Abstract
Alternative splicing involving the 3′ tandem splice site NAGNAG sequence may play a role in the structure-function diversity of proteins. However, how 3′ tandem splice site utilization is determined is not well understood. We previously demonstrated that 3′ NAGNAG-based wobble splicing occurs mostly in a tissue- and developmental stage-independent manner. Bioinformatic analysis reveals that the nucleotide preceding the AG dinucleotide may influence 3′ splice site utilization; this is also supported by an in vivo splicing assay. Moreover, we found that the intron sequence plays an important role in 3′ splice site selection for NAGNAG wobble splicing. Mutations of the region between the branch site and the NAGNAG 3′ splice site, indeed, affected the ratio of the distal/proximal AG selection. Finally, we found that single nucleotide polymorphisms around the NAGNAG motif could affect the splice site choice, which may lead to a change in mRNA patterns and influence protein function. We conclude that the NAGNAG motif and its upstream region to the branch point sequence are required for 3′ tandem splice site selection.
Introns of precursor mRNA are removed by splicing, and exons are ligated to form mature mRNAs, which can be translated to produce proteins (15, 27). In higher eukaryotes, the essential cis elements of an intron include the 5′ splice site (GT), the branch point sequence (BPS), the polypyrimidine tract (PPT), and the 3′ splice site (AG) (5). The splicing reaction involves two catalytic steps, as follows: (i) the branch point attacks the 5′ splice site to generate the splicing intermediates, and (ii) the released 5′ exon attacks at the 3′ splice site to generate the ligated exons and lariat intron. These two steps occur in the spliceosome complex containing five small nuclear RNAs (U1, U2, U4/U6, and U5) and more than 150 protein factors (12, 28, 30). Recognition of the splice sites needs to be precise and involves an interaction between the cis elements and trans-acting factors (23).
Alternative splicing has recently emerged as a major mechanism of posttranscriptional regulation in the human genome and occurs in >60% of human genes (3); the high frequency of this alternative splicing in human is supported by expressed sequence tag (EST)-based database analysis (4, 16). Recently, Wen et al. (38) reported that very short alternative splicing in the human genome might alter protein structures and thus influence protein function. A subtle variation of transcripts may come from wobble splicing at the 5′ splice site GTNGT or the 3′ splice site NAGNAG sequence, thus creating single-amino-acid insertion and deletion (InDel) isoforms (22, 35). Recently, Hiller et al. (14) and Tadokoro et al. (34) also demonstrated genome-wide distribution of the NAGNAG sequence at the 3′ splice site in human genes, which results in protein isoforms with one amino acid insertion or deletion. Such wobble splicing is predicted to exist in 30% of human genes and is active in at least 5% of genes, according to an EST database search. Although wobble splicing only subtly changes the protein sequence, it might increase the functional diversity of proteins; examples include PAX3, PAX7, and insulin-like growth factor receptor (10, 18, 37). Alternative splicing at the 5′ or 3′ tandem splice site may play important roles during the progression of diseases, as the reported cases include the human WT1 and ABCA4 genes (11, 26).
Alternative splicing involving a 5′ or a 3′ splice site choice can be regulated in different cell types or at different stages during development (24). However, wobble splicing at tandem NAGNAG motifs is independent of tissue type and may not be specifically regulated (34, 35). Although the NAGNAG motif is common in human genes, only a small number of the genes have been confirmed to generate wobble splicing isoforms (12, 34, 35). In most cases, one AG in the 3′ tandem splice site is constitutively used, suggesting that the NAGNAG motif is not sufficient to determine wobble splicing sites. Therefore, the answer to how the 3′ splice site is determined in wobble splicing remains elusive.
According to the linear scanning mechanism model (7, 9, 32, 33), the spliceosome recognizes the branch point and scans downstream for the first AG. Although the distance between two adjacent AGs is less than 6 nucleotides, the distal 3′ splice site is primarily selected (9). A recent report indicates that several features of wobble splicing are different from those of constitutive splicing, such as high conservation of the intron sequence upstream of the tandem splice site and the abundance of cis elements near the tandem splice site (1). Because the regulation of NAGNAG wobble splicing has not been well characterized in the past, we experimentally addressed this question and demonstrated that the NAGNAG motif is not sufficient for determining wobble splicing and that the intronic sequence may make a significant contribution.
MATERIALS AND METHODS
RNA extraction and cDNA preparation.
cDNA was prepared from human and mouse cell lines as described previously (17). Total RNA (10 μg) from cell lines was used as the template for cDNA preparation in a 20-μl reaction mixture. In addition to RNA extracted from cell lines, RNA samples from normal human adult and fetal tissues and normal mouse adult tissue samples (brain, liver, lung, heart, and kidney) were obtained from Clontech (Palo Alto, CA). Reverse transcription was carried out using 2.5 μg of poly(A)+ RNA, oligo(dT)15, and SuperScript II reverse transcriptase according to the manufacturer's instructions (Invitrogen, Carlsbad, CA).
Plasmid constructs.
The genomic DNA of ING4 exons 4 to 5 (exon 4-5) was amplified by PCR from the AZ-521 cell line genomic DNA (obtained from the ATCC) using the primer pair ING4-exon 4-F (forward) and ING4-exon 5-R (reverse). The amplified fragments were cloned into the pGEM-T easy vector (Promega, Madison, WI). Following cloning, several clones were randomly selected, and their sequences were determined by an autosequencer. Subsequently, the minigene construct was generated by subcloning the genomic DNA of ING4 exon 4-5 into the EcoRI site of the pEGFP-C1 vector (Clontech). The intron-exchanging minigene constructs were generated by overlapping PCR experiments as follows: the plasmid human minigene was used as a template for the first PCR with primers ING4-exon 4-F/ING4-exon 4-R or ING4-exon 5-F/ING4-exon 5-R, and these two PCR products were combined. The second PCR was performed using the mouse minigene as template and subcloned into the pGEM-T easy vector and pEGFP-C1 expression vector. The mutant constructs were generated by PCR with primers (see Table S1 in the supplemental material) containing the desired mutations using human and mouse minigenes as templates. The PCR fragments containing mutations were then subcloned into the pEGFP-C1 expression vector. The sequences of all the plasmids were verified using an autosequencer.
