Abstract
The 90S preribosomal particle is required for the production of the 18S rRNA from a pre-rRNA precursor. Despite the identification of the protein components of this particle, its mechanism of assembly and structural design remain unknown. In this work, we have combined biochemical studies, proteomic techniques, and bioinformatic analyses to shed light into the rules of assembly of the yeast 90S preribosome. Our results indicate that several protein subcomplexes work as discrete assembly subunits that bind in defined steps to the 35S pre-rRNA. The assembly of the t-UTP subunit is an essential step for the engagement of at least five additional subunits in two separate, and mutually independent, assembling routes. One of these routes leads to the formation of an assembly intermediate composed of the U3 snoRNP, the Pwp2p/UTP-B, subunit and the Mpp10p complex. The other assembly route involves the stepwise binding of Rrp5p and the UTP-C subunit. We also report the use of a bioinformatic approach that provides a model for the topological arrangement of protein components within the fully assembled particle. Together, our data identify the mechanism of assembly of the 90S preribosome and offer novel information about its internal architecture.
The formation of eukaryotic ribosomes involves the production and correct assembly of four rRNAs and ≈80 ribosomal proteins. Due to its amenability for genetic and proteomic analyses, Saccharomyces cerevisiae is the organism where the different steps of this pathway have been best characterized (4, 11, 13, 18, 32, 34). Thus, it is known that three of the four mature rRNAs that form the ribosome structure are generated from a common 35S pre-rRNA polycystronic precursor. After being transcribed in the nucleolus, this precursor is chemically modified and cleaved at three positions (known as the A0, A1, and A2 sites) of its 5′-terminal end to generate the intermediate 33S, 32S, 27SA2, and 20S pre-rRNA precursors (see Fig. S1 in the supplemental material). The 20S and 27SA2 pre-rRNAs then follow two independent maturation routes that lead to the generation of either the 18S rRNA (a component of the 40S ribosomal subunit) or the 5.8S and 25S rRNAs (two components of the 60S ribosomal subunit), respectively (see Fig. S1 in the supplemental material). These pre-rRNA maturation steps require the involvement of ≈170 nonribosomal proteins and 70 small nucleolar ribonucleoproteins (snoRNPs) (4, 11, 13, 32). Different subsets of these molecules form large ribonucleoprotein complexes with specific pre-rRNA precursors that, according to their specific Svedberg coefficients in gradient ultracentrifugation experiments, were initially referred to as 90S, 66S, and 43S preribosomal particles (31, 33). The 90S particle, also known as the “small-subunit processome,” contains the 35S pre-rRNA and assembly/processing factors needed for the early cleavage of the 35S pre-rRNA precursor at A0, A1, and A2 sites, which is strictly required for the production of 40S ribosomal subunits. The 66S and 43S particles contain 35S pre-rRNA cleavage derivatives and proteins that mediate the downstream maturation steps of rRNA biosynthesis. Recent studies have shed light on the complex structure of preribosomal particles. In the case of the 90S preribosome, the combination of several biochemically, genetically, and proteomically based studies has shown that it contains the 35S pre-rRNA, the U3 small nucleolar RNA (snoRNA), and ≈50 nonribosomal proteins. These include enzymes that participate in RNA-processing functions and a large cohort of proteins containing protein-protein and RNA-binding motifs but lacking known catalytic domains (3, 7, 13, 17, 22, 29, 34, 36).
The mechanism by which all these proteins are assembled together to form the final 90S particle is unknown. However, available evidence suggests that yeast cells might minimize the complexity of this process by preassembling 90S particle components in smaller building blocks prior to the binding to the 35S pre-rRNA. In favor of this model, electron microscopy studies have revealed that early preribosomal particles undergo time-dependent changes in size and shape upon binding to the primary pre-rRNA precursor, suggesting that their components are sequentially assembled (26). Other studies have revealed the presence of discrete 90S particle subcomplexes, such as the U3 snoRNA-containing U3 snoRNP, the Mpp10p complex, and the Pwp2p subunit. Some of these subcomplexes seem to represent true 90S preribosomal subunits, because they are detected as stable biochemical entities independently of the 90S particle (6, 17, 19, 36, 37). Recent high-throughput proteomic studies have also reported the presence in yeast cells of three small 90S particle subcomplexes that were designated U three protein (UTP)-A, UTP-B, and UTP-C (22). It is believed that these subcomplexes might represent stable preassembly or postassembly 90S modules because they are detected in cell lysates depleted of 90S particles by an ultracentrifugation step (22). This hypothesis is further supported by the observation that the UTP-B subcomplex is identical in protein composition to the Pwp2p subunit (and, as such, it will be referred to as the Pwp2p/UTP-B subunit hereafter). Finally, it has also been shown that t-UTP, an UTP-A-like subset of 90S preribosomal proteins, influences 35S rDNA transcription independently of other particle proteins (14).
In addition to the number of subunits, there are other questions related to 90S particle assembly that remain unanswered, such as those pertaining the order of incorporation of components onto the 35S pre-rRNA and the interconnections established by them, both during and after the formation of the particle. Given the large number of molecules and protein-protein interactions involved, it is likely that the only way to obtain a comprehensive view of the 90S particle architecture and assembly process is by studying the reciprocal connections among multiple components using different experimental techniques. In this work, we have used this strategy to delve in the 90S particle substructure and the mechanism of its assembly. This experimental avenue allowed us to characterize discrete 90S particle subunits, to dissect the hierarchy of assembly of those subunits onto the 35S pre-rRNA, to identify several assembly intermediaries of the 90S particle, and to gain clues for deciphering the internal 90S particle architecture.
MATERIALS AND METHODS
Yeast strains and genetic methods.
Strains with conditional mutations in genes under the control of the GAL1 promoter were generated by the one-step PCR strategy. This rendered in-frame fusions of a KANMX6-GAL1-HA cassette upstream of the ATG of the corresponding gene. The one-step integration of PCR cassettes was also used to generate yeast strains with MYC carboxyl-terminally tagged alleles. The genotypes and names of these yeast strains are indicated in Table S1 in the supplemental material. Yeast cells were cultured at 30°C in either galactose (YP-Gal [0.4% yeast extract, 0.8% peptone, 0.1 mM adenine, and 2% galactose])- or glucose (YPD) (0.4% yeast extract, 0.8% peptone, 0.1 mM adenine, and 2% glucose)-containing medium to allow the overexpression and repression of the appropriate hemagglutinin (HA)-tagged protein, respectively. The incubation times in glucose were those determined as optimal for the complete depletion of the conditionally expressed protein in each strain: 18 h in glucose for GAL::HA-pwp2 cells, 16 h in glucose for GAL::HA-rrp7 and GAL::HA-nan1 cells, and 14 h in glucose for GAL::HA-rrp5 cells.
Sucrose gradient analysis.
Polysome analysis and fractionation of lysates through 7 to 50% sucrose gradients were performed as described previously (6). Extracts equivalent to 15 absorption units at the 260-nm wavelength were layered onto each gradient. After centrifugation, fractions were collected and subjected to either Western or Northern blot analyses.
Coimmunoprecipitation experiments.
Cell cultures were grown to an A600 of between 0.8 and 1.0. Cells equivalent to 20 A600 units were harvested and disrupted using glass beads (Sigma) in IP2 buffer (20 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 150 mM potassium acetate, 1 mM dithiothreitol, and 0.2% Triton X-100) supplemented with both a vanadyl ribonucleoside complex (5 mM; New England Biolabs) and a mixture of protease inhibitors (Cφmplete; Roche Applied Science). Precleared lysates were incubated with anti-MYC antibodies (Roche Applied Science) at 4°C for 2 h under gentle rotation in the presence of RNasin (400 U/ml; Promega). After incubation with Gammabind Sepharose beads (GE Healthcare), immunoprecipitates were washed five times with IP2 buffer at 4°C. For protein analyses, half of the immunoprecipitated material was resuspended in 80 μl of sodium dodecyl sulfate (SDS) loading buffer, boiled, fractionated on 8% acrylamide gels, and analyzed by Western blotting. For RNA analyses, the other half of the sample was resuspended in 400 μl of 50 mM sodium acetate, 10 mM EDTA (pH 5.2), and 1% SDS and processed for RNA extraction.
RNA analysis.
RNAs from total cellular lysates, gradient fractions or coimmunoprecipitations were prepared by the hot-phenol method (1). After ethanol precipitation, RNAs were resuspended in either formaldehyde loading buffer (to detect high-molecular-weight RNAs) or acrylamide-urea loading buffer (to detect snoRNAs), separated electrophoretically on either 1.2% agarose-formaldehyde (in the case of high-molecular-weight pre-rRNA precursors) or 8% acrylamide-8 M urea (in the case of snoRNAs) gels, and analyzed by Northern blotting. We used the following 32P-labeled oligonucleotides to detect the 35S pre-rRNA precursor and other downstream pre-rRNA intermediaries: probe 4 (region D-A2), 5′-TTAAGCGCAGGCCCGGCT-3′; probe 5 (region A2-A3), 5′-TGTTACCTCTGGGCC-3′; and probe 8 (region E-C2), 5′-GGCCAGCA ATTTCAAGTTA-3′. These regions of the 35S pre-rRNA molecule are depicted in Fig. S1 in the supplemental material. The U3 snoRNA was detected using a labeled 5′-GGATTGCGGACCAAGCTAA-3′ probe. Oligonucleotide labeling, Northern blotting, and filter hybridizations were performed as previously described (6).
Mass spectrometry analysis.
Purifications of Pwp2p-MYC-, Rrp7p-MYC, Nan1p-MYC-, and Utp4p-MYC-containing complexes in conditional mutant strains were performed using a large-scale anti-MYC coimmunoprecipitation approach, exactly as indicated before (6). All protein bands observed after staining of the gels with Sypro Ruby (Molecular Probes) were processed and analyzed for their identification by mass spectrometry in the Genomics and Proteomics Unit of the Centro de Investigación del Cáncer of Salamanca, using an Ultraflex matrix-assisted laser desorption ionization-time-of-flight apparatus (Bruker). Complexes were purified in several independent experiments, and the patterns of proteins associated with each bait protein, in each experimental condition, were found to be the same every time. A protein was considered positively associated with a given bait protein when it was reproducibly detected as a strongly stained band in the corresponding gels of the immunoprecipitations performed with that bait. If so, it was included in Fig. 5 below. A protein was considered “not present” in the bait immunocomplexes when one of the following occurred: (i) it was not detected in the Sypro Ruby-stained SDS-polyacrylamide gels, (ii) the electrophoretic band identified by mass spectrometry did not fit the mass spectrometer fingerprint of the missing protein (these two cases occurred in ≈81% of the “not-present” proteins reported in Fig. 5), or (iii) the relative amount of that protein was drastically reduced in the bait immunoprecipitates compared to the levels of coimmunoprecipitation detected for other bait's partners (this occurred in ≈19% of the negative cases reported in this work). In addition to those cases, we also detected some strongly stained protein bands that could not be identified by mass spectrometry (6% of all stained bands analyzed). Due to their abundance and molecular size, we regarded those proteins as mixtures of degradation products or denatured immunoglobulin G chains released from the anti-MYC beads after the boiling step. These are nonidentifiable bands because the default parameters set up for the mass spectrometer protein identification software filters them out automatically. Finally, we identified proteins by mass spectrometry that had reported functions totally unrelated to ribosomal biogenesis. These proteins, representing ≈14% of all stained bands, always showed up associated to the chosen bait regardless of the experimental context and/or genetic and culture conditions used, suggesting that they likely represent contaminants carried over by the immunocomplexes. The identities of these bands will be provided upon request.
FIG. 5.
Proteomic analysis of partially assembled subunits of the 90S preribosomal particle. The MYC-tagged bait proteins used for the immunopurifications are indicated in the first row from the top. The proteins that have been depleted in the lysates used for the immunopurifications are indicated in the second row from top. The prey proteins identified in these analyses, their open reading frame names, and the type of UTP subcomplex they belong to are indicated in the second, third, and first columns on the left, respectively. Shaded areas indicate a positive association between the appropriate bait and 90S particle proteins.
Bioinformatics.
Several computational methods were applied to generate a high-confidence interaction network for the 32 preribosomal proteins selected for the study. As main data source, we used the Agile Protein Interaction Data Analyzer (APID) (http://bioinfow.dep.usal.es/apid), a web-based bioinformatic tool that combines information from five independent protein interaction databases (BIND, DIP, HPDR, IntAct, and MINT) (27). In addition, we included in our analyses the information derived from the present work and two additional proteomic data sets that had not been included initially in APID (21, 22). The combined information was taken to set up a pairwise interactomic table (see Fig. S6 in the supplemental material). A novel quantitative method (see below) was then used to calculate the “interactomic profile” of each protein within a protein-protein interaction data set. Based on these profiles, the degree of association among the proteins under analysis was calculated and protein subcomplexes predicted. To this end, we first generated a symmetric matrix (Mnn) whose rows and columns are the n proteins of interest. Each element in the matrix (Aij) is given a value corresponding to the number of published experiments that have demonstrated the pairwise interaction between protein i and protein j. According to this, the matrix for n proteins is a set of n quantitative vectors Vi = [(Ai·)n] that provides a “quantitative rated protein interaction profile” specific for each analyzed protein (protein i) that is derived from the complete protein interaction network under study. These profiles are much more informative than simple presence-absence profile matrices, which are based only on 1 and 0 numbers (5, 16). For the Aii elements (i.e., values corresponding to the interaction of a protein i with itself) an estimated total interactomic score of 10 was given, which is higher than the maximum observed Aij (in our case, 8). Using all the Aij vectors, a distance matrix (Dnn) was then generated that gave quantitative values to the interaction strength existing for each protein-protein pair (i.e., protein i versus protein j) of our protein interaction landscape. In this matrix, each value for a given protein pair (i,j) was calculated as the “anticorrelation” (or complement of the correlation coefficient) among each pair of protein vectors Vi = [(Ai·)n] and Vj = [(Aj·)n] using the function Acij = 1 − r(Vi,Vj), with r being the Pearson correlation coefficient. Since r has a range of −1 to +1 (with +1 representing an absolute direct correlation, −1 an absolute reverse correlation, and 0 an absolute noncorrelation), ACij distributes between a (0, 2) interval (where 0 represents a minimal distance and maximum direct correlation, whereas 2 reflects a maximal distance and maximum “anticorrelation” coefficient). Finally, the distance matrix (Dnn) was analyzed using the unweighted pair group method with arithmetic mean (UPGMA) algorithm to generate protein clusters according to the distances calculated by the quantitative rated protein interaction profile. To assess the stability of each of the branches generated in the hierarchical tree, we utilized a bootstrap method (8). This analysis was carried out by substituting a percentage of the observations (that is, the columns of the initial matrix Mnn) for other randomly selected columns. The percentage applied each time was a 10%, and this was done 1,000 times, generating the corresponding trees and branches. The ratio of presence of a specific branch in the bootstrapped trees is a measure of the confidence about the actual presence of that branch in the original data. With the exception of the UPGMA algorithm, which was applied using the Phylip package (12), all the described bioinformatic steps were carried out using R (http://www.r-project.org/).
RESULTS
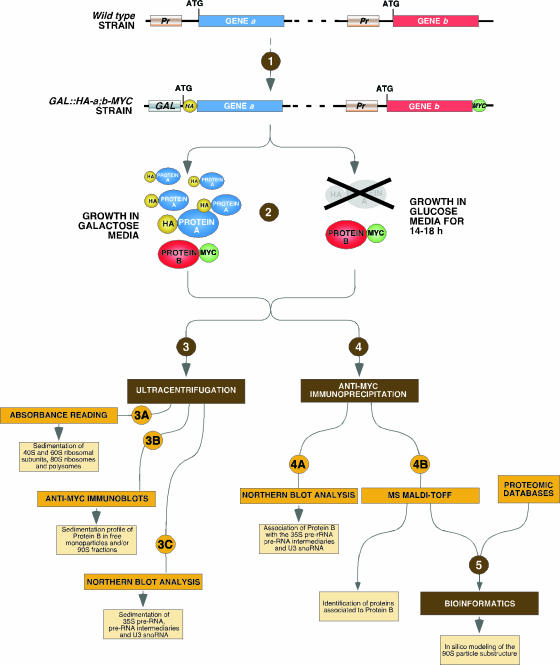
The original basis of this work stemmed from the hypothesis that the 90S preribosomal particle was composed of several subunits that followed specific rules of assembly either prior to or during the binding of the 35S pre-rRNA precursor. If that hypothesis was correct, we surmised that we could identify the subunits and their order of assembly onto the 35S pre-rRNA precursor by eliminating the expression of certain subunit components and monitoring the binding of the rest of the putative subunits to the pre-rRNA molecules. Since the 90S preribosomal substructure is not yet well characterized, we decided to analyze Nan1p, Rrp7p, and Pwp2p, three 90S preribosomal proteins assigned by previous proteomic experiments to the UTP-A/t-UTP, UTP-C, and Pwp2p/UTP-B subcomplexes, respectively (6, 17, 22). These subcomplexes were considered good candidates for being 90S particle subunits (or fragments of larger subunits) because (i) They have been detected as discrete proteomic subcomplexes in 90S preribosome-depleted yeast extracts (6, 22); (ii) some of them are found bound to the 5′ end of the pre-rRNA in the absence of the 90S particle, suggesting that they are present during the assembly or after the disassembly of the 90S preribosome (6, 22); and (iii) one of them (t-UTP) also has functions unrelated to 35S pre-rRNA cleavage (14). To conduct a comprehensive analysis of the assembly behavior of each of the selected proteins in the presence or absence of the other two, these proteins were subjected to a multistep study that combined sucrose gradient fractionation, immunoprecipitation experiments coupled to Western blot and Northern blot analyses, and, finally, mass spectrometry techniques. Figure 1 shows a schematic representation of our experimental strategy, outlining the techniques employed and the aims pursued in each one of the steps involved. First, we generated yeast strains expressing two types of epitope-tagged proteins (Fig. 1, step 1): in the case of the proteins to be depleted, their loci were modified by homologous recombination techniques to encode HA-tagged versions of the proteins under the regulation of the inducible GAL1 promoter. This strategy allowed us to induce either the overexpression or repression of the selected HA-tagged protein under study by culturing the yeast strains in the presence of galactose or glucose, respectively. To monitor the behavior of other 90S particle factors in the conditional mutant strains, the corresponding endogenous loci were modified to express C-terminally MYC-tagged versions of the proteins under their cognate promoters. Once generated, these cells were maintained in the presence of galactose and, when appropriate, shifted to glucose-containing medium for 14 to 18 h to monitor the behavior of the MYC-tagged protein in the presence or absence of the HA-tagged protein (Fig. 1, step 2). As a control for the effect of the overexpression of the HA-tagged protein, we also used yeast strains expressing the MYC-tagged protein of interest with a nonmodified locus for the other protein under study (not indicated in Fig. 1). Lysates from each experimental condition were then analyzed using a battery of complementary techniques. On the one hand (Fig. 1, step 3), lysates were subjected to sucrose gradient fractionation experiments to determine the following parameters: (i) The generation of 40S and 60S ribosomal subunits, of 80S ribosomes, and polysomes by the continuous reading of the A254 of the collected gradient fractions after the ultracentrifugation step (step 3A); (ii) the sedimentation pattern of the MYC-tagged protein in the presence or absence of the selected HA-tagged protein by analyzing aliquots of the gradient fractions by anti-MYC immunoblotting (step 3B); and (iii) the production and/or sedimentation profiles of different pre-rRNA precursors and the U3 snoRNA using Northern blot analysis with appropriate 32P-labeled oligonucleotide probes (step 3C). On the other hand (Fig. 1, step 4), lysates were subjected to analytical (step 4A) or preparative (step 4B) anti-MYC immunoprecipitations in order to detect (i) the association of the 35S pre-rRNA and the indicated downstream precursors with the MYC-tagged protein under study (step 4A), (ii) the binding of the U3 snoRNA to the MYC-tagged protein under analysis (step 4A), and (iii) the proteins and/or protein complexes associated with the MYC-tagged protein using mass spectrometry techniques (step 4B). By repeating these experiments with all the possible combinations of the proteins under study (Nan1p, Pwp2p, Rrp7p, and, subsequently, Rrp5p), this experimental strategy allowed us to obtain a comprehensive view of the behavior of these proteins in terms of association in free monoparticles, the order of the assembly of each protein onto the 35S pre-rRNA and the 90S preribosomal particle, and its participation in the formation of partially assembled intermediaries of the 90S particle. In addition, the data obtained using proteomic techniques were used subsequently, in combination with other previously described protein-protein interaction data available in public databases, to set up an in silico approach useful for shedding light on the 90S particle substructure (Fig. 1, step 5).
FIG. 1.
Schematic depiction of the experimental strategy used in this work. The experimental basis of this study relied on the analysis of how depletion of specific markers for putative 90S preribosome subunits affected the assembly of other 90S preribosomal particle proteins onto the 35S pre-rRNA. A description of the steps outlined in this figure, including the techniques employed and the aims pursued, is given in the text. The results corresponding to the experiments of each step are shown in Fig. 2 to 7 as follows. (i) Polysome profiles (step 3A) are shown in the upper panels of Fig. 2A and C, 3A and C, 4A and C, and 6A and Fig. S2, S3, and S5 in the supplemental material. (ii) The sedimentation patterns of MYC-tagged proteins (step 3B) are shown in the second panels (from the top) of Fig. 2A and C, 3A and C, 4A and C, and 6A and Fig. S2, S3, and S5 in the supplemental material. (iii) The sedimentation of pre-rRNA precursors and the U3 snoRNA (step 3C) are shown in the third (in the cases of the 35S and 27S pre-rRNA precursors), fourth (in the cases of the 35S, 27SA2, and 23S pre-rRNA precursors), and fifth (in the cases of the U3 snoRNA) panels (from the top) of Fig. 2A and C, 3A and C, 4A and C, and 6A and Fig. S2, S3, and S5 in the supplemental material. (iv) The results of coimmunoprecipitation experiments (step 4A) are shown in Fig. 2B and D, 3B and D, and 4B and D. (v) Proteins associated with MYC-tagged proteins identified by mass spectrometry (step 4B) are shown in Fig. 5 and 6B to D and Fig. S4 in the supplemental material. vi) The network of pairwise interactions and the hierarchical clustering of 90S preribosome components generated by bioinformatic analysis are shown in Fig. 7A and B. GAL, GAL1 promoter; Pr, endogenous promoter; MS, mass spectrometry.
Independent assembly of UTP-C and Pwp2p/UTP-B components on the 35S pre-rRNA.
We initiated this study by testing the mutual dependency of UTP-C and Pwp2p/UTP-B markers for their docking onto the 35S pre-rRNA. To this end, we tracked the behavior of Rrp7p-MYC, an UTP-C protein, in the presence or absence of the Pwp2p/UTP-B-specific Pwp2p. As previously described (6), sucrose gradient fractionation analyses showed that cells show normal levels of the mature ribosomal 40S and 60S subunits, of 80S ribosomes, and of polysomes under conditions of HA-Pwp2p overproduction (Fig. 2A). In addition, we observed normal profiles in the sedimentation of the 35S pre-rRNA, other maturation intermediates (27S and 23S pre-rRNAs), and the U3 snoRNA (Fig. 2A). Normal profiles for all these components have been observed in the other conditional mutant yeast strains used in this work under permissive conditions (see below, Fig. 2 to 4 and 6), indicating that the overexpression and epitope-tagging strategies used in our experimental approach did not alter the functionality of the proteins under study. Under the same conditions, we observed that Rrp7p-MYC showed a biphasic distribution in the gradients, being detected in small ≈15 to 20S sedimentation monoparticles and in an ≈90S complex (Fig. 2A). The 90S-associated Rrp7p-MYC fraction cofractionated with the 35S pre-rRNA and other 90S particle components such as the U3 snoRNA (Fig. 2A). A similar distribution of Rrp7p-MYC was observed in cells expressing wild-type levels of Pwp2p (see Fig. S2A in the supplemental material), indicating that this bimodal sedimentation profile is not due to HA-Pwp2p overexpression. The elimination of Pwp2p expression led to the expected blockage in the processing of the 35S pre-RNA, as evidenced by the accumulation of the primary transcript of the precursor (Fig. 2A and B), the disappearance of the transitional 27SA2 RNA generated upon cleavage of the 35S pre-rRNA at the A2 site (Fig. 2A; see Fig. S1 in the supplemental material), the concentration of the U3 snoRNA in small-sedimentation-coefficient fractions (Fig. 2A), and the lack of 40S ribosomal subunits (Fig. 2A). Despite the dramatic effects of the Pwp2p depletion on ribosomal biosynthesis, we observed that the sedimentation profile of Rrp7p-MYC was very similar to that observed in both wild-type and Pwp2p-overexpressing cells (compare Fig. 2A and Fig. S2A in the supplemental material), suggesting that the Rrp7 subunit could bind the 35S pre-rRNA in the absence of Pwp2p and the U3 snoRNP. In agreement with this, coimmunoprecipitation experiments showed that Rrp7p associated with the 35S pre-rRNA independently of Pwp2p (Fig. 2B). In contrast, and consistent with the accumulation of the U3 snoRNA in free monoparticles in Pwp2p-depleted cells (Fig. 2A), the detection of the U3 snoRNA in the Rrp7p immunoprecipitates was lost upon Pwp2p elimination (Fig. 2B). Control experiments confirmed the specificity of these RNA-protein interactions (Fig. 2B).
FIG. 2.
Independent association of Pwp2p- and Rrp7p-containing complexes with the 35S pre-rRNA. (A and C) Cellular extracts from the indicated yeast strains (top) grown in either galactose (left panels)- or glucose (right panels)-containing medium were subjected to sucrose gradient sedimentation analysis. After the ultracentrifugation step, fractions were automatically collected and polysome profiles recorded by continuous reading of the A254 (upper panels). The distribution of Rrp7p-MYC (A) or Pwp2p-MYC (C) in aliquots of the collected fractions was analyzed by anti-MYC immunoblotting (second panels from the top). In parallel, total RNAs were obtained from aliquots of the collected fractions and subjected to Northern blot analysis with 32P-labeled oligonucleotide probes to the 35S pre-rRNA EC2 (third panels from top) and A2-A3 (fourth panels from top) regions or to the U3 snoRNA (bottom panels) (see Materials and Methods and Fig. S1 in the supplemental material). The number of each gradient fraction is indicated at the bottom of the first and fifth panels from top. Gal, galactose; Glu, glucose; WB, Western blot; NB, Northern blot. (B and D) Total cellular lysates (lanes 1 to 4) and anti-MYC immunoprecipitates (lanes 5 to 8) obtained for the indicated yeast strains and growth conditions (top) were analyzed by anti-MYC immunoblotting (upper panels), anti-HA immunoblotting (second panels from top), and Northern blot analyses using 32P-labeled oligonucleotide probes to the 35S pre-rRNA DA2 region (third panels from the top; see Fig. S1 in the supplemental material) and the U3 snoRNA (bottom panel). TCL, total cellular lysates; IP, immunoprecipitation.
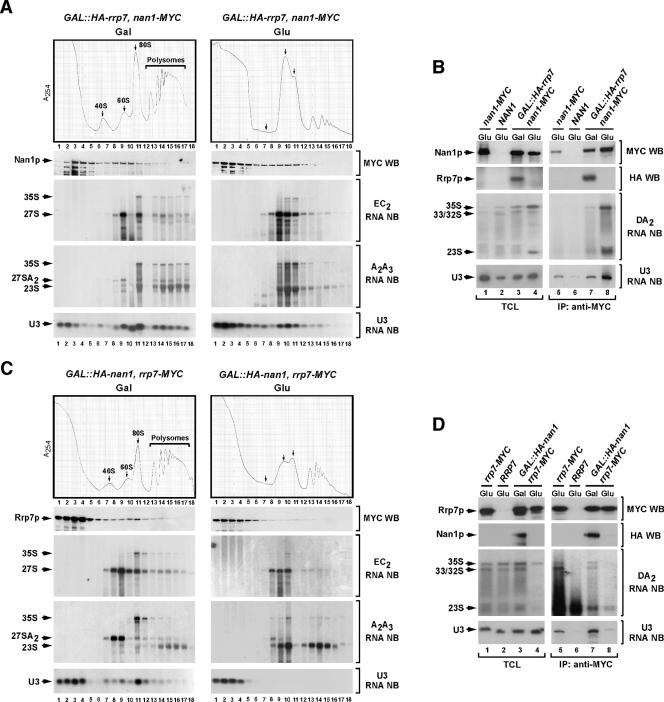
FIG. 4.
Asymmetrical dependency of Nan1p- and Rrp7p-containing complexes for their assembly onto the 35S pre-rRNA. (A and C) Cellular extracts from cultures of the indicated yeast strains (top) grown in either galactose (left panels)- or glucose (right panels)-containing medium were subjected to sucrose gradient sedimentation analysis. The distribution of Nan1p-MYC (A) and Rrp7p-MYC (C) in aliquots of the collected fractions was analyzed by anti-MYC immunoblotting (second panels from the top). In parallel, the sedimentation profiles of ribosomal subunits (top panels), pre-rRNA precursor molecules (third and fourth panels from top), and the U3 snoRNA (bottom panels) were analyzed in aliquots of the same gradient fractions, as indicated in Fig. 2. WB, Western blot; NB, Northern blot. (B and D) RNAs and proteins from either total cellular lysates (lanes 1 to 4) or anti-MYC immunoprecipitates (lanes 5 to 8) obtained for the indicated yeast strains and growth conditions (top) were probed with anti-MYC (upper panels) and anti-HA (second panels from the top) antibodies or 32P-labeled oligonucleotides hybridizing to sequences within the 35S pre-RNA DA2 region (third panels from the top; see Fig. S1 in the supplemental material) and the U3 snoRNA (bottom panel). TCL, total cellular lysates; IP, immunoprecipitation.
FIG. 6.
Rrp5p is required for the association of Rrp7p, but not of Pwp2p-, Nan1p-, or U3 snoRNP-containing complexes, with the 35S pre-rRNA precursor. (A) Cellular extracts from the indicated yeast strain (top) grown in either galactose (left panels)- or glucose (right panels)-containing medium were subjected to sucrose gradient sedimentation analysis. The distribution of Rrp7p-MYC in the gradient fractions was analyzed using anti-MYC immunoblots (second panels from the top). In parallel, the sedimentation profiles of ribosomal subunits (top panels), pre-rRNA precursor molecules (third and fourth panels from top), and the U3 snoRNA (bottom panels) were analyzed in aliquots of the gradient fractions, as indicated in Fig. 2. The distinctive accumulation of an aberrant 12S′ pre-rRNA species is observed in Rrp5-depleted cells, as previously described (35). WB, Western blot; NB, Northern blot. (B to D) Identification by mass spectrometry of molecules associated with the indicated MYC-tagged bait proteins (top) purified from yeast cells lacking Rrp5p. Large-scale extracts from yeast cells that had been depleted of Rrp5p were incubated with anti-MYC antibodies covalently bound to a solid matrix. After incubation and washes, the proteins bound to the MYC-tagged bait were released from the matrix by boiling in SDS-polyacrylamide gel electrophoresis sample buffer, fractionated electrophoretically, stained with Sypro Ruby, and identified by mass spectrometry. The identified proteins are indicated on the left of the original gel used for their purification. The proteins belonging to the bait's subunit are indicated in blue. The migration of molecular mass markers is indicated on the right.
We next performed the reciprocal experiments in order to assess whether the assembly of Pwp2p and the U3 snoRNA onto the pre-rRNA precursor was Rrp7p dependent. Analysis of yeast extracts by use of sucrose gradients showed wild-type sedimentation profiles for Pwp2p-MYC and the U3 snoRNA regardless of the presence or absence of Rrp7p (compare Fig. 2C and Fig. S2B in the supplemental material). Accordingly, the majority of both components was detected in gradient fractions that also contained the 35S pre-rRNA (Fig. 2C; see Fig. S2B in the supplemental material). Consistent with these results, we observed that Pwp2p-MYC coimmunoprecipitated with the 35S and 23S pre-rRNA molecules in an Rrp7p-independent manner (Fig. 2D). Control experiments confirmed the specificity of these interactions (Fig. 2D). The lack of effect of Rrp7p on the binding of Pwp2p and the U3 snoRNP to the 35S pre-rRNA was not due to deficient repression of Rrp7p, because these cells showed the expected defects in the production of 40S ribosomal subunits and in the cleavage of the 35S pre-rRNA precursor at the A2 site (2) (Fig. 2C and D). This cleavage defect was demonstrated by the accumulation of the 35S pre-rRNA, the enrichment in the 23S RNA species, and in the normal generation of 33/32S RNA intermediates (product of cleavages of the precursor at both A0 and A1 sites [see Fig. S1 in the supplemental material]) (Fig. 2D). These results are consistent with the idea that Pwp2p and Rrp7p are part of preformed 90S particle modules that dock independently onto the pre-rRNA precursor.
The UTP-A/t-UTP component Nan1p is required for the association of the Pwp2p/UTP-B element Pwp2p with the 35S pre-rRNA but not vice versa.
We next analyzed the relationship between proteins of the UTP-A/t-UTP and Pwp2p/UTP-B subcomplexes for their docking onto the primary pre-rRNA precursor. We observed that Nan1p-MYC, a component of the UTP-A/t-UTP subcomplex, cofractionated almost exclusively with the 35S pre-rRNA and large U3 snoRNA-containing complexes under conditions of Pwp2p overexpression (Fig. 3A). In contrast, no low-sedimentation-coefficient monoparticles containing Nan1p were observed. This is probably due to the overexpression of HA-Pwp2p, because Nan1p-MYC is detected in ≈15 to 20S monoparticles in cells expressing wild-type levels of Pwp2p (see Fig. S2C in the supplemental material). Under conditions of Pwp2p depletion, Nan1p-MYC showed a sedimentation profile similar to that observed in wild-type cells (Fig. 3A). Parallel analyses confirmed that the biosynthesis of 40S ribosomal subunits (Fig. 3A) and the processing of the 35S pre-rRNA (Fig. 3B) were blocked upon Pwp2p elimination. We could not confirm in this case the physical association between Nan1p and the 35S pre-rRNA in the absence of Pwp2p, because Nan1p immunoprecipitates were consistently inefficient in pulling down that precursor (Fig. 3B, see also Fig. 4B below). This may be a reflection of either a rather feeble/indirect interaction or, alternatively, a conformation of the complex that does not protect the 35S pre-RNA from degradation during the immunoprecipitation step. However, we do not consider that this could be an artifactual problem derived from the inclusion of the C-terminal MYC epitope, because cells expressing this protein showed normal cleavage of the 35S pre-rRNA at A0-A2 sites and optimal levels of 40S ribosomal subunits (Fig. 3A; see Fig. 4A and see Fig. S2C in the supplemental material). Despite the lack of confirmation by coimmunoprecipitation, the sedimentation analysis of Nan1p indicates that its binding to the 35S pre-rRNA occurs independently of both Pwp2p and the U3 snoRNA. This is consistent with previous results indicating that t-UTP proteins can bind the primary pre-rRNA precursor in a U3 snoRNA-independent manner (14).
FIG. 3.
A Nan1p-containing complex is required for the interaction of Pwp2p with the 35S pre-rRNA but not vice versa. (A and C) Cellular extracts from cultures of the indicated yeast strains (top) grown in either galactose (left panels)- or glucose (right panels)-containing medium were subjected to sucrose gradient sedimentation analysis. The distribution of Nan1p-MYC (A) and Pwp2p-MYC (C) in aliquots of the collected fractions was visualized by anti-MYC immunoblotting (second panels from the top). In parallel, the sedimentation profiles of ribosomal subunits (top panels), pre-rRNA precursor molecules (third and fourth panels from top), and the U3 snoRNA (bottom panels) were analyzed in aliquots of each of the gradient fractions, as indicated in Fig. 2. WB, Western blot; NB, Northern blot. (B and D) RNAs and proteins from either total cellular lysates (lanes 1 to 4) or anti-MYC immunoprecipitates (lanes 5 to 8) obtained for the indicated yeast strains and growth conditions (top) were analyzed by Western blotting using anti-MYC (upper panels) and anti-HA (second panels from top) antibodies or by Northern blot analyses using 32P-labeled oligonucleotide probes to the 35S pre-rRNA DA2 region (third panels from the top; see Fig. S1 in the supplemental material) and the U3 snoRNA (bottom panel). TCL, total cellular lysates; IP, immunoprecipitation.
By contrast, we found that the association of Pwp2p and the U3 snoRNA with the 35S pre-rRNA is Nan1p dependent, because those molecules were no longer detectable in 90S fractions and concentrated in fractions corresponding to free Pwp2p/UTP-B subunits and free U3 snoRNA complexes upon Nan1p depletion (Fig. 3C). Likewise, we observed that the association of Pwp2p-MYC with the 35S pre-rRNA, other rRNA precursors, and the U3 snoRNA was Nan1p dependent (Fig. 3D). Since Nan1p has been shown to be involved in rDNA gene transcription (14), the above results could be attributed to the physical absence of the 35S pre-rRNA precursor in the lysates. Prima facie, we considered this possibility unlikely, because Northern blot analyses indicated that Nan1p-deficient cells still had detectable levels of the pre-rRNA precursor (Fig. 3D) as well as significant levels of 60S ribosomal subunits (Fig. 3C). Notwithstanding these observations, we decided to rule out this possibility more firmly by analyzing under the same conditions the sedimentation profile of Nop7p, a protein involved in the independent processing branch of the 35S pre-rRNA that leads to the production of 60S ribosomal subunits (25) (see Fig. S1 in the supplemental material). We reasoned that if the shifts in the sedimentation of Pwp2p and the U3 snoRNA were due to low levels of rDNA transcription, Nop7p had to show a Pwp2p/U3 snoRNA-like change in its sedimentation pattern in the absence of Nan1p. However, that was not the case, as large preribosomal particles containing Nop7 were readily detectable in Nan1p-depleted cells (see Fig. S3 in the supplemental material). Taken together, these results indicate that the expression of the UTP-A/t-UTP component Nan1p is a conditio sine qua non for the association of Pwp2p and the U3 snoRNA with the 35S pre-rRNA.
The UTP-A/t-UTP component Nan1p is required for binding of Rrp7p to the 35S pre-rRNA but not vice versa.
To finish the initial part of this work, we evaluated the assembly relationships established between UTP-A/t-UTP and UTP-C components. The repression of the UTP-C-specific Rrp7p resulted in no substantial modifications in the sedimentation profile of Nan1p-MYC (Fig. 4A). In addition, and consistent with our previous results (Fig. 2C), we observed that the U3 snoRNA was also detected in 90S sedimentation fractions, indicating that the binding of the U3 snoRNP to the 35S pre-rRNA is Rrp7p independent (Fig. 4A). To further confirm these results, we assessed the physical association of Nan1p-MYC with specific RNA molecules of the 40S ribosomal subunit biosynthetic route. Unlike the results obtained under conditions of Rrp7 expression (Fig. 3B and 4B), we did find in this case significant amounts of both 35S and 23S pre-rRNAs bound to Nan1p when Rrp7p was absent from cells (Fig. 4B). These results confirm that Nan1p assembles onto the 35S pre-rRNA in an Rrp7-independent manner and, at the same time, suggest that Rrp7p conditions the type of structure established between the primary pre-rRNA precursor and some of its associated protein complexes. The cleavage of the 35S pre-rRNA at the A2 site (Fig. 4B) and the biosynthesis of the mature 40S ribosomal subunit (Fig. 4A) were both impaired under conditions of Rrp7p depletion, indicating that the presence of Nan1p protein in 90S fractions in the absence of Rrp7p was not due to the inefficient elimination of that protein. In contrast to the above results, we found in reciprocal experiments that the depletion of Nan1p led to the accumulation of Rrp7p in monoparticles with small sedimentation coefficients and to a lack of association of Rrp7p with both the 35S pre-rRNA and the U3 snoRNA (Fig. 4C and D).
Taken together, these results indicate that individual constituents of the UTP-A/t-UTP, Pwp2p/UTP-B, and UTP-C subcomplexes associate with the 35S pre-rRNA in three distinct assembly steps. Nan1p, a component of the UTP-A/t-UTP subcomplex, interacts with the 35S pre-rRNA independently of Pwp2p and Rrp7p, two proteins belonging to the Pwp2p/UTP-B and UTP-C subcomplexes, respectively. In addition, Pwp2p and Rrp7p depend on Nan1p but do not require each other to associate with the 35S pre-rRNA. These findings suggest that a subset of factors, which include the protein Nan1, form an assembly module that is required for the subsequent incorporation of two independent sets of proteins: one set includes Rrp7p and associated proteins, and the other set encompasses Pwp2p- and U3 snoRNA-containing complexes.
Characterization of partially assembled preribosomal particles.
The above studies suggested the presence of discrete building blocks of the 90S preribosomal particle that can be detected as free small complexes in vivo and/or forming part of both partially assembled and fully assembled 90S particles. However, our experiments could not identify unambiguously the components of each subunit or its physical interactions with proteins of other 90S particle subunits. Due to this, we decided to conduct a proteomic-based approach to identify the proteins associated with Pwp2p-MYC, Rrp7p-MYC, and Nan1p-MYC. To make this analysis more comprehensive, we also included in these experiments the identification of proteins bound to Utp4p-MYC, another component of the previously described UTP-A/t-UTP subcomplex (14, 22). We expected that, by comparing the spectra of proteins bound to these proteins under the experimental conditions used for the above sections (i.e., presence or absence of other 90S particle proteins), these experiments could shed light on the intrinsic components of each subunit and, at the same time, reveal close interactions with neighboring subunits when in the context of partially assembled preribosomal particles. Consistent with this hypothesis, we found that the selected bait proteins associated with different sets of proteins depending on the type of protein eliminated in the original yeast strain. In the case of Pwp2p, we observed that it associated with intrinsic components of the Pwp2p/UTP-B subunit and an integral component of the Mpp10p complex (Mpp10p) in the absence of Nan1p (Fig. 5; see Fig. S4A in the supplemental material). By contrast, in the absence of Rrp7p, Pwp2p was found associated with additional 90S proteins, including the UTP-A/t-UTP complex (Fig. 5; see Fig. S4B in the supplemental material). This differential proteomic pattern is fully consistent with our prior experiments showing that Pwp2p is present as both free and 35S pre-RNA-associated complexes in the absence of Rrp7p, but it is only detected as free 15 to 20S monoparticles in the absence of Nan1p (Fig. 2C and D and 3C and D). A similar script was found with Rrp7, since this protein bound primarily to UTP-C components in the absence of Nan1p (Fig. 5; see Fig. S4C in the supplemental material) but also associated with UTP-A/t-UTP components and three additional 90S particle proteins when Pwp2p was the protein missing in the lysates (Fig. 5; see Fig. S4D in the supplemental material). Very importantly, UTP-A/t-UTP components, together with Rrp5p, were the only proteins commonly found associated with Rrp7p and Pwp2p when these proteins were isolated from Pwp2p- and Rrp7p-deficient cells, respectively, further confirming that these two subunits bind independently to the UTP-A/t-UTP·35S pre-RNA complex (Fig. 5). In agreement with the idea of two independent assembling branches for the generation of the 90S particle, the spectra of Nan1p-binding proteins were also different depending on whether Nan1p was immunopurified from Pwp2p- or Rrp7p-deficient cells. In the former case, we found that Nan1p could bind only other UTP-A/t-UTP proteins and Rrp5p (Fig. 5; see Fig. S4E in the supplemental material). However, we did not detect the UTP-A-specific Pol5p, indicating that the Nan1p subunit that we are detecting is probably the t-UTP subcomplex (Fig. 5; see Fig. S4E in the supplemental material). Surprisingly, we did not find any UTP-C protein associated with Nan1p in these immunopurifications, suggesting that some Nan1p-associated proteins might be lost during the purification procedure. Consistent with this, we previously observed that Nan1p immunoprecipitates are very inefficient in pulling the 35S pre-rRNA precursor (Fig. 3B and 4B). When we performed the same set of experiments with another UTP-A/t-UTP component (Utp4p), we found the same set of Nan1p-binding proteins, but, in addition, we could recover substoichiometric amounts of Utp22p, an integral component of the UTP-C complex (Fig. 5; see Fig. S4F in the supplemental material). In the absence of Rrp7, we found that Nan1p associated with its t-UTP complex counterparts and, additionally, with a wider collection of factors that included Pwp2p/UTP-B and other 90S particle components (Fig. 5; see Fig. S4G in the supplemental material), a result consistent with the previous observations indicating that Nan1p, Pwp2p, and the U3 RNA remained associated with the 35S pre-rRNA upon Rrp7p elimination (Fig. 2C and D and 4A and B). Taken collectively, these results allowed us to correlate unequivocally the 90S particle subunits detected in our gradient fractionation experiments with the previously reported UTP complexes, to confirm the rules of engagement of those subunits in the context of the 90S particle, and to elucidate the compositions of two multisubunit intermediaries during the assembly of that preribosomal particle.
Rrp5p is another building block of the 90S particle required for the docking of UTP-C components onto the primary pre-RNA transcript.
An intriguing result derived from the aforementioned proteomic experiments was the observation that Rrp5p, a protein not previously assigned to any of the reported 90S particle subcomplexes, was a recurrent binding partner for proteins of the three UTP complexes when they were present in partially assembled 90S particle structures (Fig. 5; see Fig. S4 in the supplemental material). Rrp5p is a 90S particle component essential for the cleavage of the 35S pre-rRNA precursor at the A0, A1, A2, and A3 sites and, consequently, for the production of both 18S and 5.8S pre-RNAs (35) (see Fig. S1 in the supplemental material). We used a strategy similar to that described in previous sections to evaluate the effect of the Rrp5p depletion on the association of the identified 90S particle subunits with the 35S pre-rRNA. We observed that the expression of Rrp5p was essential for the interaction of Rrp7p with the 35S pre-rRNA (Fig. 6A). In contrast, this protein was irrelevant for the proper assembly of the U3 snoRNA (Fig. 6A; see Fig. S5A and B in the supplemental material), Nan1p (see Fig. S5A in the supplemental material), and Pwp2p (see Fig. S5B in the supplemental material) onto large preribosomal particles. Consistent with these results, we observed that the spectra of binding partners of Pwp2p and Nan1p were highly similar when they were immunopurified from either Rrp7p- or Rrp5p-depleted cells (Fig. 5 and 6B and C; see Fig. S4 in the supplemental material). In contrast, Rrp7p could bind only to UTP-C components and not to UTP-A/t-UTP or Pwp2p/UTP-B proteins in the absence of Rrp5p expression (Fig. 5 and 6D). These results indicate that the UTP-C complex requires Rrp5p for its incorporation into early preribosomal particles and, in addition, further confirm the UTP-C-independent association of the Pwp2p/UTP-B-U3 snoRNA branch with the primary pre-rRNA precursor.
An in silico view of the 90S preribosomal particle and its constitutive building blocks.
We finally resorted to a bioinformatic-based analysis to extract more information about the internal architecture and topology of the 90S preribosomal particle. To this end, we used several computational methods to generate a high-confidence network of the protein-protein interactions established among the 27 90S proteins identified in our proteomic analyses and, for the sake of completeness, of five additional ones not detected in our immunopurifications that belonged to the UTP-C (Cka2, Ckb1, and Ckb2), Pwp2p/UTP-B (Utp21p), and Mpp10p (Imp3) subcomplexes. With this strategy, we could probe bioinformatically over 60% (32 proteins) of all known 90S particle components. To strengthen the significance of the analysis, the network was built by compiling the protein-protein interaction data generated in the present report with additional information derived from previously published proteomic analyses (see Material and Methods). This approach led to the detection of 234 independent pairwise interactions (see Fig. S6 in the supplemental material), which were used subsequently to develop a final interactome network of 90S particle components (Fig. 7). This analysis pinpointed a subgroup of 15 heavily interconnected 90S proteins that included Utp22p (a UTP-C member), Utp20p, and all components of the t-UTP and Pwp2p/UTP-B subunits (Fig. 7A). Given the extensive number of interactions established by these first-order interactors, it is likely that they form a particle core that stabilizes the rest of the 90S particle components. The remaining set of analyzed proteins showed up in the analysis as second-order interactors, since they establish many fewer binary associations and are seen at the periphery of the 90S core structure (Fig. 7A). These peripheral molecules include five UTP-C proteins that are linked to the core elements through Utp22p (Fig. 7A) and, in addition, a heterogeneous subset of proteins connected to the particle core via associations with different first-order interactors (Fig. 7A). This subset of proteins includes, among others, the Mpp10p complex (Fig. 7A).
FIG. 7.
A bioinformatic-based model depicting the hierarchical interactions established among 90S preribosome particle components. (A) Network of pairwise interactions established by components of t-UTP (labeled in yellow), Pwp2p/UTP-B (labeled in green), UTP-C (labeled in purple), the Mpp10p complex (labeled in blue), and nine additional 90S preribosomal proteins (labeled in gray). Blue lines indicate interactions between protein pairs that have been detected in at least two independent proteomic experiments. First-order interactors are underlined. (B) Hierarchical clustering analysis of the interaction data available for the 32 proteins shown in panel A. Eight specific branching points of the dendrogram that gave bootstrap values of ≥745 (optimal score = 1,000) are depicted as closed circles and identified by arabic numerals. The specific stability value of each branching point is indicated in parentheses.
We next used a combination of a distance-based matrix that calculated the correlation of each pairwise protein interactome profile and the UPGMA clustering method to generate a physical proximity-based map of the 90S particle (see Fig. S6 in the supplemental material and Materials and Methods). The dendrogram obtained for the 32 proteins under analysis indicated the presence of several independent subunits that, in turn, cosegregated topologically within the 90S particle according to statistically significant neighborhood criteria. Thus, it was observed that t-UTP, Pwp2p/UTP-B, and Utp20p, a 90S particle protein not previously assigned to any of the previously published proteomic subcomplexes, colocalize in a single branch of the physical proximity dendrogram (Fig. 7B). This cosegregation pattern is consistent with the previous observations indicating that all proteins of these subunits establish a high number of pairwise contacts among themselves to form the particle core (Fig. 7A) and, in addition, with the assembly of all these proteins onto the 35S pre-rRNA in an Rrp5p- and UTP-C-independent manner (Fig. 5 and 6B and C; see Fig. S4 in the supplemental material). In contrast, UTP-C, Rrp5p, and Has1p are pooled together in a dendrogram branch clearly separated from the one containing t-UTP, Pwp2p/UTP-B, and Utp20p (Fig. 7B). Finally, Mpp10p complex proteins cosegregate in a fourth cluster that also contains seven additional 90S particle components (Fig. 7B). All these proteins had been characterized as second-order interactors that are located peripherally to the particle core in our previous bioinformatic studies (Fig. 7A). Bootstrapping controls conducted with 1,000 randomly picked protein pairwise combinations confirmed the statistical significance of the main branching points detected in this clustering analysis (Fig. 7B). This in silico model therefore recapitulates the multimodular structure of the 90S preribosome and the two-branch assembly mechanism deduced from our genetic and proteomic experiments. In addition, it provides clues to place 90S particle proteins not previously assigned to proteomic subcomplexes and/or functional groups within defined topological regions of the 90S preribosome.
DISCUSSION
The results presented in this work indicate that the 90S preribosomal particle is a multisubunit structure that follows a hierarchical and stepwise protocol of assembly. In agreement with the multimodular structure, we have demonstrated that, in addition to the already known U3 snoRNP, Mpp10p complex, and Pwp2p/UTP-B subunit, the 90S preribosome can be further subdivided into at least three additional building blocks, each containing one of the proteins monitored in our studies (Nan1p, Rrp5p, and Rrp7p). Using proteomic techniques, we have shown that the Nan1p-containing building block identified in our experiments is the previously described t-UTP and not UTP-A, since it contains the t-UTP-associated Utp5 protein instead of the UTP-A-specific Pol5p component (14, 22). In addition, we identified the Rrp7p-containing subunit as the previously described UTP-C subcomplex (22). We have not attempted to characterize the Rrp5p subunit due to the additional roles of this protein in the biosynthesis of the 60S ribosomal subunit, a property that precludes the isolation of pure, 90S particle-specific complexes in Rrp5p immunoprecipitations. At this point, therefore, we cannot formally discriminate whether this protein acts alone or in combination with other proteins in the formation of the 90S preribosome. Despite this, we currently favor the former possibility because Rrp5p was the only protein coimmunopurified with t-UTP components in Pwp2p-depleted cells (Fig. 5). Several independent observations indicate that the subunits characterized here do exist as separate functional entities in vivo. First, we have detected the Nan1p/t-UTP and Rrp7p/UTP-C subunits as stable small particles in gradient ultracentrifugation experiments. Second, we have seen that the relative amounts of these free subunits may fluctuate according to specific genetic and/or growth conditions. For example, we have observed that the levels of free Nan1p/t-UTP subunits vary very significantly depending on the total levels of Pwp2p in yeast cells. Third, we demonstrated that some of these subunits are present exclusively as monoparticles when the 90S preribosome assembly is compromised by the depletion of other 90S components. All these properties are identical to those of bona fide 90S preribosomal subunits such as the U3 snoRNP and the Pwp2p/UTP-B subunit (6, 17, 36). Finally, we have demonstrated that components of each of these subunits assemble onto the 35S pre-rRNA in steps separable from the rest of subunits.
Our results also indicate that the 90S preribosome is a ribonucleoprotein complex that assembles in a highly hierarchical and topologically defined manner (Fig. 8). The t-UTP subunit is at the pinnacle of this hierarchical process, since it is required for the subsequent assembly of the rest of identified subunits, including essential components of the 90S preribosome such as the U3 snoRNP. Because t-UTP does not depend on other subunits for the binding to the 35S pre-rRNA, we propose that its assembly is a very early and autonomous step (Fig. 8). The rest of the 90S particle modules bind to the t-UTP-primed 35S pre-rRNA following two different, and mutually independent, assembly branches (Fig. 8). In one of these branches, the U3 snoRNP and the Pwp2p/UTP-B subunit bind to the 35S pre-rRNA and the t-UTP subunit to form a highly stable intermediate of the 90S preribosomal particle (Fig. 8). This assembly branch contains additional proteins and/or subunits, because parallel proteomic experiments indicate that nine additional proteins (Mpp10p, Imp4p, Utp20p, Bms1p, Kre33p, Enp2p, Noc4p, Krr1p, and Nop1p) assemble onto preribosomes in conjunction with the Pwp2p/UTP-B subunit in an UTP-C- and Rrp5p-independent manner (Fig. 8). Although we still do not know whether this whole cohort of proteins incorporates onto the 35S pre-rRNA at the same time or in subsequent assembly steps, previous evidence suggests that at least some of them (Mpp10p and Imp4p) do assemble concurrently with the Pwp2p/UTP-B subunit and the U3 snoRNP (6, 15). In the other assembly branch, the growth of the particle requires the initial incorporation of Rrp5p that facilitates the subsequent binding of the UTP-C module (Fig. 8). Unlike the case of the coassembly of Pwp2p/UTP-B, U3 snoRNP, and the Mpp10p subunits, our proteomic experiments indicate that Rrp5p and UTP-C do not have to assemble simultaneously, because large amounts of Rrp5p are found in the partially assembled 90S particle intermediates formed in cells lacking the UTP-C component Rrp7p. As a consequence of this bifurcated incorporation of building blocks onto the t-UTP-primed 35S pre-rRNA precursor, we could identify intermediary assembly states of the 90S particle that contain either t-UTP·Pwp2p/UTP-B·U3 snoRNP·Mpp10p or, alternatively, the t-UTP·Rrp5p·UTP-C subunits bound to the 35S pre-rRNA. Importantly, components of these partially assembled complexes are readily detected in association with the full-length 35S pre-rRNA transcript, ruling out the possibility that they could represent either free preassembly stages or postcleavage 90S particle by-products. Although this hierarchical assembly evokes a temporal order of incorporation of 90S subunits onto the 35S pre-rRNA, it should be noted that our data are compatible with other scenarios. For example, it is possible that the upstream position of t-UTP in this assembly protocol is due not to an earlier incorporation of this subunit onto the 35S pre-rRNA but, rather, to the generation of a highly stable t-UTP/UTP-A·Pwp2p/UTP-B·UTP-C complex that maintains its integrity during the sedimentation and immunoprecipitation experiments.
FIG. 8.
A model for the stepwise assembly of the 90S preribosomal particle. The 90S preribosome is formed by independent building blocks that follow hierarchical rules of assembly onto the 35S pre-rRNA. The binding of the t-UTP subunit to the pre-rRNA precursor (step 1) is required for the subsequent assembly of other 90S preribosome components. Experimental evidence suggests that the assembly of t-UTP might occur at an early and independent step. However, it cannot be excluded that the recruitment of this subunit is simultaneous with the assembly events shown as immediately subsequent in this model (step 2 and/or step3) (see Discussion). The other building modules associate with the 35S pre-rRNA through two mutually independent assembly branches. In one of the branches (step 2), the Pwp2p/UTP-B subunit, the U3 snoRNP, the Mpp10 complex, and seven other proteins stably associate with the pre-rRNA. Previously published evidence suggests that the Pwp2p/UTP-B subunit and the U3 snoRNP coassemble with the 35S pre-rRNA precursor in one cooperative event (6). Whether the Mpp10p complex and the other proteins of the same assembly branch (Utp20, Bms1p, Kre33p, Nop14p, Enp2p, Noc4p, and Krr1p) also incorporate into the particle in a cooperative manner remains to be determined. In the second assembly branch (step 3), Rrp5 binds to the 35S pre-rRNA prior to the UTP-C subunit. Further details about this model are given in the text.
To further define the architectural plan of the 90S preribosome, we developed new bioinformatic tools that, by compiling the frequency at which a protein is copurified with others in independent proteomic experiments, allowed us to identify the main interactors of the 90S preribosome and, at the same time, to position them topologically according to their calculated physical proximity to other 90S components. This bioinformatic analysis revealed the presence within the 90S particle of 15 “first-order” interactors that likely form the structural core of the preribosome. These molecules include a UTP-C subunit protein (Utp22p), Utp20p, and all the t-UTP and Pwp2p/UTP-B subunit proteins. These proteins have in common the presence of either RNA or protein interaction domains in their structures and their ascription to the category of non-energy-consuming assembly factors for the 90S preribosomal particle (4). The rest of the 90S particle proteins establish fewer interactions and behave as satellites to different components of the core. These “second-order” interactors include elements proposed to be essential for wrapping up the 35S pre-rRNA (i.e., proteins of the U3 snoRNP and Mpp10p complex) (15, 30, 37) and other ancillary molecules such as a regulatory GTPase (Bms1p) (20), an RNA helicase (Has1p) (9), the bifunctional protein Rrp5 (10, 35), putative chaperone molecules and/or assembly factors for ribosomal proteins (Rrp7p and Krr1p) (2, 28), and proteins implicated in the maturation and transport of 40S subunits (Nop14p and Noc4p) (23, 24). Taken together, these results suggest that the major stabilization factor for the formation of the 90S particle is the association of its core components with the 35S pre-rRNA precursor, an step that confers the structural stability required for a conformational change of the pre-rRNA precursor that allows the exposure of specific domains and the subsequent incorporation of other 90S proteins with specialized functions in the modification and cleavage of the precursor. This computer-assisted modeling of the 90S preribosomal particle also revealed information on hitherto poorly characterized components of the 90S preribosomal particle. For example, the bioinformatic data show that Utp20p, a 90S preribosomal protein not previously ascribed to any of the characterized UTP subcomplexes, might be an important component of the structural core of the 90S particle that is in close physical proximity to the Pwp2p/UTP-B subunit and the Mpp10p complex. According to these data, it is possible that this protein may aid in the previously described coassembly of the Mpp10p complex, the Pwp2p/UTP-B subunit, and the U3 snoRNP on the 5′ end of the 35S pre-rRNA precursor (6) (Fig. 8).
Our data showing the stepwise assembly of the 90S particle are reminiscent of the changes observed in the 90S preribosome in chromatin spreads by electron microscopy. Due to the megadalton size of 90S particles, these structures can be visualized using this technique as “knobs” present at the 5′ ends of the nascent pre-rRNA transcripts (7, 26). Interestingly, it has been observed that such knobs are rather dynamic structures, because they change in size and shape as transcription of the 35S pre-rRNA proceeds (26). Although these changes are evocative of some of the assembly steps described here, it should be noted that there are important differences between the results obtained using these two experimental avenues that preclude any side-by-side comparisons. For instance, it has been shown that the early small knobs detected by electron microscopy disappear upon U3 snoRNP depletion (26), indicating that they cannot correspond to the initial t-UTP assembly stage proposed here. It is possible, therefore, that the putative Nan1p/t-UTP·35S pre-RNA complex is too small to be detected in electron micrographs or, more likely, that it lacks enough stability to survive the highly stringent conditions used for the preparation of the chromatin spreads. The future analysis of each of our conditional mutant strains using electron microscopy will clarify these discrepancies and, at the same time, will provide a more mechanistic interpretation of the dynamic changes undergone by the terminal knobs during 35S pre-rRNA transcription.
In addition to the obvious advantage of reducing the levels of freedom involved in the assembly of the numerous components that form part of the mature 90S preribosome, the subdivision of this particle into smaller building blocks offers further functional advantages. For instance, it is tempting to speculate that the stepwise assembly of the 90S subunits could be potentially used to separate in time and space different folding, maturation, or site-specific modification events that take place on the pre-rRNA. The existence of preassembled subunits can also facilitate the participation of specific 90S protein subsets in 90S particle-independent processes such as 60S ribosomal subunit biogenesis (as could be the case for the putative Rrp5p/Has1p complex found in this study) or rDNA-related transcriptional events (as is the case for the t-UTP subunit). These and other functional possibilities can be addressed in the future by further studying the preribosome subcomplexes and assembly intermediates described in this study.
Supplementary Material
Acknowledgments
We thank the Proteomics Unit personnel for their enthusiastic work in the determination of protein complexes by mass spectrometry.
M.D.'s work is supported by the Spanish Ministry of Education and Science (MEC) (BFU2005-06421) and the Castilla-León Autonomous Government (SA040/02). X.R.B.'s work is supported by grants from the U.S. NCI/NIH (5RO1-CA73735-10), MEC (SAF2003-00028), and Red Temática de Investigación Cooperativa en Cáncer (RTICC) (RD06/0020/0001, Fondo de Investigaciones Sanitarias [FIS], Carlos III Institute, Spanish Ministry of Health). J.D.L.R.'s work is possible due to a grant from the Spanish Ministry of Health (PI030920). J.P.-F. was supported by a postdoctoral fellowship from the Spanish Cooperative Research Network of Cancer Centers (C03/10, Spanish Ministry of Health) and the RTICC (RD06/0020/0001). All Spanish funding is cosponsored by the European FEDER Program.
Footnotes
Published ahead of print on 21 May 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, NY.
- 2.Baudin-Baillieu, A., D. Tollervey, C. Cullin, and F. Lacroute. 1997. Functional analysis of Rrp7p, an essential yeast protein involved in pre-rRNA processing and ribosome assembly. Mol. Cell. Biol. 17:5023-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein, K. A., J. E. Gallagher, B. M. Mitchell, S. Granneman, and S. J. Baserga. 2004. The small-subunit processome is a ribosome assembly intermediate. Eukaryot. Cell 3:1619-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de la Cruz, J., D. Kressler, and P. Linder. 2003. Ribosomal subunit assembly, p. 262-290. In M. O. J. Olson (ed.), The nucleolus. Landes Bioscience, Georgetown, United Kingdom.
- 5.De Las Rivas, J., J. J. Lozano, and A. R. Ortiz. 2002. Comparative analysis of chloroplast genomes: functional annotation, genome-based phylogeny, and deduced evolutionary patterns. Genome Res. 12:567-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dosil, M., and X. R. Bustelo. 2004. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 279:37385-37397. [DOI] [PubMed] [Google Scholar]
- 7.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Efron, B., E. Halloran, and S. Holmes. 1996. Bootstrap confidence levels for phylogenetic trees. Proc. Natl. Acad. Sci. USA 93:13429-13434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emery, B., J. de la Cruz, S. Rocak, O. Deloche, and P. Linder. 2004. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 52:141-158. [DOI] [PubMed] [Google Scholar]
- 10.Eppens, N. A., S. Rensen, S. Granneman, H. A. Raue, and J. Venema. 1999. The roles of Rrp5p in the synthesis of yeast 18S and 5.8S rRNA can be functionally and physically separated. RNA 5:779-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521-565. [DOI] [PubMed] [Google Scholar]
- 13.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 14.Gallagher, J. E., D. A. Dunbar, S. Granneman, B. M. Mitchell, Y. Osheim, A. L. Beyer, and S. J. Baserga. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18:2506-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerczei, T., and C. C. Correll. 2004. Imp3p and Imp4p mediate formation of essential U3-precursor rRNA (pre-rRNA) duplexes, possibly to recruit the small subunit processome to the pre-rRNA. Proc. Natl. Acad. Sci. USA 101:15301-15306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glazko, G. V., and A. R. Mushegian. 2004. Detection of evolutionarily stable fragments of cellular pathways by hierarchical clustering of phyletic patterns. Genome Biol. 5:R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 18.Granneman, S., and S. J. Baserga. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43-50. [DOI] [PubMed] [Google Scholar]
- 19.Granneman, S., J. E. Gallagher, J. Vogelzangs, W. Horstman, W. J. van Venrooij, S. J. Baserga, and G. J. Pruijn. 2003. The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res. 31:1877-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karbstein, K., S. Jonas, and J. A. Doudna. 2005. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol. Cell 20:633-643. [DOI] [PubMed] [Google Scholar]
- 21.Krogan, N. J., G. Cagney, H. Yu, G. Zhong, X. Guo, A. Ignatchenko, J. Li, S. Pu, N. Datta, A. P. Tikuisis, T. Punna, J. M. Peregrin-Alvarez, M. Shales, X. Zhang, M. Davey, M. D. Robinson, A. Paccanaro, J. E. Bray, A. Sheung, B. Beattie, D. P. Richards, V. Canadien, A. Lalev, F. Mena, P. Wong, A. Starostine, M. M. Canete, J. Vlasblom, S. Wu, C. Orsi, S. R. Collins, S. Chandran, R. Haw, J. J. Rilstone, K. Gandi, N. J. Thompson, G. Musso, P. St Onge, S. Ghanny, M. H. Lam, G. Butland, A. M. Altaf-Ul, S. Kanaya, A. Shilatifard, E. O'Shea, J. S. Weissman, C. J. Ingles, T. R. Hughes, J. Parkinson, M. Gerstein, S. J. Wodak, A. Emili, and J. F. Greenblatt. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440:637-643. [DOI] [PubMed] [Google Scholar]
- 22.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 23.Liu, P. C., and D. J. Thiele. 2001. Novel stress-responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol. Biol. Cell 12:3644-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milkereit, P., D. Strauss, J. Bassler, O. Gadal, H. Kuhn, S. Schutz, N. Gas, J. Lechner, E. Hurt, and H. Tschochner. 2003. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 278:4072-4081. [DOI] [PubMed] [Google Scholar]
- 25.Oeffinger, M., A. Leung, A. Lamond, and D. Tollervey. 2002. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA 8:626-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osheim, Y. N., S. L. French, K. M. Keck, E. A. Champion, K. Spasov, F. Dragon, S. J. Baserga, and A. L. Beyer. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol. Cell 16:943-954. [DOI] [PubMed] [Google Scholar]
- 27.Prieto, C., and J. De Las Rivas. 2006. APID: Agile Protein Interaction DataAnalyzer. Nucleic Acids Res. 34:W298-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasaki, T., E. A. Toh, and Y. Kikuchi. 2000. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol. Cell. Biol. 20:7971-7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schafer, T., D. Strauss, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma, K., and D. Tollervey. 1999. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 19:6012-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapman, J., J. Retel, and R. J. Planta. 1975. Ribosomal precursor particles from yeast. Exp. Cell Res. 90:95-104. [DOI] [PubMed] [Google Scholar]
- 32.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 33.Udem, S. A., and J. R. Warner. 1972. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J. Mol. Biol. 65:227-242. [DOI] [PubMed] [Google Scholar]
- 34.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 35.Venema, J., and D. Tollervey. 1996. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15:5701-5714. [PMC free article] [PubMed] [Google Scholar]
- 36.Watkins, N. J., V. Segault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Luhrmann. 2000. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 37.Wehner, K. A., J. E. Gallagher, and S. J. Baserga. 2002. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol. 22:7258-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.