Abstract
Cholera toxin (CT) produced by Vibrio cholerae causes the devastating diarrhea of cholera by catalyzing the ADP-ribosylation of the α subunit of the intestinal Gs protein (Gsα), leading to characteristic water and electrolyte losses. Mammalian cells contain ADP-ribosyltransferases similar to CT and an ADP-ribosyl(arginine)protein hydrolase (ADPRH), which cleaves the ADP-ribose-(arginine)protein bond, regenerating native protein and completing an ADP-ribosylation cycle. We hypothesized that ADPRH might counteract intoxication by reversing the ADP-ribosylation of Gsα. Effects of intoxication on murine ADPRH−/− cells were greater than those on wild-type cells and were significantly reduced by overexpression of wild-type ADPRH in ADPRH−/− cells, as evidenced by both ADP-ribose-arginine content and Gsα modification. Similarly, intestinal loops in the ADPRH−/− mouse were more sensitive than their wild-type counterparts to toxin effects on fluid accumulation, Gsα modification, and ADP-ribosylarginine content. Thus, CT-catalyzed ADP-ribosylation of cell proteins can be counteracted by ADPRH, which could function as a modifier gene in disease. Further, our study demonstrates that enzymatic cross talk exists between bacterial toxin ADP-ribosyltransferases and host ADP-ribosylation cycles. In disease, toxin-catalyzed ADP-ribosylation overwhelms this potential host defense system, resulting in persistence of ADP-ribosylation and intoxication of the cell.
Cholera toxin (CT), the ADP-ribosyltransferase secreted by Vibrio cholerae, is a 84-kDa protein comprising one catalytic A subunit associated noncovalently with five B subunits (11, 31, 44). Through its B subunit pentamer, the toxin binds the cell surface receptor monosialoganglioside GM1, which facilitates entry of the A subunit and its access to intracellular ADP-ribosyltransferase substrates, β-NAD and the α subunit of the Gs protein (Gsα) (8). Like other G proteins, Gsα is active with bound GTP. Its intrinsic GTPase activity converts the active GTP-bound protein to the inactive GDP-bound species (34). Because the Gsα catalytic arginine is critical for GTP hydrolysis to GDP and thereby inactivation, ADP-ribosylation of that arginine inhibits GTPase activity and prolongs the lifetime of active, GTP-bound Gsα (3, 9, 42). The resulting persistent activation of adenylyl cyclase increases cyclic AMP (cAMP) accumulation, along with its effects on water and electrolyte transport that cause the diarrhea of cholera (7, 33).
In addition to CT, other bacterial toxin ADP-ribosyltransferases utilize arginine residues in proteins as ADP-ribose acceptors (8). Escherichia coli heat-labile enterotoxin acting by a mechanism very similar to that of CT is responsible for some cases of traveler's diarrhea (22). Pseudomonas aeruginosa exoenzyme S, a cytotoxin introduced into cells via a type III secretory system, modified free arginine and arginine in the signaling protein Ras (5, 10). Another P. aeruginosa cytotoxin, exoenzyme T, modified arginine 20 in Crk (CT10 regulator of kinase) (6), affecting the actin cytoskeleton. Clostridial C2 toxin ADP-ribosylates an arginine in actin (40), resulting in displacement of the equilibrium toward depolymerization, with adverse effects on cytoskeletal organization and chemotaxis. Other bacterial protein products, such as Clostridium perfringens iota toxin (41) and Salmonella enterica SpvB cytotoxin (14), also use an arginine in actin as an ADP-ribose acceptor. Of note, bacteriophage T4 modifies arginine (36). Based on these observations, modification of arginine in critical cellular proteins appears to be a common mechanism for toxin disruption of signal transduction pathways and cytoskeletal regulation.
Mammalian cells contain ADP-ribosyltransferases that, like CT and other cytotoxins, catalyze the stereospecific transfer of ADP-ribose from β-NAD to the guanidino moiety of arginine to generate α-ADP-ribosylarginine (26, 32). Several NAD:arginine ADP-ribosyltransferases have been purified from avian or mammalian tissues, and their cDNAs have been cloned (12, 29, 30, 39, 45). In mammalian cells, ADP-ribosylation appears to be a reversible posttranslational modification of proteins (23, 30, 43). An ADP-ribosylarginine hydrolase (ADPRH) that cleaves the ribosylarginine linkage, generating the unmodified protein (23, 25, 27), can complete a potentially regulatory ADP-ribosylation cycle. Such a cycle is thus far best documented in the photosynthetic bacterium Rhodospirillum rubrum, where it controls the activity of dinitrogenase reductase, a key enzyme in nitrogen fixation (20). Mammalian cells may similarly use transferase and hydrolase enzymes for controlling their content of ADP-ribose-arginine in proteins.
ADPRHs in humans and rodents are ∼39-kDa proteins that share some amino acid identity (37). Only one hydrolase has been identified in mammalian tissues, consistent with the existence of only one ADP-ribose-arginine hydrolase gene (2). Its function, presumably, is to reverse the action of the ADP-ribosyltransferase and thereby regulate cellular levels of proteins containing ADP-ribose-arginine (30, 43). We asked whether the hydrolase can also counteract the effects of CT on cells, by removing ADP-ribose from the modified Gsα protein. To evaluate this possibility, we studied the small intestine (and other cells) from knockout (KO) mice that lacked hydrolase activity. Sensitivity to CT was assessed by determining the extent of modification of Gsα and the levels of ADP-ribosyl(arginine)protein. The toxin effects on fluid accumulation in intestinal loops were much greater in KO than wild-type mice. Data from experiments with cells grown from mouse embryos were similarly consistent with the conclusion that the hydrolase can, indeed, moderate the effects of CT both in vitro and in vivo.
MATERIALS AND METHODS
Materials.
Rabbit antibodies against full-length recombinant mouse ADPRH were from Cocalico Biological, Inc. (Reamstown, PA); rabbit antibodies against Gsα were kindly provided by Lee Weinstein (NIDDK, NIH).
Animal studies.
Animal protocols were approved by the National Heart, Lung, and Blood Institute, National Institutes of Health Animal Care and Use Committee (9-PCCM-4R, 2-PCCMB-30, and 3-PCCMB-30). ADPRH mice were backcrossed seven times using C57BL/6J mice (Jackson Laboratory, Bar Harbor, MI).
Preparation of targeting vector and ES cells.
A targeting vector was prepared to delete a 1.0-kb DNA fragment of the mouse ADPRH gene between XhoI and BamHI sites, which includes exon 2; the deleted region includes D60 and D61 critical for catalytic activity (18). A ∼13-kb SalI genomic fragment was cloned from a genomic library of 129/SvJ mice in a Lambda FIX II vector (Stratagene, La Jolla, CA), and from it, a ∼7-kb EcoRI fragment containing exons 1 and 2 was subcloned into pGEM 3Z. A 4.5-kb fragment comprising the 5′ flanking region, exon 1, and part of intron 1 was excised from the 7-kb EcoRI fragment with EcoRI and XhoI and subcloned into the scrambler A region of pKO Scrambler vector (Stratagene). A 1.8-kb fragment containing part of intron 2 was amplified by PCR (primers are listed in Table 1) with the 7-kb EcoRI fragment in pGEM 3Z as a template and subcloned into scrambler B region of the pKO Scrambler vector. Neomycin resistance and thymidine kinase genes in the targeting vector were used, respectively, as positive and negative selection markers of homologous recombination. The unique SalI site was used to linearize the pKO vector before electroporation of embryonic stem (ES) cells. Clones resistant to both G418 (Sigma, St. Louis, MO) and ganciclovir (Sigma) were analyzed by Southern blotting. Wild-type and targeted ADPRH alleles were found in, respectively, 11- and 6-kb bands and 6.7- and 5.8-kb bands by Southern blotting of the ES cell genomic DNA digested with PstI and hybridized with a 5′ probe (490-bp; −360 to 130, including exon 1), and SacI and hybridized with 3′ probe (360-bp; positions 5053 to 5412, including exon 3) (Table 1).
TABLE 1.
Nucleotide primers and probes for mouse ADPRH gene
| Primer/probe function or product | Name | Sequence |
|---|---|---|
| PCR genotyping (genomic DNA) | TP | 5′-(1826)-CCAAAGCGAATATCAGGTGAT-(1846)-3′ |
| LP | 5′-(2610)-TTACTCTCCATCTACAGTCTTAGCG-(2586)-3′ | |
| NP | 5′-GTTCTTTTTGTCAAGACCGACCTG-3′ | |
| Southern blotting (genomic DNA) | ||
| 5′ exon 1 probe (490 bp) | Forward | 5′-(−360)-CTAAGCTCCCGTTTGAAACCCTTCTG-(−335)-3′ |
| Reverse | 5′-(130)-CTGCTGAGGCGGAGCTGAGAGCGT-(107)-3′ | |
| 3′ exon 3 probe (360 bp) | Forward | 5′-(5053)-GGTGCTTGCATGCAGAACGCC-(5073)-3′ |
| Reverse | 5′-(5412)-CCAGTATTGAAGATTCTCCTTCAC-(5389)-3′ | |
| Northern blotting probe (291 bp) | Forward | 5′-(181)-GGGGCTGATTGAGAGGTATGT-(201)-3′ |
| Reverse | 5′-(471)-TCCATGTCTCCCATGCAGTCC-(451)-3′ | |
| Short arm (1.8-kb fragment) | Forward | 5′-CGCTAGACTGTAGATGAGAGT-3′ |
| Reverse | 5′-GGATAGAAGATGCTAAGACG-3′ | |
| Double mutant (D60A, D61A) | Forward | 5′-(63)-CGATGGAGAGTCAGTGCCGCCACCGTCATGCACCTG-(98)-3′ |
| Reverse | 5′-(98)-CAGGTGCATGACGGTGGCGGCACTGACTCTCCATCG-(63)-3′ | |
| ADPRH wild-type cDNA (coding) to pGEX2T and pcDNA4/V5-His | Forward | 5′-(1)-ATGGGTGGGGGGCTGATT-(18)-3′ |
| Reverse | 5′-(1089)-CATGGGATCTAATACAGGGCT-(1069)-3′ |
Generation and genotyping of ADPRH KO mice and cultured primary cells.
Chimeric mice were generated by microinjecting ES cells (129/SvJ) into blastocysts from C57BL/6J mice following standard procedures (17). Heterozygous (ADPRH+/−) mice were used for breeding to produce homozygous (ADPRH−/−), heterozygous (ADPRH+/−), and wild-type (ADPRH+/+) animals. Genotypes of mice and primary cultured cells were confirmed by PCR and Southern blot analyses. Genomic DNA was extracted from mouse tails or primary cells using a Wizard genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer's protocol. A set of three primers (Table 1) was used for PCR amplification of the ADPRH wild-type genomic DNA fragment (bp 1826 to 2610). The same forward primer, TP, based on the ADPRH sequence, was used for both wild-type and mutant alleles. Two reverse primers, NP and LP, respectively, based on the ADPRH and neomycin resistance cassette sequences, were used in 35 to 40 cycles of PCR (30 s at 95°C, 30 s at 68°C, and 5 min at 68°C) with the Advantage 2 PCR system (BD Clontech, Palo Alto, CA). PCR products were subjected to electrophoresis in 1% agarose gel containing ethidium bromide. With primers TP and LP, a 1.8-kb fragment and a 0.8-kb fragment were amplified from mutant and wild-type cDNAs, respectively; TP and NP were used to amplify a specific 1.0-kb mutant DNA fragment. All genotypes were confirmed by both Southern blotting and PCR.
Southern blot analysis.
Samples (5 to 10 μg) of genomic DNA prepared from ES cells or mouse tails using a Wizard genomic DNA purification kit (Promega) according to standard protocols were digested with PstI or SacI (Roche, Indianapolis, IN), subjected to electrophoresis in 1.0% agarose gel, and transferred (Turboblotter system; Schleicher & Schuell, Keene, NH) to nylon membrane (S & S Nyrtan; Schleicher & Schuell). The 5′ and 3′ cDNA probes corresponding to bases in the mouse ADPRH genomic DNA had been amplified from a 129/SvJ mouse genomic library in a Lambda FIX II vector (Stratagene), using specific primers (Table 1). Membranes were incubated for 1 h at 68°C in ExpressHyb (BD Clontech, Palo Alto) with [α-32P]dATP (Perkin Elmer, Boston, MA)-labeled probes (Table 1) located outside the 5′- and 3′-flanking regions of the targeting vector (Fig. 1). Membranes were washed twice for 20 min at room temperature with 2× SSC-0.1% sodium dodecyl sulfate (SDS) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and twice for 30 min at 60°C with the same solution before exposure to film. 32P was detected by exposure of X-ray film (Kodak, Rochester, NY).
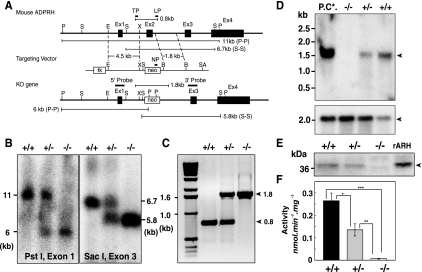
FIG. 1.
Preparation of ADPRH gene. (A) Diagram of mouse ADPRH gene locus with exons 1 to 4 as boxes. Targeting vector (middle) was generated by insertion of the neomycin resistance cassette (neo), which is flanked by XhoI (X) and BamHI (B) sites, to replace exon 2 and flanking sequences in the vicinity of XhoI (X). The unique SalI site was used to linearize the targeting vector before electroporation. The recombinant ADPRH KO allele is shown below. The position of the exon 1 5′ probe is shown above. After digestion of genomic DNA with PstI (P), this probe is predicted to hybridize with an 11-kb fragment from the wild-type allele and a 6-kb fragment from the ADPRH KO allele. The position of exon 3 as a 3′ probe was used to confirm correct insertion of the targeting vector after digestion with SacI (S). This probe is predicted to hybridize with a 6.7-kb genomic fragment in the wild-type allele and a 5.8-kb genomic fragment in the ADPRH mutant allele. (B) Genotyping using Southern blot analysis of an ADPRH KO mouse derived from mating of F1 ADPRH heterozygous mice. Genomic DNA was digested with PstI and SacI and hybridized with 5′ and 3′ probes. The genotype of individual animals is indicated above each lane. (C) Genotyping of representative pups using two sets of PCR primers simultaneously and DNA from tail cuts. TP and LP primers amplified a 0.8-kb fragment from the wild-type allele and a 1.8-kb fragment from the KO allele (arrowheads). +/+, ADPRH wild-type; +/−, ADPRH heterozygous; −/−, ADPRH homozygous. (D) Northern blot analysis of intestine from ADPRH KO, heterozygous, and wild-type mice using exon 2 probes. Poly(A)+ RNA (5 μg) was isolated from intestines of ADPRH mice (+/+, +/−, and −/−) with poly(A)+ RNA (5 μg) from mouse brain as a positive control (P.C.*). Northern blot analyses were performed in triplicate (upper arrowhead, ADPRH 1.7-kb; lower arrowhead, β-actin 2.1-kb). (E) Western blotting. Intestinal lysates (20 μg protein) from +/+, +/−, and −/− mice with recombinant ADP-ribosylarginine hydrolase protein (2 μg) as a positive control (rARH) were separated by SDS-PAGE and probed with antibodies raised against full-length recombinant mouse ADPRH protein, showing 39-kDa ADPRH. (F) ADPRH activities in lysates (100 μg protein) of intestine from +/+, +/−, and −/− mice. Data are means ± SEM of values from triplicate assays. *, P = 0.0038; **, P = 0.0039; and ***, P = 0.000012. Results were similar in three experiments.
Northern blot analysis.
Samples (5 μg) of poly(A)+ RNA, prepared from ES cells and mouse tissues using, respectively, a Micro-FastTrack 2.0 kit or FastTrack 2.0 kit (Invitrogen, Carlsbad, CA) as described by the manufacturer, were separated in 1.2% agarose gel containing formaldehyde and transferred to nylon membranes (S & S Nyrtan). A 291-bp fragment corresponding to bases 181 to 471 (Table 1) in the ADPRH coding region that had been amplified from a mouse brain cDNA library (BD Clontech) using specific primers (Table 1) and labeled with [α-32P]dATP (Perkin Elmer) was used as a probe. After hybridization for 1 h at 68°C in ExpressHyb (BD Clontech) with heat-denatured salmon sperm DNA (100 mg/ml) and [α-32P]dATP-labeled 291-bp probes, membranes were washed twice with 2× SSC-0.1% SDS at room temperature for 20 min and twice with 0.2× SSC-0.1% SDS at 60°C for 20 min before exposure to film. 32P was detected by exposure to X-ray film (Kodak). After the blot was stripped to remove the labeled probe, the blot was hybridized with a β-actin cDNA probe (Ambion, Austin, TX).
Construction of wild-type and mutant mouse ADPRH expression vectors.
ADPRH wild-type cDNA was amplified from mouse brain cDNA library (BD Clontech) with forward and reverse primers (Table 1). Fragments were excised with HindIII and BamHI or with BamHI and SmaI, respectively, digested for ligation into a pcDNA4/V5-His (Invitrogen) or pGEX-2T (Amersham Bioscience, Piscataway, NJ) vector to produce the recombinant wild-type plasmid. Pfu polymerase (Stratagene) was used for PCR (Perkin-Elmer Thermal Cycler 9600) amplification according to the manufacturer's protocol for 35 cycles of 95°C for 45 s, 60°C for 45 s, and 72°C for 2.5 min. Mutant ADPRH (D60A, D61A) that lacks hydrolase activity was generated using a QuikChange site-directed mutagenesis kit (Stratagene) with oligonucleotide primers (Table 1) according to the manufacturer's protocol. After amplification, supercoiled double-stranded DNA was digested (2 h at 37°C; total volume, 50 μl) with DpnI endonuclease (2.5 units), which is specific for methylated and hemimethylated DNA. DNA isolated from almost all E. coli strains is dam-methylated and therefore susceptible to DpnI digestion. The nicked vector DNA, containing the mutant hydrolase, was transfected into Epicurian Coli XL1-Blue supercompetent cells, which were plated on LB-ampicillin agar plates and incubated at 37°C overnight. For each mutant, 20 colonies were grown overnight individually in 3 ml of LB-ampicillin at 37°C. Plasmid cDNA was isolated by a QIAprep spin miniprep kit (QIAGEN, Studio City, CA), and the entire coding region was sequenced (ABI PRISM 377; Perkin-Elmer). Finally, pcDNA4/V5-His and pGEX-2T vectors, each containing wild-type and mutant ADPRH cDNAs, were prepared.
Production of GST fusion wild-type and double-mutant ADPRH proteins by pGEX-2T.
Wild-type and mutant recombinant ADPRH (rADPRH) proteins were synthesized in E. coli BL21 during incubation at 30°C for 12 h in the presence of 0.2 mM isopropyl-β-d-thiogalactopyranoside (total volume, 100 ml) (18, 25). Cells were harvested, washed with ice-cold phosphate-buffered saline (PBS), pH 7.4, and suspended in 5 ml of lysis buffer (10 mM sodium phosphate, 10 mM EDTA, pH 8.0). After repeated freezing (on dry ice for 5 min) and thawing at room temperature (three times) and sonification for 1 min on ice, the cell lysate was centrifuged (4,000 × g for 20 min), and the supernatant was incubated (gentle agitation at room temperature for 30 min) with 2 ml of glutathione-Sepharose 4B (50% slurry in PBS). The matrix was washed with 20 ml of PBS before elution of glutathione S-transferase (GST)-bound proteins in 4 ml of 10 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0, which was concentrated to a final volume of 1 ml using a p30 Microcon (Millipore, Billerica, MA). Samples (0.15 ml) were mixed with 0.05 ml of 4× SDS sample buffer (4% SDS, 10% glycerol, 0.08% bromophenol blue, 2% β-mercaptoethanol, 125 mM Tris-HCl, pH 6.8), boiled for 5 min, and subjected to electrophoresis in a 12% Tris-glycine gel (Invitrogen), followed by staining with 0.05% Coomassie blue. Fusion proteins were estimated to be 95% pure by SDS-polyacrylamide gel electrophoresis (PAGE) (data not shown). The expression of wild-type and mutant ADPRH was confirmed with Western blot analysis and ADPRH assay.
Transfection and clonal selection of ADPRH−/− cells.
ADPRH−/− cells from a 14.5-day embryo were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% (newborn calf serum) NBCS and transformed with pcDNA4/V5-His, pcDNA4/V5-His wild-type, or mutant vectors using Fugene 6 (Roche) transfection reagent. After culture for 2 to 3 weeks with zeocin (1 mg/ml; Invitrogen) as a selection reagent, single colonies were grown in 96-well plates to confirm the presence of rADPRH wild-type or mutant proteins with Western blot analyses and ADPRH assays. ADPRH−/− cells expressing ADPRH wild-type cDNA are designated ADPRH−/−* and ADPRH−/− cells expressing the ADPRH mutant protein are designated ADPRH−/−**. After selection, these reconstituted (rescued) ADPRH cells were grown in DMEM supplemented with 10% NBCS, penicillin (10,000 units/ml), streptomycin (10 mg/ml), and zeocin (1 mg/ml) (Invitrogen) at 37°C in a humidified atmosphere with 5% CO2.
Western blot analysis.
ADPRH protein was quantified by Western blotting. Protein (20 μg) from each cell lysate was subjected to SDS-PAGE, transferred to nitrocellulose membranes, and reacted with rabbit anti-ADPRH polyclonal antibodies, or rabbit anti-Gsα polyclonal antibodies. A detection kit, SuperSignal West Pico Chemiluminescent Substrate (Pierce), was used for visualization of immunocomplexes. Blots were stripped according to the manufacturer's instructions before another reaction with antibodies. An anti-glyceraldehyde-3-phosphate dehydrogenase (Chemicon, Temecula, CA) blot was treated in parallel to monitor loading and effects of stripping. Antibodies were detected by exposure of X-ray film (Kodak) with detection using an LAS-3000 bioimaging system (Fuji, Tokyo).
ADPRH assay.
ADPRH activity of embryonic cells, transformed cells, and mouse tissues was assayed, as described previously (18). The substrate was synthesized in a reaction catalyzed by 50 μg of CT A subunit (List Biological Laboratory), with 2 mM β-NAD, 2 mM [14C]arginine (50 μCi; Sigma), 20 mM dithiothreitol, 50 mM potassium phosphate (pH 7.5), and 30 μg of ovalbumin in a total volume of 0.3 ml during incubation at 30°C overnight. ADP-ribosyl-[14C]arginine was purified by high-performance liquid chromatography (Hewlett-Packard series 1100 system equipped with a diode-array spectrophotometric detector set at 260 nm) using an anion exchange column (Zorbax SAX; 4.6-mm inner diameter; Rockland Technologies, Newport, DE) and elution with a linear gradient of NaCl (0 to 1 M) in 20 mM sodium phosphate buffer, pH 4.5, for 30 min at a flow rate of 1 ml/min. ADP-ribosyl-[14C]arginine eluted at 11 to 12 min and separated from arginine and nicotinamide (which eluted between 3 and 6 min), NAD (17 to 19 min), and ADP-ribose (38 to 39 min). ADP-ribosyl-[14C]arginine was lyophilized, dissolved in 50 μl of H2O, and stored at −20°C. Samples (100 μg) of homogenized tissue (ADPRH+/+, ADPRH+/−, and ADPRH−/−), proteins (40 μg) from homogenized cells (ADPRH+/+, ADPRH+/−, and ADPRH−/− embryonic cells and ADPRH−/−* and ADPRH−/−** transformed cells), or purified recombinant mouse ADPRH proteins (50 ng) (wild-type and mutant ADPRH) synthesized as GST-fusion proteins were assayed in 50 mM potassium phosphate, pH 7.5, containing 5 mM dithiothreitol, 10 mM MgCl2, 50 μM ADP-ribosyl-[14C]arginine (6,000 cpm) (total volume, 100 μl). After 1 h at 37°C, a sample (90 μl) was applied to a column (0.8 by 4 cm) of Affi-Gel boronate (Bio-Rad, Hercules, CA), equilibrated, and eluted with 5 ml of 0.1 M glycine, pH 9.0, 0.1 M NaCl, and 10 mM MgCl2. Total eluate was counted by a liquid scintillation counter. For calculation of ADPRH activity, ADP-ribose values were corrected for the activity present at time zero and in replicate assays without enzyme.
Culture of mouse embryo cells.
Fibroblasts were prepared from embryonic day 14.5 embryos as described previously (38). In brief, a skin sample (5 to 7 mg) was washed in 7.5 ml of PBS with gentle shaking in a 15-ml polypropylene tube. Subcutaneous tissue was removed from the skin sample by scraping the dermal surface using pairs of forceps before it was incubated in 0.25% trypsin-PBS for 30 min at 37°C. The skin samples were then placed on a 100-mm dish, epidermis was removed by scraping, and tissue was washed in the same way before being cut into 2- to 4-mm squares on the 100-mm tissue culture dish. Skin pieces (5 to 7 pieces) were placed in each well of a six-well plate; 5 min later, DMEM (Invitrogen) supplemented with 10% NBCS was added carefully to avoid to disturbing the tissue, followed by incubation at 37°C in a humidified atmosphere of 5% CO2. Fibroblast outgrowth was monitored every 2 days using an inverted phase-contrast microscope, and when cells were confluent, cells were washed twice with ice-cold PBS, harvested using 0.05% trypsin-EDTA, and plated on collagen I-coated 100-mm dishes (passage 1 cells). Cell culture was continued in the same way for more than 21 passages in DMEM with 10% NBCS in an atmosphere of 5% CO2. Samples of cells for stock at passages 2 to 15 were frozen in Cell Banker (Wako Chemical USA, Richmond, VA) and stored at −80°C. For experiments, thawed cells were grown in DMEM supplemented with 10% NBCS, penicillin (10,000 units/ml), and streptomycin (10 mg/ml) (Invitrogen) at 37°C in a humidified atmosphere of 5% CO2.
Preparation of proteins from cultured cells and mouse tissues.
For experiments with CT, cultured cells (passages 6 to 10) from exponentially dividing stock cultures (3.0 × 105 cells per 150-mm dish) were grown to ca. 80% confluence and incubated in DMEM medium without serum for 24 h. Cells were incubated with 100 ng/ml CT (List Biological Laboratory, Campbell, CA) in 10 ml of DMEM containing 10% NBCS for 6 h; then, medium was discarded, and cells were rinsed twice with ice-cold PBS, pH 7.4, before the addition of 5 ml of 20% ice-cold trichloroacetic acid (TCA) (Sigma). After 20 min at 4°C, contents of dishes were scraped with a Cell Lifter (Corning, Corning, NY) into 1.5-ml tubes, which were centrifuged (20,000 × g for 30 min at 4°C). Supernatants were discarded, and pellets were washed once with 20% ice-cold TCA and twice with ether; residual ether was removed under vacuum, and the pellets were stored at −80°C.
Animal tissues were rapidly excised and immediately frozen in liquid nitrogen and pulverized as described previously (16). After evaporation of liquid nitrogen, 2 ml of ice-cold 20% (wt/vol) TCA was added, and the tissue was homogenized using 30 strokes of a Dounce all-glass, hand homogenizer (Kontes, Vineland, NJ) on ice. Insoluble material was pelleted by centrifugation (20,000 × g at 30 min for 4°C), and supernatants were discarded; pellets were washed once with cold 20% TCA and twice with ether; residual ether was removed under vacuum, and the powder was stored at −80°C.
Solubilization of TCA-precipitated proteins.
TCA-precipitated proteins in 2 ml of TENDS buffer (20 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM NaN3, 2 mM dithiothreitol, 0.25 M sucrose) with 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 0.5 mM benzamidine, and a 10 μg/ml concentration of leupeptin, aprotinin, and trypsin inhibitor were homogenized on ice in a 4°C room using 30 strokes of a Dounce all-glass, hand homogenizer (Kontes). Protein was quantified using a Micro-BCA protein assay reagent kit (Pierce, Rockford, IL), according to the manufacturer's protocol.
In vivo CT treatment and in vitro rADPRH treatment.
TCA-precipitated proteins from cultured cells in CT experiments were homogenized in TENDS buffer on ice in a 4°C room, using 30 strokes of a Dounce all-glass hand homogenizer (Kontes). After homogenization, samples (total, 30 μg of protein) were incubated with 0.5 μg of rADPRH protein (0.5 μg/μl) or 0.5 μg of bovine serum albumin (0.5 μg/μl) at 30°C for 1 h in vitro. After incubation, 5 μl of 4× SDS sample buffer was added directly to each sample tube for Western blot analysis.
Fluid accumulation induced by CT.
Fluid accumulation induced by CT was quantified using mouse intestinal loops, as described previously (13). ADPRH+/+, ADPRH+/−, and ADPRH−/− (male and female) mice (20 to 25 g; 6 to 8 weeks old) had access to water ad libitum but no food for 12 h before experiments. Mice were anesthetized by inhalation of 1 to 3% isoflurane, and the intestine was exteriorized through a midline incision. In each mouse, 2 or 3 intestinal segments (ca. 4 cm) were separated by ligation with nylon suture. PBS (0.2 ml) without or with CT (0.5 μg) or 5 mM 8-bromo-cAMP was injected into each loop. The abdomen was closed, and 0.5% bupivacaine (2 to 3 drops) was applied to the incision while the mouse recovered from anesthesia on a 37°C mat. Mice were monitored until euthanized with CO2 via gas cylinders after being placed in a desiccator jar or equivalent. Weight and length of each intestinal loop with its contents were recorded. Fluid accumulation in the ligated loops, reflecting CT action, is reported as weight per length (mg/cm); data are presented as means ± standard errors of the means (SEMs) of values from six experiments.
Assay of ADP-ribosylarginine in cells.
ADP-ribosylarginine content of cultured cells, intestinal loops, and their epithelial cells was quantified using TCA-precipitated proteins that had been stored at −80°C. TCA-precipitated proteins (4.0 mg) were dissolved in 2 ml of 6 M guanidinium chloride, 50 mM morpholinepropanesulfonic acid, and 10 mM EDTA, pH 4.0, with 30 strokes of a Dounce all-glass hand homogenizer (Kontes) on ice. A 0.5-ml aliquot of each sample was subjected to analysis as described previously (16). All assays were performed in triplicate, and data represent means of values from six experiments.
Densitometry and statistics.
The bands were analyzed by the densitometric quantification of proteins on blots. We used a LAS 3000 Bioimaging system with the image analysis software Image Gauge, version 4.0 (Fuji). Data are presented as means ± SEMs of values from the indicated number of experiments or samples. Student's t test was used to evaluate differences, with a P value of <0.05 considered to be significant.
RESULTS
Generation of ADPRH KO mice.
The ADPRH targeting vector contained a neomycin resistance gene that replaced all of exon 2 (Fig. 1A), which includes the critical catalytic site residues D60 and D61 (18). After G418 selection, 250 independent ES clones were analyzed by Southern blot analyses with probes corresponding to an upstream genomic fragment, the neomycin cassette, and a region downstream of the targeting vector (Table 1). Six clones with homologous recombination were obtained. Two ES clones with correct karyotypes were injected into blastocysts from C57BL/6J (Jackson laboratory) mice, and both transmitted the mutant allele through the germ line. Germ line transmission of the ADPRH KO allele was achieved by mating male chimeras with C57BL/6J females. ADPRH mutant genes in mouse DNA were identified by Southern blotting (Fig. 1B) (see Materials and Methods). Inactivation of the ADPRH gene from genomic DNA was confirmed by PCR (Fig. 1C) (see Materials and Methods).
ADPRH proteins and activity in KO and heterozygous mice and embryonic cells.
ADPRH mRNA was not detected by Northern blot analysis of ADPRH−/− mice (Fig. 1D) with an exon 2-specific probe (Table 1). Absence of the ADPRH gene product was confirmed by immunoblotting and activity measurements. By Western analysis, no immunoreactive protein was detected in KO mouse tissues. Amounts of the 39-kDa ADPRH were less in lysates of intestine from heterozygous than from wild-type mice (Fig. 1E). No hydrolase activity was detected in intestinal lysates from ADPRH−/− mice, and activity was significantly lower in those from heterozygous than from wild-type mice (Fig. 1F).
Mice were interbred to generate litters with ADPRH+/+, ADPRH+/−, and ADPRH−/− mice from the heterozygous intercrosses (ADPRH+/+:ADPRH+/−:ADPRH−/− in males 46:80:54 and in females, 44:84:48) with a typical litter size of 8 to 10 pups, indicating that the ADPRH mutation is not embryonic lethal.
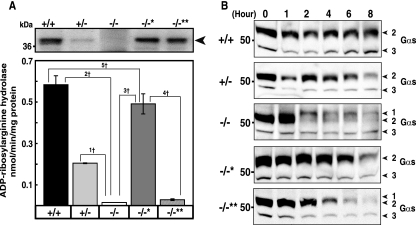
The embryonic ADPRH−/− cells were transfected with cDNA to overexpress wild-type ADPRH or a mutant in which D60 and D61, amino acids that are critical for hydrolase activity, were replaced by alanine. Immunoblotting with antibodies against ADPRH revealed considerably less 39-kDa hydrolase in ADPRH+/− than ADPRH+/+ cells and none in ADPRH−/− cells. Both ADPRH−/−* and ADPRH−/−** cells overexpressing wild-type and inactive mutant ADPRH, respectively, showed immunoreactive 39-kDa hydrolase bands. ADPRH−/− cells had no measurable hydrolase activity, and ADPRH+/− cells exhibited ∼30% of the activity of wild-type ADPRH+/+ cells (Fig. 2A). After transfection, activity of ADPRH−/−* cells expressing wild-type hydrolase was 80% of wild-type cells, whereas overexpression of the inactive mutant enzyme in ADPRH−/−** cells did not increase significantly the activity of ADPRH−/− cells (Fig. 2A).
FIG. 2.
ADPRH activity in cells grown from +/+, +/−, and −/− embryos. (A) Western blot (top) of proteins (20 μg) in lysates of cells with the indicated ADPRH alleles showing a 39-kDa ADPRH. −/−* and −/−** indicate, respectively, KO cells overexpressing wild-type or mutant ADPRH. ADPRH was assayed as described in Materials and Methods. Data are means ± SEM of values from five experiments with triplicate assays. 1†, P = 0.0006; 2†, P = 0.0001; 3†, P = 0.0003; 4†, P = 0.0004; 5†, P = 0.023. Results were similar in five experiments. (B) Western blot of Gsα in cultured cells with the indicated ADPRH alleles that had been incubated with CT for the indicated time before separation of proteins and immunoblotting with antibodies against Gsα. Numbered arrowheads are as follows: 1, ADP-ribosylated Gsα; 2, 52-kDa Gsα; and 3, 45-kDa Gsα band.
Effect of CT on Gsα in ADPRH−/− embryonic cells and ADPRH−/− cells reconstituted with wild-type or mutant ADPRH.
To determine the effects of ADPRH activity on intoxication of cells by CT, wild-type, heterozygous, KO, and KO cells overexpressing wild-type or inactive mutant ADPRH genes were incubated with CT before evaluation by immunoblotting to detect alterations in Gsα, the major intracellular toxin substrate. These cells contained both 45- and 52-kDa forms of Gsα. CT-catalyzed ADP-ribosylation decreases the mobility of modified Gsα, seen in the KO cells after 2 h of toxin exposure, where the amount of 52-kDa Gsα was also markedly reduced (Fig. 2B). Continued accumulation of ADP-ribose Gsα was not seen, presumably because the modified protein is degraded, as reported previously (4). The data are consistent with release of the ADP-ribose from Gsα by ADPRH in wild-type and heterozygous cells but not in ADPRH−/− cells at a rate sufficient to largely reverse CT-catalyzed ADP-ribosylation. The mobility of Gsα was similar in all cells not exposed to toxin, whether KO or KO cells reconstituted with wild-type or mutant hydrolase, and amounts of Gsα were in general greater than those in toxin-treated cells.
The loss of Gsα following incubation of cells with CT was dependent on time of incubation of cells with toxin and occurred earlier in the KO than in the wild-type cells, consistent with their more extensive modification (Fig. 2B). Cells reconstituted with the mutant hydrolase exhibited an intermediate response. This observation is consistent with the finding that the mutant possesses some hydrolase activity. The amount of hydrolase per cell basis is similar to the amount of ADP-ribose-arginine in the cells, perhaps explaining why a relatively inactive mutant can be effective in modulating ADP-ribosylation (data not shown).
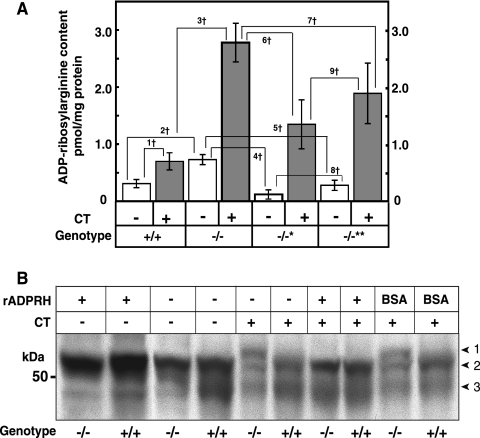
Effect of CT on ADP-ribosylarginine content of ADPRH−/− embryonic cells and ADPRH−/− cells reconstituted with wild-type or mutant ADPRH.
Consistent with the greater extent of modification of Gsα by toxin in ADPRH−/− cells, we found that the KO cell content of ADP-ribose-arginine protein was significantly higher than that of wild-type cells, both before and after incubation with CT (Fig. 3A). These data show that the more extensive modification of Gsα by toxin in KO cells was associated with enhanced accumulation of total ADP-ribose-(arginine)protein; absence of the hydrolase also resulted in elevated basal ADP-ribose-(arginine)protein. Lower levels of both basal and CT-catalyzed ADP-ribose-(arginine)protein were associated with overexpression of wild-type or mutant ADPRH in KO cells (Fig. 3A). Similar to the extent of modification of Gsα by CT in the heterozygous cells, KO cells overexpressing the mutant hydrolase had an ADP-ribose-(arginine)protein content intermediate between those of KO and wild-type cells. It was more similar to that of the wild-type than the KO cells, perhaps because the mutant hydrolase does have catalytic activity, albeit less than 0.01% that of the wild-type enzyme.
FIG. 3.
Effects of CT on ADP-ribosylated Gsα and other proteins in cultured cells of ADPRH+/+ and ADPRH−/− mice. (A) ADP-ribosylarginine content of cultured cells with indicated ADPRH alleles that had been incubated for 6 h without (open bar) or with (shaded bar) 1.0 μg of CT in 10 ml of DMEM. Data are means ± SEM of values from three experiments with triplicate assays. 1†, P = 0.0001; 2†, P = 0.0001; 3†, P = 0.0006; 4†, P = 0.000003; 5†, P = 0.00004; 6†, P = 0.0004; 7†, P = 0.018; 8†, P = 0.016; 9†, NS, not significant. (B) Effect of recombinant mouse ADPRH on Gsα from cultured cells incubated with CT. The cells were treated with CT (0.1 μg/ml) for 2 h in 5 ml of DMEM medium containing 10% fetal calf serum before homogenization. Samples (30 μg of protein) of homogenates from TCA-treated cells were incubated at 30°C for 1 h without or with 0.5 μg of ADPRH or bovine serum albumin followed by the addition of 5 μl of 4× SDS sample buffer, separation of protein by SDS-PAGE, and immunoblotting with antibodies against Gsα. Numbered arrowheads are as follows: 1, ADP-ribosylated Gsα; 2, 52-kDa Gsα; and 3, 45-kDa Gsα band.
Effect of rADPRH protein on CT-catalyzed modification of Gsα in ADPRH KO and wild-type cells.
It was possible that the effects on the electrophoretic mobility of Gsα seen in KO cells did not result from ADP-ribosylation of arginine but from some other alteration of the protein. To explore this possibility, lysates of CT-treated KO and wild-type cells were incubated with purified recombinant ADPRH synthesized in E. coli. After incubation with hydrolase, the mobility of Gsα from KO cells resembled that of the protein from untreated wild-type cells (Fig. 3B), consistent with the conclusion that ADP-ribosylation was responsible for the observed changes in migration of protein on SDS-PAGE.
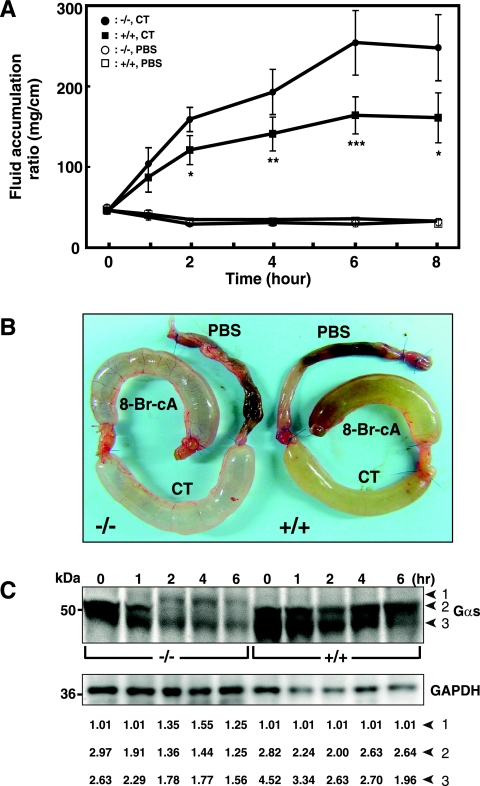
Effect of Gsα and fluid accumulation in CT and ADPRH genotype on intestinal loops.
The mouse intestinal loop model has been used extensively to investigate effects of CT and other enteric toxins (13). Accumulation of fluid following injection of toxin into a loop results in increased weight and distention of the intestinal segment. The effect of CT on fluid accumulation was quantified every 2 h and clearly increased with time; differences between wild-type and KO intestinal loops were evident (Fig. 4A). Differences in the behavior of Gsα in KO and wild-type intestinal loops after CT treatment were also notable. The extensive modification of the Gsα seen in KO lysates was not apparent in the wild-type (Fig. 4C), indicating that the CT effects were greatly magnified in KO mice.
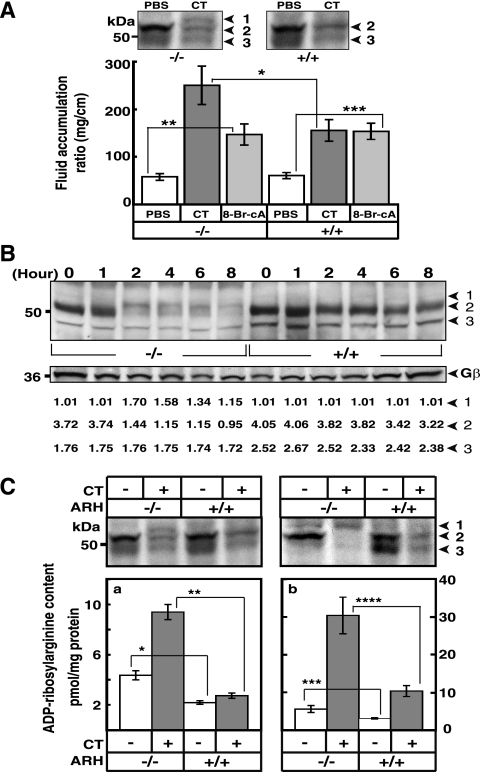
FIG. 4.
Effect of CT on accumulation of fluid and ADP-ribosylation of Gsα in intestinal loops of ADPRH wild-type and KO mice. (A) In each wild-type (square) or KO (circle) mouse, intestinal loops were injected with 0.2 ml of PBS without (□ or ○, respectively) or with (▪ or •, respectively) CT (0.5 μg) and were separately excised at the indicated time thereafter. The weight and length of each loop reported as milligrams per centimeter were used to measure of fluid accumulation. Data are means ± SEM (n = 6) of values for fluid accumulation in intestinal loops of ADPRH+/+ and ADPRH−/− mice. For the difference between two alleles incubated with CT, the P values are as follows: *, P = 0.004; **, P = 0.007; ***, P = 0.002. (B) Appearance of intestinal loops 6 h after injection of 0.2 ml of PBS without or with 0.5 μg of CT or 5 mM 8-bromo-cAMP (8-Br-cA). (C) The top blot shows Gsα in KO (−/−) or wild-type (+/+) intestinal loops after exposure to CT (0.5 μg) for 0 to 8 h as indicated. The bottom blot shows immunoreactive glyceraldehyde-3-phosphate dehydrogenase (GAPDH) on the same blot. Data are ratios of the densitometric value of the indicated Gsα band to the background, set at 1.0 (LAS 3000 Bioimaging system with Image Gauge, version 4.0, software). Numbered arrowheads are as follows: 1, ADPR-Gsα; 2, 52-kDa Gsα; and 3, 45-kDa Gsα.
Effects of CT and cAMP on fluid accumulation in the intestinal loops of ADPRH wild-type and KO animals.
As shown in Fig. 4A and B and 5A, CT enhanced fluid accumulation in intestinal loops of KO mice more than it did in those of wild-type mice. The effects of cAMP, however, on fluid accumulation were similar in both wild-type and KO mice (Fig. 5A), indicating that responsiveness to the downstream signaling molecule was apparently unchanged.
FIG. 5.
Effects of CT on ADP-ribosylated Gsα and other proteins in intestinal loops of ADPRH wild-type and KO mice. (A) Above, Gsα in lysates of epithelial cells from representative intestinal loops of mice with the indicated ADPRH alleles (−/− or +/+) after 6 h without (PBS) or with CT (0.5 μg). Numbered arrowheads are as follows: 1, ADP-ribosylated Gsα; 2, 52-kDa Gsα; 3, 45-kDa Gsα. Data below are means ± SEM (n = 6) of values for fluid accumulation (mg/cm) in intestinal loops of ADPRH+/+ and ADPRH−/− mice exposed for 6 h to PBS alone or containing 0.5 μg of CT or 5 mM 8-bromo-cAMP (8-Br-cA). *, P = 0.002 (for difference between two alleles incubated with CT); **, P = 0.0001; ***, P = 0.00006 (both, for difference between PBS and 8-bromo cAMP in −/− and +/+ mice). (B) Gsα is present in epithelial cells from intestinal loops of ADPRH−/− or ADPRH+/+ mice after exposure to 0.5 μg of CT for 0 to 8 h (top). Arrowheads are as defined for panel A. The immunoreactive β subunit of G (Gβ) is shown on the same blot (center). Data are the ratios of densitometric values of the indicated Gsα band to the background, set at 1.0 (LAS 3000 Bioimaging system with Image Gauge, version 4.0, software) of the immunoblot bands (bottom). (C) Proteins from whole intestine (left) or epithelial cells (right) of loops, incubated without or with 0.5 μg of CT for 6 h were immunoblotted for Gsα (top). Arrowheads are as defined for panel A. The graphs shows ADP-ribosylarginine content of proteins from whole intestine (left) or epithelial cells from loops (right) incubated for 6 h without (−) or with (+) 0.5 μg CT. Data are means ± SEM of values from two loops of two mice in each of three experiments. *, P = 0.006; **, P = 0.0006; ***, P = 0.014; ****, P = 0.005.
Effect of CT on ADP-ribosylarginine content of intestinal loops of ADPRH wild-type and KO animals.
Because CT appears to affect the epithelial cells on the inner surface of the intestine, this superficial layer of lining cells was removed and analyzed separately from the remainder of the intestinal tissue. As shown in Fig. 5A and B, electrophoretic mobility of Gsα in epithelial cells from KO loops incubated with toxin for 6 h was less than that of Gsα in wild-type tissue. The amount of Gsα detected by immunoblotting decreased significantly with time of exposure of tissue to CT (Fig. 5B). To determine whether the lower mobility resulted from ADP-ribosylation or some other modification, membranes after toxin treatment were incubated with recombinant hydrolase in the presence of Mg2+ and/or dithiothreitol. As seen in the KO cell experiments (Fig. 3B), incubation with hydrolase converted the modified Gsα from the KO loops to a protein with mobility similar to that of the protein from control loops (data not shown). The ADP-ribose-(arginine)protein content of loops from KO mice without toxin exposure was significantly greater than that of the loops from the wild-type mice (Fig. 5C). Similarly, following incubation with toxin, the ADP-ribose-(arginine)protein content in loops from KO mice exceeded that of loops from the wild-type animals (Fig. 5C). Further, the ADP-ribose-(arginine)protein content of the epithelial cells isolated from mutant loops, incubated either with or without CT, greatly exceeded that of intact mutant loops (Fig. 5C). Thus, the observations with the KO and wild-type loops are in agreement with the findings from the related cultured cells. The studies support a role for hydrolase in determining cell sensitivity to CT.
DISCUSSION
Two vicinal aspartate residues at positions 60 and 61 (in exon 2 of the open reading frame) had been shown to be critical for hydrolase activity in vitro; mutant enzyme in which both residues had been replaced with alanine exhibited <0.01% of the activity of the wild-type enzyme (18). Taking advantage of this fact, exon 2 was replaced in the targeting vector used to generate the KO mouse. If our hypothesis regarding the potential role of hydrolase were correct, the hydrolase KO cells and the small intestine of a KO mouse would exhibit enhanced sensitivity to CT. The data demonstrate that sensitivity to CT was enhanced in both the small intestine and cells cultured from ADPRH KO mice. The ADP-ribose-(arginine)protein content, both basal and following incubation with CT, and the extent of modification of Gsα seen by Western blotting were greater in KO than wild-type cells. Both were decreased in KO cells by overexpression of wild-type hydrolase but not an inactive mutant. In the intestinal loop assay, CT effects, as quantified by fluid accumulation, were greater in the KO than the wild-type mice, but the responses to cAMP, the “second messenger” that is elevated by CT as it is by physiological agonists that activate adenylyl cyclase, were identical in KO and wild-type tissues.
CT increases adenylyl cyclase activity, leading to an increase in cellular cAMP through ADP-ribosylation and activation of Gsα, which, in turn, activates the adenylyl cyclase catalytic unit. The effects of CT may differ in KO and wild-type cells, since in wild-type cells ADPRH is present and can cleave the ADP-ribose-(arginine)Gsα modification to some extent, reversing CT effects. This reaction does not occur in ADPRH−/− cells, and hence the response to CT is greater. Since cAMP is downstream of ADP-ribose Gsα, its effects are independent of ADPRH.
CT has NAD glycohydrolase as well as NAD:arginine ADP-ribosyltransferase activity (24, 28). In addition to modifying a specific arginine in its “true” substrate, i.e., the one that is responsible for its pathogenicity, the toxin can ADP-ribosylate guanidine and other arginine moieties in a number of proteins (28). In this regard, CT is similar to mammalian transferases that can ADP-ribosylate arginine in a variety of proteins (30). In cultured cells and membrane preparations, Gsα appears to be the protein that is predominantly modified (8), as demonstrated here for both the intestinal loops and cells.
Specific intracellular protein substrates for the hydrolase have not been defined. In vitro, hydrolase exhibits a broad substrate specificity and can cleave ADP-ribose attached to arginines in a number of proteins, including Gsα (21, 30, 43). In the latter instance, however, although recombinant hydrolase was shown to cleave ADP-ribose-Gsα, the enzyme was present in ∼8- to 20-fold molar excess over substrate, and under these extreme conditions, only a fraction of the ADP-ribosylated Gsα was cleaved. In our studies as well, a large excess of hydrolase was necessary to observe cleavage of ADP-ribosyl-Gsα. These in vitro data would suggest that ADP-ribosylated Gsα is a poor hydrolase substrate. The in vivo results are, therefore, surprising in that the KO genotype had a marked effect on tissue content of ADP-ribose-Gsα and ADP-ribose-arginine as well as fluid accumulation, consistent with a major role for hydrolase in the wild-type mouse. Indeed, in vitro activity appears to have been an unreliable predictor of in vivo findings. In vitro results cannot be extrapolated to in vivo effects of hydrolase, perhaps due to the localization of hydrolase in the intestinal epithelial cells or the activation of the hydrolase in the cellular environment. It remains to be seen whether the action of other bacterial toxin ADP-ribosyltransferases may be modified by specific cellular hydrolases.
The hydrolase enzymatic activity is dependent on two aspartate residues at positions 60 and 61 (18); their replacement with alanine results in a dramatic reduction in activity to <0.01% of that the intact enzyme, although the mutant protein appears structurally intact and retains its ability to bind ADP-ribose. It can, therefore, serve as a control for hydrolase function in overexpression experiments. KO cells overexpressing the mutant hydrolase had an ADP-ribose-(arginine)protein content intermediate between that of KO and wild-type cells. It seemed surprising that they were more similar to the wild-type than to the KO cells. In fact, the differences between cells expressing mutant and wild-type hydrolase were not statistically significant, whereas the difference between KO cells and cells expressing mutant hydrolase were statistically significant (Fig. 3A). Thus, the low residual activity of the mutant may be sufficient to alter the responsiveness of the cells to CT. On reflection, however, although mutant enzyme activity is less than 0.01% that of the wild-type hydrolase, the amount of overexpressed protein is similar on a molar basis to the ADP-ribose-(arginine)protein content of the cell. Thus, even a single turnover could be adequate to reverse some amount of CT-catalyzed modification and delay the accumulation of ADP-ribose-(arginine)protein in intoxicated cells.
Because only one ADPRH gene has thus far been identified in human and rodent tissues, it was not surprising that hydrolase KO mice exhibited enhanced sensitivity to CT, although cells do contain enzymes that might metabolize ADP-ribosylated proteins in different reactions. ADP-ribose is subject to cleavage of the AMP-phosphoribosyl bond by pyrophosphatases, which would generate phosphoribosylated protein (1, 19, 35). Phosphoribose can be hydrolyzed by phosphatases, which would yield a ribosylarginine(protein). Degradation of ADP-ribosylarginine in proteins could occur as a result of ADPRH activity or the combined action of pyrophosphatases and phosphatases. Alternatively, disappearance of ADP-ribose Gsα may result from rapid degradation by a proteolytic pathway. In KO cells and tissues with increased amounts of ADP-ribosylated proteins, the destruction of Gsα was apparently accelerated. These findings are consistent with prior findings (4) that CT-catalyzed ADP-ribosylation of Gsα resulted in degradation of the protein. Presumably, the decrease in Gsα content alters the responses of cells to hormones and the function of other signaling pathways that intersect or are influenced by those dependent on Gsα.
The accumulation of ADP-ribosylated Gsα in the KO mice was presumably a result of failure, in the absence of ADPRH, to cleave the ribosyl-guanidino bond at a rate equal to that of its synthesis by CT. It appears that the hydrolase was important in counteracting CT-catalyzed ADP-ribosylation and that the greater persistence of CT-modified Gsα in the KO than wild-type cells resulted in accelerated loss of Gsα. The plasma membrane of epithelial cells contains substantial numbers of ganglioside GM1 molecules, which bind toxin and thus facilitate ADP-ribosylation of Gsα (15). In disease, presumably, the capacity of cells to counteract intoxication by hydrolysis of the ADP-ribose-arginine linkage is overwhelmed. The data support the conclusion that the hydrolase might serve as a modifier gene in cholera and perhaps other diseases caused by toxin ADP-ribosyltransferases.
Acknowledgments
We thank Lee Weinstein for his generous gift of anti-Gsα antibodies. We also thank Martha Vaughan and Vincent Manganiello for helpful discussions and critical review of the manuscript.
This research was supported by the Intramural Research Program at the National Heart, Lung, and Blood Institute, National Institutes of Health.
Footnotes
Published ahead of print on 25 May 2007.
REFERENCES
- 1.Abdelraheim, S. R., D. G. Spiller, and A. G. McLennan. 2003. Mammalian NADH diphosphatases of the Nudix family: cloning and characterization of the human peroxisomal NUDT12 protein. Biochem. J. 374:329-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoki, K., J. Kato, M. T. Shoemaker, and J. Moss. 2005. Genomic organization and promoter analysis of the mouse ADP-ribosylarginine hydrolase gene. Gene 351:83-95. [DOI] [PubMed] [Google Scholar]
- 3.Cassel, D., and Z. Selinger. 1977. Mechanism of adenylate cyclase activation by cholera toxin: inhibition of GTP hydrolysis at the regulatory site. Proc. Natl. Acad. Sci. USA 74:3307-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, F. H., and H. R. Bourne. 1989. Cholera toxin induces cAMP-independent degradation of Gs. J. Biol. Chem. 264:5352-5357. [PubMed] [Google Scholar]
- 5.Coburn, J., and D. M. Gill. 1991. ADP-ribosylation of p21ras and related proteins by Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 59:4259-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng, Q., J. Sun, and J. T. Barbieri. 2005. Uncoupling Crk signal transduction by Pseudomonas exoenzyme T. J. Biol. Chem. 280:35953-35960. [DOI] [PubMed] [Google Scholar]
- 7.Field, M., D. Fromm, Q. al-Awqati, and W. B. Greenough III. 1972. Effect of cholera enterotoxin on ion transport across isolated ileal mucosa. J. Clin. Investig. 51:796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishman, P. H. 1990. Mechanism of action of cholera toxin, p. 127-140. In J. Moss, and M. Vaughan, (ed.), ADP-ribosylating toxins and G proteins: insights into signal transduction. ASM Press, Washington, DC.
- 9.Freissmuth, M., and A. G. Gilman. 1989. Mutations of Gs alpha designed to alter the reactivity of the protein with bacterial toxins. Substitutions at ARG187 result in loss of GTPase activity. J. Biol. Chem. 264:21907-21914. [PubMed] [Google Scholar]
- 10.Ganesan, A. K., D. W. Frank, R. P. Misra, G. Schmidt, and J. T. Barbieri. 1998. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J. Biol. Chem. 273:7332-7337. [DOI] [PubMed] [Google Scholar]
- 11.Gill, D. M. 1976. The arrangement of subunits in cholera toxin. Biochemistry 15:1242-1248. [DOI] [PubMed] [Google Scholar]
- 12.Glowacki, G., R. Braren, K. Firner, M. Nissen, M. Kuhl, P. Reche, F. Bazan, M. Cetkovic-Cvrlje, E. Leiter, F. Haag, and F. Koch-Nolte. 2002. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 11:1657-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hitotsubashi, S., Y. Fujii, H. Yamanaka, and K. Okamoto. 1992. Some properties of purified Escherichia coli heat-stable enterotoxin II. Infect. Immun. 60:4468-4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochmann, H., S. Pust, G. von Figura, K. Aktories, and H. Barth. 2006. Salmonella enterica SpvB ADP-ribosylates actin at position arginine-177-characterization of the catalytic domain within the SpvB protein and a comparison to binary clostridial actin-ADP-ribosylating toxins. Biochemistry 45:1271-1277. [DOI] [PubMed] [Google Scholar]
- 15.Holmgren, J., I. Lonnroth, J. Mansson, and L. Svennerholm. 1975. Interaction of cholera toxin and membrane GM1 ganglioside of small intestine. Proc. Natl. Acad. Sci. USA 72:2520-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobson, M. K., D. M. Payne, R. Alvarez-Gonzalez, H. Juarez-Salinas, J. L. Sims, and E. L. Jacobson. 1984. Determination of in vivo levels of polymeric and monomeric ADP-ribose by fluorescence methods. Methods Enzymol. 106:483-494. [DOI] [PubMed] [Google Scholar]
- 17.Joyner, A. L. 2000. Gene targeting: a practical approach. Oxford University Press, New York, NY.
- 18.Konczalik, P., and J. Moss. 1999. Identification of critical, conserved vicinal aspartate residues in mammalian and bacterial ADP-ribosylarginine hydrolases. J. Biol. Chem. 274:16736-16740. [DOI] [PubMed] [Google Scholar]
- 19.Lin, S., L. Gasmi, Y. Xie, K. Ying, S. Gu, Z. Wang, H. Jin, Y. Chao, C. Wu, Z. Zhou, R. Tang, Y. Mao, and A. G. McLennan. 2002. Cloning, expression and characterisation of a human Nudix hydrolase specific for adenosine 5′-diphosphoribose (ADP-ribose). Biochim. Biophys. Acta 1594:127-135. [DOI] [PubMed] [Google Scholar]
- 20.Lowery, R. G., and P. W. Ludden. 1990. Endogenous ADP-ribosylation in prokaryotes, p. 459-478. In J. Moss, and M. Vaughan, (ed.), ADP-ribosylating toxins and G proteins: insights into signal transduction. ASM Press, Washington, DC.
- 21.Maehama, T., H. Nishina, and T. Katada. 1994. ADP-ribosylarginine glycohydrolase catalyzing the release of ADP-ribose from the cholera toxin-modified alpha-subunits of GTP-binding proteins. J. Biochem. (Tokyo) 116:1134-1138. [DOI] [PubMed] [Google Scholar]
- 22.Moss, J., S. Garrison, N. J. Oppenheimer, and S. H. Richardson. 1979. NAD-dependent ADP-ribosylation of arginine and proteins by Escherichia coli heat-labile enterotoxin. J. Biol. Chem. 254:6270-6272. [PubMed] [Google Scholar]
- 23.Moss, J., M. K. Jacobson, and S. J. Stanley. 1985. Reversibility of arginine-specific mono(ADP-ribosyl)ation: identification in erythrocytes of an ADP- ribose-l-arginine cleavage enzyme. Proc. Natl. Acad. Sci. USA 82:5603-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss, J., V. C. Manganiello, and M. Vaughan. 1976. Hydrolysis of nicotinamide adenine dinucleotide by choleragen and its A protomer: possible role in the activation of adenylate cyclase. Proc. Natl. Acad. Sci. USA 73:4424-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moss, J., S. J. Stanley, M. S. Nightingale, J. J. Murtagh, Jr., L. Monaco, K. Mishima, H. C. Chen, K. C. Williamson, and S. C. Tsai. 1992. Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J. Biol. Chem. 267:10481-10488. [PubMed] [Google Scholar]
- 26.Moss, J., S. J. Stanley, and N. J. Oppenheimer. 1979. Substrate specificity and partial purification of a stereospecific NAD- and guanidine-dependent ADP-ribosyltransferase from avian erythrocytes. J. Biol. Chem. 254:8891-8894. [PubMed] [Google Scholar]
- 27.Moss, J., S. C. Tsai, R. Adamik, H. C. Chen, and S. J. Stanley. 1988. Purification and characterization of ADP-ribosylarginine hydrolase from turkey erythrocytes. Biochemistry 27:5819-5823. [DOI] [PubMed] [Google Scholar]
- 28.Moss, J., and M. Vaughan. 1977. Mechanism of action of choleragen. Evidence for ADP-ribosyltransferase activity with arginine as an acceptor. J. Biol. Chem. 252:2455-2457. [PubMed] [Google Scholar]
- 29.Okazaki, I. J., H. J. Kim, and J. Moss. 1996. Cloning and characterization of a novel membrane-associated lymphocyte NAD:arginine ADP-ribosyltransferase. J. Biol. Chem. 271:22052-22057. [DOI] [PubMed] [Google Scholar]
- 30.Okazaki, I. J., and J. Moss. 1998. Glycosylphosphatidylinositol-anchored and secretory isoforms of mono-ADP-ribosyltransferases. J. Biol. Chem. 273:23617-23620. [DOI] [PubMed] [Google Scholar]
- 31.O'Neal, C. J., E. I. Amaya, M. G. Jobling, R. K. Holmes, and W. G. Hol. 2004. Crystal structures of an intrinsically active cholera toxin mutant yield insight into the toxin activation mechanism. Biochemistry 43:3772-3782. [DOI] [PubMed] [Google Scholar]
- 32.Oppenheimer, N. J. 1978. Structural determination and stereospecificity of the choleragen-catalyzed reaction of NAD+ with guanidines. J. Biol. Chem. 253:4907-4910. [PubMed] [Google Scholar]
- 33.Peterson, J. W., and L. G. Ochoa. 1989. Role of prostaglandins and cAMP in the secretory effects of cholera toxin. Science 245:857-859. [DOI] [PubMed] [Google Scholar]
- 34.Price, S. R., A. Barber, and J. Moss. 1990. Structure-function relationships of Guanine nucleotide-binding proteins, p. 397-424. In J. Moss, and M. Vaughan, (ed.), ADP-ribosylating toxins and G proteins: insights into signal transduction. ASM Press, Washington, DC.
- 35.Ribeiro, J. M., A. Carloto, M. J. Costas, and J. C. Cameselle. 2001. Human placenta hydrolases active on free ADP-ribose: an ADP-sugar pyrophosphatase and a specific ADP-ribose pyrophosphatase. Biochim. Biophys. Acta 1526:86-94. [DOI] [PubMed] [Google Scholar]
- 36.Rohrer, H., W. Zillig, and R. Mailhammer. 1975. ADP-ribosylation of DNA-dependent RNA polymerase of Escherichia coli by an NAD+: protein ADP-ribosyltransferase from bacteriophage T4. Eur. J. Biochem. 60:227-238. [DOI] [PubMed] [Google Scholar]
- 37.Takada, T., K. Iida, and J. Moss. 1993. Cloning and site-directed mutagenesis of human ADP-ribosylarginine hydrolase. J. Biol. Chem. 268:17837-17843. [PubMed] [Google Scholar]
- 38.Takahashi, A. 1998. Establishment of fibroblast cutures, p. 2.1.1-12. In J. S. Bonifacino, M. Dasso, J. B. Harford, J. Lippincotte-schwartz, K. M. Yamada, (ed.), Current protocols in cell biology, vol. I. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 39.Terashima, M., H. Osago, N. Hara, Y. Tanigawa, M. Shimoyama, and M. Tsuchiya. 2005. Purification, characterization and molecular cloning of glycosylphosphatidylinositol-anchored arginine-specific ADP-ribosyltransferases from chicken. Biochem. J. 389:853-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandekerckhove, J., B. Schering, M. Barmann, and K. Aktories. 1988. Botulinum C2 toxin ADP-ribosylates cytoplasmic beta/gamma-actin in arginine 177. J. Biol. Chem. 263:696-700. [PubMed] [Google Scholar]
- 41.Vandekerckhove, J., B. Schering, M. Barmann, and K. Aktories. 1987. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett. 225:48-52. [DOI] [PubMed] [Google Scholar]
- 42.Van Dop, C., M. Tsubokawa, H. R. Bourne, and J. Ramachandran. 1984. Amino acid sequence of retinal transducin at the site ADP-ribosylated by cholera toxin. J. Biol. Chem. 259:696-698. [PubMed] [Google Scholar]
- 43.Williamson, K. C., and J. Moss. 1990. Mono-ADP-ribosyltransferases and ADP-ribosylarginine hydrolase: a mono-ADP-ribosylation cycle in animal cells, p. 493-510. In J. Moss, and M. Vaughan, (ed.), ADP-ribosylating toxins and G proteins: insights into signal transduction. ASM Press, Washington, DC.
- 44.Zhang, R. G., D. L. Scott, M. L. Westbrook, S. Nance, B. D. Spangler, G. G. Shipley, and E. M. Westbrook. 1995. The three-dimensional crystal structure of cholera toxin. J. Mol. Biol. 251:563-573. [DOI] [PubMed] [Google Scholar]
- 45.Zolkiewska, A., M. S. Nightingale, and J. Moss. 1992. Molecular characterization of NAD:arginine ADP-ribosyltransferase from rabbit skeletal muscle. Proc. Natl. Acad. Sci. USA 89:11352-11356. [DOI] [PMC free article] [PubMed] [Google Scholar]