Splicing analysis in vivo.
The minigene plasmids were introduced into human AZ-521 and mouse B16-F10 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications. Forty-eight hours after transfection, total RNA was extracted, and the wobble splicing products were analyzed by capillary electrophoresis (35) using primer pair EGFP-F/specific gene primers as described above (see Table S2 in the supplemental material).
Capillary electrophoresis analysis.
PCRs were performed to amplify cDNA in a 20-μl mixture containing 10× PCR buffer, fluorescently labeled primers (6-carboxyfluorescein [FAM])/complementary primers, deoxynucleoside triphosphates, and Takara Taq DNA polymerase (Takara Shuzo Company, Shiga, Japan). PCR was conducted at 94°C for 5 min followed by 26 cycles at 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, and extended at 72°C for 10 min. The samples were then cooled at 4°C. One microliter of the PCR mixture was diluted to 10 μl with formamide (Applied Biosystems, Foster City, CA), and 1 μl of ROX 350 fluorescent size standard (Applied Biosystems) was added. The mixture was then denatured at 95°C for 5 min and cooled at 4°C. Amplified PCR products were separated on an ABI 3100-avant DNA analyzer using polymer 3100 POP4 (Applied Biosystems) and then quantified with GeneScan 3.7 software. Ratios of the wobble splicing isoforms was determined as the peak area for the individual isoform/total area for wobble splicing isoforms × 100.
Sequence validation of cDNA single nucleotide polymorphisms.
Genomic DNA was prepared from 11 human cancer cell lines, including AGS, AZ521, KatoIII, NUGC, TSGH, Hep3B, HepG2, HEL299, huh7, LNCap, and HeLa. The splice junction fragments of the LAP1B, C14orf105, TLR3 and AP1GBP1 genes were amplified by PCR using genomic DNA as a template with individual primer pairs (LAP1B, LAP1B-1F/LAP1B-R; C14orf105, C140rf105-F/C14orf105-R; TLR3, TLR3-1F/TLR3-R; and AP1GBP1, AP1GBP1-F/AP1GBP1-R). Gene-specific PCR was conducted at 94°C for 5 min and then 35 cycles of 94°C for 20 s, 58°C for 30 s, and 72°C for 30 s, and a final extension phase at 72°C for 10 min using a PCR thermocycler and Takara Taq polymerase (Takara; Shiga, Japan). Following PCR amplification, the products were purified using a PCR clean-up kit (QIAGEN, Hilden, Germany) and subsequently sequenced by an autosequencer.
RESULTS
The tandem splice site involves wobble splicing.
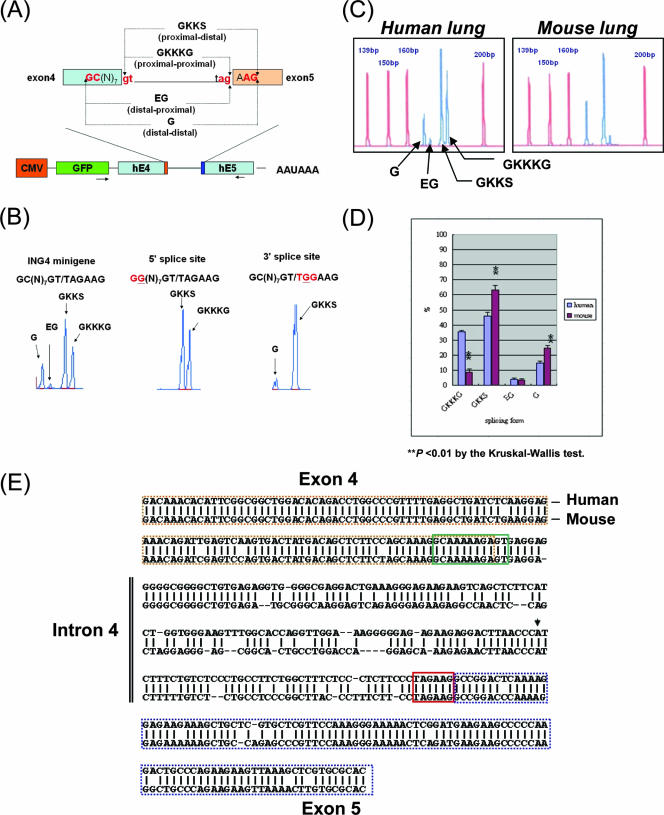
The ING4 gene contains the 5′ GC(N)7GT and the 3′ TAGAAG tandem splice sites in intron 4, which may generate four wobble splicing isoforms (35, 36). To investigate the molecular mechanism of such wobble splicing, we used a minigene approach. The reporter constructs contained the genomic sequence of exon 4, intron 4, and exon 5 of the human ING4 gene, which were introduced into AZ521 cells. The transcripts produced from the minigene were analyzed by capillary electrophoresis using primers specific for the minigenes (Fig. 1A). As expected, the wild-type ING4 minigene generated four splicing isoforms by wobble splicing. When a mutation was introduced at the distal GC dinucleotide at the 5′ splice site, 5′ wobble splicing was abolished, and two isoforms, GKKKG and GKKS, were generated through tandem splicing only at the 3′ splice sites (Fig. 1B). The minigene containing a mutant 3′ tandem splice site generated the isoforms GKKS and G by wobble splicing at the wild-type 5′ splice site (Fig. 1B, right panel). These results indicated that the tandem splice site is essential for wobble splicing. In the case of ING4, the 5′ and 3′ tandem splicing events seemed to be independent of one another.
FIG. 1.
ING4 minigene splicing assay. (A) Schematic illustration of the pGFP-C1-ING4 minigene in which the ING4 genomic DNA fragment from exon 4 to exon 5 is placed downstream of green fluorescent protein (GFP) and is controlled by the cytomegalovirus (CMV) promoter. The PCR primers (EGFP-F/ING4-R) are indicated by arrows. Four different isoforms of ING4 (ING4GKKKG, ING4GKKS, ING4EG, and ING4G) are caused by alternative splicing at the 5′ and 3′ tandem motifs. (B) The 5′ and 3′ tandem splice sites were mutated in the human ING4 minigenes. After transfection into AZ521 cells, total RNA was isolated and analyzed by PCR and capillary electrophoresis. The splicing profiles of the wild-type (left panel), 5′ GC(N)7GT→GG(N)7GT (middle panel), and 3′ TAGAAG→TGGAAG (right panel) are shown by blue peaks. (C) A representative capillary electrophoresis scan shows the percentages of four ING4 wobble splicing isoforms (GKKKG, 182 bp; GKKS, 179 bp; EG, 173 bp; G, 170 bp) in human and mouse lung tissues (blue peaks). The reference peaks of the GS350 ROX internal size standard (Applied Biosystems) are indicated in red. (D) The ING4 splicing pattern in adult human tissues (brain, liver, lung, heart, and kidney) was compared to that in adult mouse tissues. The values are the means ± standard deviations obtained from three independent experiments (**, P < 0.01 using the Kruskal-Wallis test). (E) Alignment of the human and mouse genomic sequences of ING4 exon 4-5. Exon 4 is indicated by a dashed brown outline; exon 5 is indicated by a dashed blue outline. The putative branch point (AACCCAT) is indicated by an arrowhead. The 5′ (GC(N)7GT) and 3′ (TAGAAG) tandem splice sites are boxed with a solid green line and a red line, respectively.
The nucleotide preceding AG of the 3′ tandem splice site could affect 3′ wobble splicing.
Through a search of an EST database, we obtained 441 genes that may undergo wobble splicing at the 3′ NAGNAG splice site. As shown in Table S3 in the supplemental material, we found that the CAGCAG sequence is the most common (170 genes), followed by TAGCAG (83 genes), CAGAAG (63 genes), TAGAAG (33 genes), and AAGCAG (33 genes). By comparing the individual “N” nucleotides in the NAGNAG tandem motif, we found that the frequency of the “N” proximal to the BPS was as follows: C (59.4%) > T (29.0%) > A (10.2%) > G (1.4%), whereas the distal “N” site was C (66.0%) > A (23.1%) > G (6.1%) > T (4.8%). Interestingly, GAG appeared at a very low frequency at both the proximal and distal 3′ splice sites (1.4% and 6.1%, respectively), and the distal site had a low preference for TAG (4.8%). However, bioinformatic results could not completely represent the influence of the “N” nucleotide preceding AG, since the splice site selection is interfered with by neighboring adjacent sequences. Also, the coverage of the EST database might not be comprehensive enough. Moreover, Hiller et al. (14) reported that the tandem acceptors are biased toward intron phase 1 (40% for phase 0, 43% for phase 1, and 17% for phase 2), which is significantly different from all human intron (46% for phase 0, 33% for phase 1, and 21% for phase 2). To experimentally verify the influence of the “N” nucleotide preceding AG in greater detail, we used the human ING4 E4-I4-E5 minigenes (5′ splice site mutant), in which only the “N” nucleotide preceding both the proximal and the distal AG was changed. By using 16 mutation minigene constructs with a single ING4 E4-I4-E5 environment, we demonstrated that the “N” nucleotide preceding AG plays a critical role in the 3′ wobble splicing. When A, T, or C was substituted preceding AG at either site, selection of the 3′ splice site varied among the ING4 minigenes (Table 1). Consistent with the result obtained from the database, GAG was inefficiently used. Interestingly, we observed certain difference between experimental and bioinformatics analyses of the 16 NAGNAG sequence combinations (Table 1, ING4 minigene, and see Table S3 in the supplemental material for over 400 reference genes). Thus, the neighboring adjacent sequences of the NAGNAG motif could affect the efficiency of AG selection.
TABLE 1.
Summary of subtle wobble splicing that utilizes two successive 3′ splice sites
| Tandem motifa | AG selection (%)b
|
|
|---|---|---|
| Proximal position | Distal position | |
| AAG AAG | 8.8 | 91.2 |
| AAG TAG | 3.0 | 97.0 |
| AAG CAG | 1.5 | 98.5 |
| AAG GAG | 81.2 | 18.8 |
| TAG AAG | 45.1 | 54.9 |
| TAG TAG | 20.2 | 79.8 |
| TAG CAG | 7.9 | 92.1 |
| TAG GAG | 96.8 | 3.2 |
| CAG AAG | 48.3 | 51.7 |
| CAG TAG | 26.0 | 74.0 |
| CAG CAG | 12.1 | 87.9 |
| CAG GAG | 96.9 | 3.1 |
| GAG AAG | 0 | 100 |
| GAG TAG | 0 | 100 |
| GAG CAG | 0 | 100 |
| GAG GAG | 1.9 | 98.1 |
The 5′ tandem splice site mutation mini-genes (H-H-H5′ site mutant) in which the “N” position was replaced with A, T, C, or G were transfected into human AZ521 cells, and the wobble splicing patterns were analyzed by capillary electrophoresis.
The ratio of the distal or proximal AG selection was determined by dividing the individual peak area by the total area.
The intron sequence influences wobble splicing of the ING4 intron 4.
According to our previous study, we observed that the percentage of ING4 exon 4-5 wobble splicing varies among different species (35). Compared to the level of human splicing, that of mouse ING4GKKKG splicing was low (only one-fourth that of human), but the level of mouse ING4G splicing was high (about 1.5-fold) compared to that of human splicing (Fig. 1C and D). Such wobble splicing variations of ING4 may result from genomic sequence differences between species (Fig. 1E).
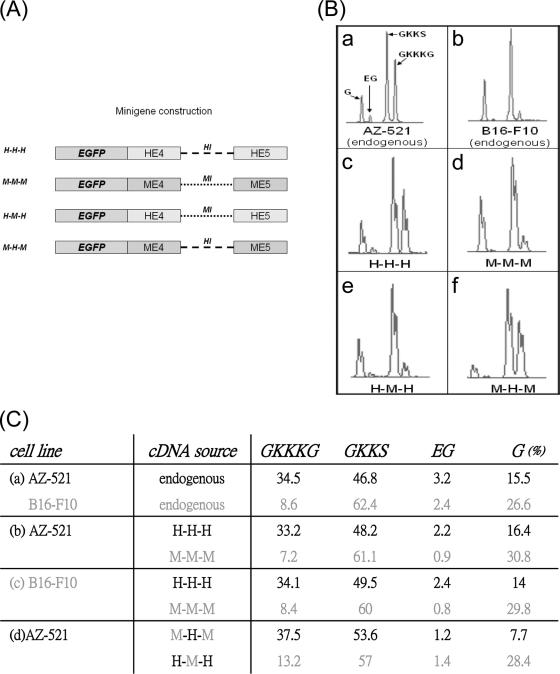
To decipher cis elements involved in ING4 wobble splicing, we constructed human and mouse ING4 minigene vectors containing exon 4, intron 4, and exon 5 (H-H-H and M-M-M, respectively). Since the intron was less conserved than the exon (Fig. 1E), we made the hybrid minigene patterns H-M-H and M-H-M, in which the human and mouse introns were exchanged reciprocally (Fig. 2A). The wobble splicing pattern of M-H-M was similar to that of H-H-H and human endogenous ING4 transcripts (Fig. 2B and C). Moreover, the H-M-H minigene generated a splicing pattern that was similar to that of M-M-M and mouse endogenous ING4 transcripts (Fig. 2B and C). This suggests that the cis elements that regulate ING4 wobble splicing are primarily located in the intron. Nevertheless, the splicing pattern of these minigenes occurred irrespective of the cell line used (Fig. 2C, panels b and c), suggesting that the variant wobble splicing pattern is not due to variation of trans-acting factors.
FIG. 2.
The splicing assay using the hybrid ING4 minigenes. (A) Schematic illustration of the human (H-H-H), mouse (M-M-M), and hybrid ING4 minigenes (H-M-H, hExon 4-mIntron 4-hExon 5; M-H-M, mExon4-hIntron 4-mExon 5). (B) Panels a and b show the distribution of the ING4 wobble splicing isoforms in AZ521 and B16-F10 cells. (c to f) Transient transfection of human AZ521 cells was performed using the ING4 minigenes. (C) Transient transfection of human AZ521 or mouse B16-F10 cells was performed using the ING4 minigenes. The relative expression level of the ING4 four isoforms was calculated using GeneScan 3.7. The values are the average of two independent experiments. The black and gray letters represent the human and mouse experiments, respectively.
The intron sequence is generally involved in 3′ splice site selection of wobble splicing.
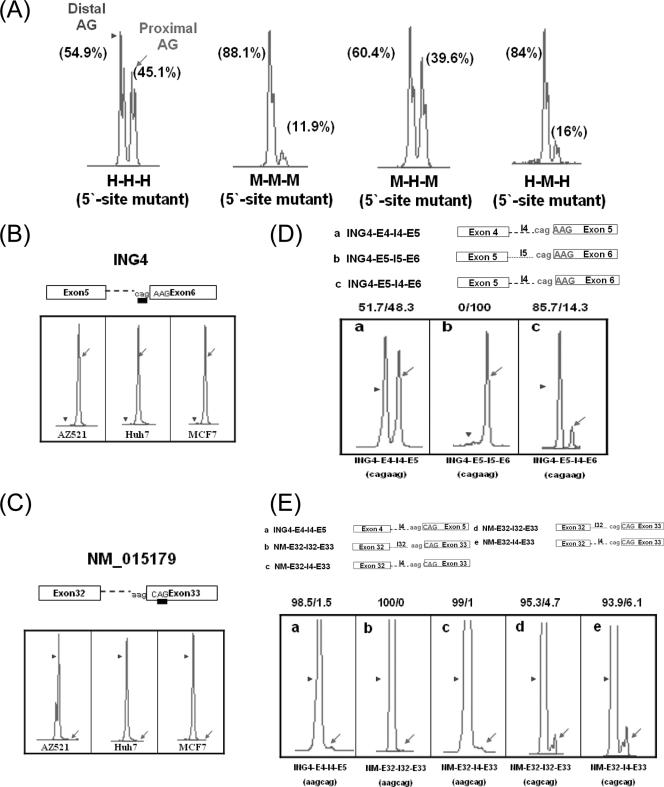
We further examined the intronic region for its roles in determining wobble splicing site choices. The ING4 wobble splicing patterns are significantly different between human and mouse, especially on 3′ splice site choice (38% GKKKG plus EG isoforms and 62% GKKS plus G in human, 11% GKKKG plus EG and 89% GKKS plus G in mouse) (35). A mutation (underlined) at the 5′ tandem splice site [GC(N)7GT→GG(N)7GT] abolished 5′ alternative splicing, and therefore wobble splicing occurred only at the 3′ splice site, generating two transcripts by wobble selection of the 3′ proximal or distal AG (Fig. 1B). Such 5′ splice site mutation minigenes allow us to better reveal the 3′ tandem NAGNAG site choice patterns. Transient expression of the H-H-H minigene in AZ521 cells showed that the H-H-H minigene with the mutated 5′ splice site used the proximal (45.1%) and distal AG (54.9%) sites equally, whereas the mutant M-M-M minigene preferentially selected the distal AG (88.1%) (Fig. 3A and Table 2). Consistent with what was observed for the endogenous splicing pattern, the result obtained from the minigenes suggested that the 3′ wobble splicing of ING4 differs between that of human and that of mouse. The hybrid minigene M-H-M with a 5′ splice site mutation showed a somewhat similar splicing pattern to that of the human (H-H-H) minigene construct, that is, ∼40% proximal AG and ∼60% distal AG usage. However, when the intron of the H-H-H minigene was replaced by the mouse sequence, the distal AG was preferred (Fig. 3A, H-M-H, and Table 2). According to the above-described result, the intron sequence may play a decisive role in 3′ splice site selection for wobble splicing.
FIG. 3.
The 3′ NAGNAG wobble splicing of the ING4 and NM_015179 minigenes is regulated by cis elements. (A) Four ING4 minigenes with 5′ splice site mutations (H-H-H, M-M-M, M-H-M, and H-M-H) were each transfected into AZ-521 cells for splicing analysis. A representative splicing profile for the 5′ mutant minigenes is shown, and the percentage of splicing resulting from distal (arrowhead) and proximal AG (arrow) is shown at the top of each peak. (B and C) The splicing pattern profiles of ING4 (exon 5-6) and NM_015179 (exon 32-33) were analyzed with different cell lines (AZ521, Huh7, and MCF7). (D and E) Schematic illustrations of the ING4 exon 4-5 (E4-I4-E5) and exon 5-6 (E5-I5-E6) and the NM_015179 (E32-I32-E33) minigenes and their fusion constructs (ING4-E5-I4-E6 and NM_015179-E32-I4-E33). The minigene constructs were introduced into AZ521 cells, and the splicing profile was analyzed by capillary electrophoresis. The arrowheads and arrows in each panel indicate the use of distal and proximal AG, respectively. The percentage of wobble splicing isoforms is shown at the top of each panel. A representative data set is shown; similar results were obtained from two independent experiments.
TABLE 2.
Swap test of the wobble splicing intron and its bilateral ING4 exons in cells
| cDNA sourcea | ING4 splicing isoforms (%)c
|
|
|---|---|---|
| Proximal position | Distal position | |
| Humanb | 38.0 | 62.0 |
| Mouseb | 11.0 | 89.0 |
| H-H-H | 45.1 | 54.9 |
| M-M-M | 11.9 | 88.1 |
| M-H-M(5′ site mutant) | 39.6 | 60.4 |
| H-M-H(5′ site mutant) | 16.0 | 84.0 |
| M-Mh-M | 36.1 | 63.9 |
| H-Hm-H | 21.7 | 78.3 |
| M-M(+3nt)-M | 15.7 | 84.3 |
| M-M(−3nt)-M | 18.9 | 81.1 |
| H-Hmut(9c/g)-H | 48.5 | 51.5 |
| H-Hmut(13t/a)-H | 50.7 | 49.3 |
| H-Hmut(15t/g)-H | 41.7 | 58.3 |
| H-Hmut(9c/g; 15t/g)-H | 41.6 | 58.4 |
The 5′ splice site mutation mini-genes of ING4 were transfected into AZ-521 cells for 24 h, and the percentages of wobble splicing transcripts at the proximal and distal AG positions were calculated. nt, nucleotide.
The endogenous ING4 wobble splicing pattern was analyzed using gene-specific primers; among the four types of transcripts identified, the “GKKKG” and “EG” forms are attributed to the proximal position, whereas “GKKS” and “G” forms are attributed to distal position.
The ratio of the ING4 wobble splicing isoforms was determined by dividing the peak areas of the individual forms by the total areas.
Next, we examined the effect of the NAGNAG boundary on wobble splicing. Because sequence analysis indicated that human ING4 intron 5 also contains a 3′ tandem splice site, we first examined the exon 5-intron 5-exon 6 (E5-I5-E6) minigene. This construct showed a preferential use of the proximal AG in all three cell lines tested (AZ521, Huh7, and MCF7) (Fig. 3B), suggesting that the NAGNAG motif is not sufficient for wobble splice site selection. When the intron sequence of the E5-I5-E6 minigene was replaced by intron 4, splicing drastically shifted to use of the distal AG (85.7%) (Fig. 3D, panel c). On the other hand, the minigene derived from another gene (NM_015179), containing the 3′ tandem splice site AAGCAG in the intron between exons 32 and 33, constitutively chose the distal AG (Fig. 3C and E). This splicing pattern did not change in the context of ING4 I4 (Fig. 3E, panel c). When the nucleotide preceding the proximal AG was changed to C (AAGCAG→CAGCAG) in both the I32 and the I4 intron, utilization of the proximal AG was considerably enhanced (Fig. 3E, panels d and e). These observations indicate that both the NAGNAG motif and the intron sequence contribute to wobble splicing.
The sequence between the BPS and the NAGNAG motif affects 3′ splice site selection.
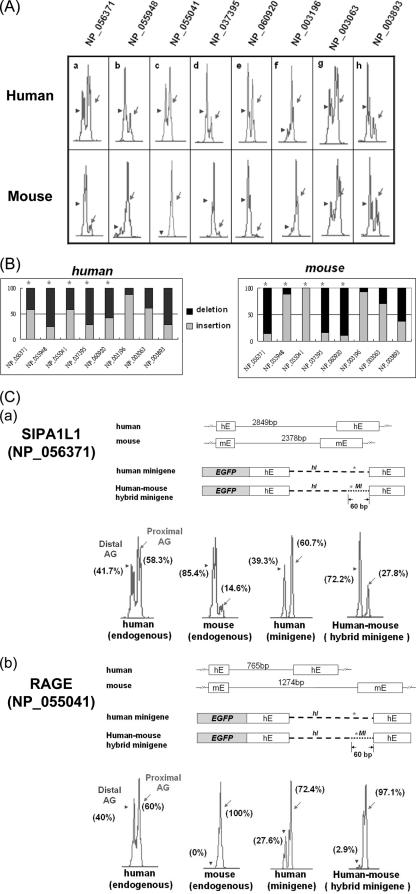
To further confirm that the intron sequence is an important factor in wobble splicing, we analyzed EST databases to evaluate additional cases of wobble splicing for which the splicing pattern is clearly different between that of human and that of mouse. We selected five genes (SIPA1L1 [NP_056371], MAPK8IP3 [NP_055948], RAGE [NP_055041], ARID1A [NP_060920], and BRD7 [NP_037395]) that showed possible species-specific expression patterns and another five genes (TCF12 [NP_003196], SMARCA4 [NP_003063], FUBP1 [NP_003893], VASP [NP_003361], and PUM1 [NP_055491]) with no noted difference by EST comparison (Table 3). Using capillary electrophoresis, we confirmed that five genes have species-specific wobble splicing and three genes do not significantly differ among species (Fig. 4A and B). In all of these cases, the exons flanking the wobble splice junction are highly conserved (>90%), whereas the intron sequences are less conserved. Thus, this result indicated that the intron sequence could indeed influence NAGNAG-based wobble splicing.
TABLE 3.
Sequence conservation between human and mouse within 10 wobble splicing intron-exon sets
| Accession no.a | EST countb
|
Exon conservation (%)c | Intron conservation (%)d | Conservation in the 60-bp region preceding NAGNAG (%)e | |
|---|---|---|---|---|---|
| Human | Mouse | ||||
| NP_056371 (SIPA1L1)* | 33/14 | 2/27 | 92 | 22 | 80 |
| NP_055948 (MAPK8IP3)* | 1/4 | 4/3 | 90 | 1 | 56 |
| NP_055041 (RAGE)* | 7/9 | 9/0 | 88 | 5 | 28 |
| NP_060920 (ARID1A)* | 14/14 | 0/18 | 93 | 46 | 49 |
| NP_037395 (BRD7)* | 8/19 | 1/36 | 82 | 27 | 70 |
| Avg | 89 | 20.2 | 56.6 | ||
| NP_003196 (TCF12) | 12/4 | 31/2 | 91 | 13 | 61 |
| NP_003063 (SMARCA4) | 77/27 | 39/10 | 92 | 1 | 100 |
| NP_003893 (FUBP1) | 48/47 | 50/48 | 92 | 40 | 70 |
| NP_003361 (VASP) | 27/3 | 20/2 | 86 | 44 | 71 |
| NP_055491 (PUM1) | 46/3 | 48/1 | 95 | 5 | 98 |
| Avg | 91.2 | 20.6 | 80 | ||
*, statistical differences were found between species-specific splicing patterns of human and mouse.
The number of insertion and deletion isoforms were counted by NCBI human and mouse EST database screening.
Conservation of exon sequences flanking the wobble splicing junction is indicated. Human/mouse alignments were analyzed, and conservation scores were calculated by ClustalW software.
Conservation of intron sequences upstream of the NAGNAG tandem motif is indicated.
Sequence conservation was observed for the 60-bp region upstream of the splice sites, including the sequence of the 3′ tandem splice site.
FIG. 4.
Analysis of 3′ wobble splicing of selected human and mouse genes. (A) Analysis of 3′ wobble splicing of eight genes (NP_056371, NP_055948, NP_055041, NP_037395, NP_060920, NP_003196, NP_003063, and NP_003893) in human and mouse lung tissue, using capillary electrophoresis. The relative percentages of the two isoforms were calculated using GeneScan 3.7 (B). (C) Schematic illustration of the human SIPA1L1 (exon 20-intron 20-exon 21) (a) and human RAGE (exon 6-intron 6-exon 7) (b) minigenes and their fusion constructs (human-mouse hybrid minigene). The minigene constructs were introduced into AZ521 cells, and the splicing profiles were analyzed by capillary electrophoresis (lower panels). Arrowheads and arrows indicate the use of distal and proximal AG, respectively. One representative data set is shown for the use of the distal and proximal AG, respectively. Stars indicate statistical differences in the splicing patterns between human and mouse.
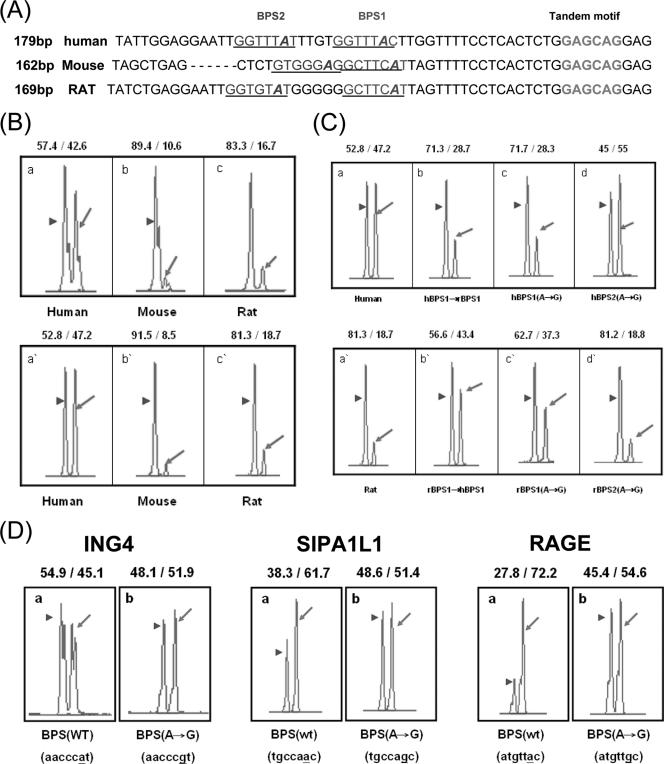
Furthermore, sequence analysis of five genes with species-specific wobble splicing (SIPA1L1, MAPK8IP3, RAGE, ARID1A, and BRD7) revealed that the intronic sequence (∼60 bp) upstream of the NAGNAG sequence was relatively poorly conserved (Table 3). To further examine the intronic sequence 60 bp upstream of the tandem NAGNAG motif, we constructed the human-mouse hybrid SIPA1L1 minigene in which this intronic region of human SIPA1L1 was replaced by the mouse equivalent sequence. As shown in Fig. 4C (panel a), the preferential AG usage was shifted from proximal to distal in this hybrid minigene. Similarly, the distal AG usage was considerably suppressed by replacing the 60-bp intronic sequence of the RAGE gene with that of the mouse (Fig. 4C, panel b). These results further demonstrated that the intronic 60-bp sequence is indeed involved in determining 3′ splice site NAGNAG usage. Therefore, we focused further on the intronic region between the BPS and the NAGNAG site to study its roles in wobble splicing regulation.
The PPT and BPS could affect 3′ site selection.
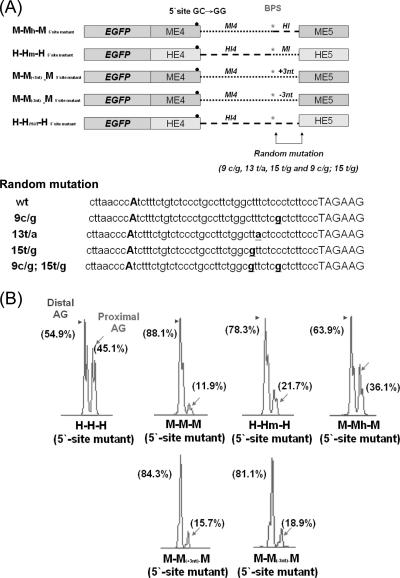
The above data indicated that the intronic sequence may play an important role in NAGNAG 3′ splice site choice, especially the 60 bp preceding the tandem motif (Fig. 3 and Fig. 4). A previous report has indicated that the average distance between the BPSand the 3′ splice site is 33 to 34 bp (19). We thus propose that the region between the BPS and the NAGNAG site is a key factor for 3′ wobble splicing regulation. To determine whether the sequence between the BPS and the NAGNAG motif plays a role in 3′ splice site choice, this region of human ING4 intron 4 was replaced with that of the mouse equivalent sequence (H-Hm-H). As shown in Fig. 5, the splicing pattern of the H-Hm-H minigene (proximal, 21.7%; distal, 78.3%), which contains the mouse BPS-to-NAGNAG region, exhibited a pattern similar to that of the mouse M-M-M minigene (Fig. 3A and Table 2). However, the M-Mh-M minigene significantly increased the level of the proximal AG isoforms (36.1%) (Fig. 5 and Table 2). Insertion of three nucleotides (three T nucleotides) into the mouse BPS-to-NAGNAG region to conform the length of human ING4 intron 4 changed the splicing pattern only slightly (Fig. 5B and Table 2). Similarly, removal of three nucleotides (three T nucleotides) did not significantly change the splicing. This result suggests that the length between the BPS and the NAGNAG region is not a key factor mediating ING4 wobble splicing. Furthermore, when we randomly mutated the BPS-to-NAGNAG region of the H-H-H minigene, 3′ NAGNAG wobble splicing was only slightly affected (Table 2). Because the BPS sequence is conserved between human and mouse, the PPT sequence may contain putative cis elements for 3′ tandem splice site selection.
FIG. 5.
Sequence of PPT within the BPS-to-NAGNAG region mediates 3′ NAGNAG wobble splicing. (A) Schematic illustration of the ING4 minigenes containing the mutated 5′ wobble splice site and the modified sequence at the 3′ end of intron 4, including M-Mh-M, H-Hm-H, M-M(+3nt)-M, M-M(−3nt)-M (where nt is nucleotide), and four random mutant constructs (9c/g, 13t/a, 15t/g, and 1c/g 15t/g). The positions of random mutant constructs are underlined (lower panel). Constructs were transfected into AZ-521 cells. (B) A representative expression profile of the minigenes is shown.
Previous studies showed that mutations in a putative BPS site could activate alternative splicing or change the exon inclusion/skipping ratio (19, 20). Therefore, the BPS may play a role in alternative splicing regulation. Interestingly, we found that the ARID1A (NP_060920) gene possesses a conserved 3′ tandem splice site, GAGCAG, and a PPT sequence in intron 12, but the splicing pattern varies among different species (Fig. 6A, B and Table 3). Therefore, we compared the sequence and location of the intron 12 BPS of the human and mouse ARID1A genes. The human BPS (GGTTTAC) is different from that of mouse and rat ( GCTTCAT); hence, we proposed that the BPS has a potential effect on wobble splicing. To further investigate this possibility, we constructed the ARID1A minigenes of human, mouse, and rat. The result of the in vivo minigene splicing assay showed that these constructs behaved similarly to the those of endogenous transcripts (Fig. 6B, compare the upper with the lower panel). When the human BPS was changed to the rat sequence, the distal AG was more preferred (Fig. 6C, panels a and b). Mutation of the putative branch point of BPS1 shifted 3′ splice site utilization from the proximal to the distal AG (Fig. 6C, panels a, c, and d). When the rat BPS was replaced by the human sequence or was mutated at the branch point adenosine, an increase in the proximal AG site utilization was observed (Fig. 6C, panels a′, b′, and c′). As shown with ARID1A in Fig. 6C, the splicing patterns of the SIPA1L1 and RAGE minigenes were significantly changed by substituting the branch point A. However, when the putative branch point of ING4 (AACCCAT→AACCCGT) was mutated, only a minor switch of the 3′ splice site from the distal to proximal AG was observed (Fig. 6D). The PPT sequence seems to be more important in determining splice site usage than the BPS in ING4. Our result indicates that the sequence of the BPS plays an important role in regulating 3′ NAGNAG-based wobble splicing in the cases we examined.
FIG. 6.
Influence of the intronic sequence on 3′ NAGNAG wobble splicing. (A) Alignment of the human, mouse and rat ARID1A (NP_060920) intron 12 sequence near the 3′ GAGCAG tandem motif. The putative BPS is predicted using the BPS predictive program (http://ast.bioinfo.tau.ac.il/). The BPS region is underlined, and the putative branch point adenosine is marked in italics. The 3′ tandem splice site is marked by bold letters. (B) Splicing pattern profiles of the human, mouse, and rat minigenes containing the genomic fragment of NP_060920 exon 12-13. The splicing assay of the human, mouse, and rat were performed using capillary electrophoresis as shown in Fig. 1. The endogenous transcripts are shown in the upper panels, and the minigenes are shown in the lower panels. (C) The splicing assay was performed using the human-derived sequences shown in the upper panels and the rat-derived sequences shown in the lower panels. (D) Minigenes of ING4, SIPA1L1, and RAGE and its putative branch point A mutant constructs were analyzed by capillary electrophoresis. The usage percent of distal and proximal AG, respectively, is indicated at the top of each panel.
The single nucleotide polymorphism in the NAGNAG motif could alter wobble splicing.
The above data implied that 3′ NAGNAG wobble splicing is controlled by intronic cis elements. Therefore, we proposed that mutations or single nucleotide polymorphisms (SNPs) around the NAGNAG tandem splice site may affect wobble splicing. By searching an SNP database, we found that 28.8% (127 of 441) of the EST-confirmed wobble splicing genes contained SNPs near the NAGNAG motif (∼60-bp region). It is noteworthy that 9 of the 127 SNPs were located within the NAGNAG motif and 78 were found in the intron sequences near NAGNAG (see Table S4 in the supplemental material).
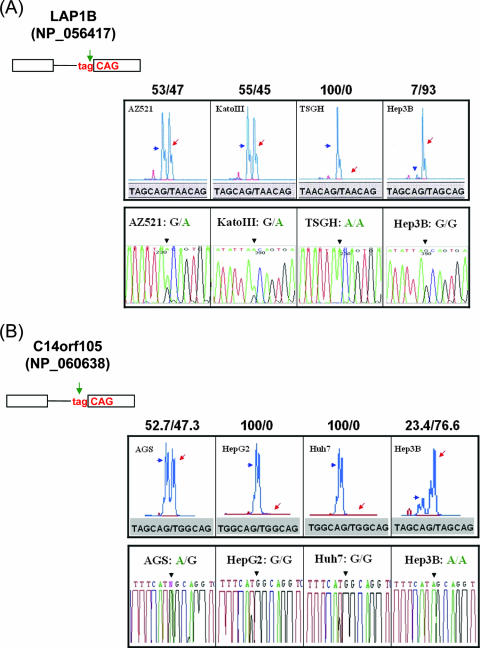
Next, we selected four samples from the above-described 127 SNPs to examine their effect on wobble splicing. The LAP1B gene (NP_056417) possesses a 3′ tandem splice site, TAGCAG, which results in one amino acid insertion or deletion in the encoded protein. Our reverse transcription (RT)-PCR analysis of four human cell lines showed that the SNPs (rs2245425) at the third position of the proximal 3′ splice site almost completely destroyed wobble splicing, particularly in homozygotic alleles with the TAACAG sequence (Fig. 7A). We obtained a similar phenomenon with the SNP (rs1152522) at a second position of the proximal 3′ splice site of the C14orf105 gene (NP_060638) which also completely abolished wobble splicing (Fig. 7B).
FIG. 7.
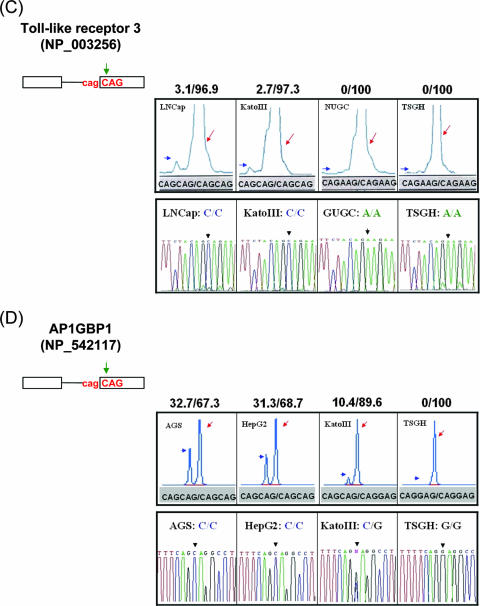
The influence of SNPs on 3′ NAGNAG wobble splicing. (A) The SNPs (rs2245425) were identified at the TAGCAG motif in the boundary between exon 2 and exon 3 of the human LAP1B gene (NP_056417). A representative splicing profile of LAP1B with different cell lines (AZ521, KatoIII, TSGH, and Hep3B) is shown (upper panel). (B) The splicing pattern of the C14orf105 (NP_060638) gene with different cells (AGS, HepG2, Huh7, and Hep3B) was analyzed. (C and D) The SNP occurred at the N nucleotide of the tandem motif. The human Toll-like receptor 3 (NP_003256) and AP1GBP1 (NP_542117) genes were analyzed in the four differently genotyped cell lines. Green arrows indicate the position of the SNP at the NAGNAG tandem motif. The percentage of wobble splicing isoforms is shown at the top of each panel. The arrowheads and arrows represent usage of the distal and proximal AG, respectively. The genomic sequence of the wobble splice sites was identified by an autosequencer. The black arrowhead indicates the splice junction (lower panel).
The Toll-like receptor 3 gene (NP_003256) contains the CAGCAG tandem sequence in the boundary of exon 1-2. