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durant, M., and B. F. Pugh. 2006. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:2791-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garbett, K. A., M. K. Tripathi, B. Cencki, J. H. Layer, and P. A. Weil. 2007. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol. Cell. Biol. 27:297-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 17.Guillemette, B., A. R. Bataille, N. Gevry, M. Adam, M. Blanchette, F. Robert, and L. Gaudreau. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3:e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassan, A. H., S. Awad, and P. Prochasson. 2006. The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes. J. Biol. Chem. 281:18126-18134. [DOI] [PubMed] [Google Scholar]
- 19.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 20.Haynes, S. R., C. Dollard, F. Winston, S. Beck, J. Trowsdale, and I. B. Dawid. 1992. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 20:2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson, B. P., M. A. Martinez-Yamout, H. J. Dyson, and P. E. Wright. 2000. Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol. 304:355-370. [DOI] [PubMed] [Google Scholar]
- 22.Huisinga, K. L., and B. F. Pugh. 2004. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13:573-585. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAF(II)250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 24.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 25.Kasten, M., H. Szerlong, H. Erdjument-Bromage, P. Tempst, M. Werner, and B. R. Cairns. 2004. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 23:1348-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keogh, M. C., T. A. Mennella, C. Sawa, S. Berthelet, N. J. Krogan, A. Wolek, V. Podolny, L. R. Carpenter, J. F. Greenblatt, K. Baetz, and S. Buratowski. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20:660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 29.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 30.Kurdistani, S. K., S. Tavazoie, and M. Grunstein. 2004. Mapping global histone acetylation patterns to gene expression. Cell 117:721-733. [DOI] [PubMed] [Google Scholar]
- 31.Ladurner, A. G., C. Inouye, R. Jain, and R. Tjian. 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 11:365-376. [DOI] [PubMed] [Google Scholar]
- 32.Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen, C. Seidel, J. Gerton, and J. L. Workman. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 102:18385-18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieb, J. D., and N. D. Clarke. 2005. Control of transcription through intragenic patterns of nucleosome composition. Cell 123:1187-1190. [DOI] [PubMed] [Google Scholar]
- 34.Liu, C. L., T. Kaplan, M. Kim, S. Buratowski, S. L. Schreiber, N. Friedman, and O. J. Rando. 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning, E. T., T. Ikehara, T. Ito, J. T. Kadonaga, and W. L. Kraus. 2001. p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol. 21:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Campa, C., P. Politis, J. L. Moreau, N. Kent, J. Goodall, J. Mellor, and C. R. Goding. 2004. Precise nucleosome positioning and the TATA box dictate requirements for the histone H4 tail and the bromodomain factor Bdf1. Mol. Cell 15:69-81. [DOI] [PubMed] [Google Scholar]
- 37.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 38.Matangkasombut, O., and S. Buratowski. 2003. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell 11:353-363. [DOI] [PubMed] [Google Scholar]
- 39.Millar, C. B., F. Xu, K. Zhang, and M. Grunstein. 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20:711-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 41.Owen, D. J., P. Ornaghi, J. C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pamblanco, M., A. Poveda, R. Sendra, S. Rodriguez-Navarro, J. E. Perez-Ortin, and V. Tordera. 2001. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 496:31-35. [DOI] [PubMed] [Google Scholar]
- 43.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122:517-527. [DOI] [PubMed] [Google Scholar]
- 44.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 46.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 47.Robert, F., D. K. Pokholok, N. M. Hannett, N. J. Rinaldi, M. Chandy, A. Rolfe, J. L. Workman, D. K. Gifford, and R. A. Young. 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16:199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 49.Ruhl, D. D., J. Jin, Y. Cai, S. Swanson, L. Florens, M. P. Washburn, R. C. Conaway, J. W. Conaway, and J. C. Chrivia. 2006. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry 45:5671-5677. [DOI] [PubMed] [Google Scholar]
- 50.Sermwittayawong, D., and S. Tan. 2006. SAGA binds TBP via its Spt8 subunit in competition with DNA: implications for TBP recruitment. EMBO J. 25:3791-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma, V. M., R. S. Tomar, A. E. Dempsey, and J. C. Reese. 2007. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol. Cell. Biol. 27:3199-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, E. R., A. Eisen, W. Gu, M. Sattah, A. Pannuti, J. Zhou, R. G. Cook, J. C. Lucchesi, and C. D. Allis. 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95:3561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 55.Yang, X. J. 2004. Lysine acetylation and the bromodomain: a new partnership for signaling. Bioessays 26:1076-1087. [DOI] [PubMed] [Google Scholar]
- 56.Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu, S. J. Altschuler, and O. J. Rando. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309:626-630. [DOI] [PubMed] [Google Scholar]
- 57.Zanton, S. J., and B. F. Pugh. 2004. Changes in genomewide occupancy of core transcriptional regulators during heat stress. Proc. Natl. Acad. Sci. USA 101:16843-16848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zanton, S. J., and B. F. Pugh. 2006. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 20:2250-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, H., D. N. Roberts, and B. R. Cairns. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123:219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.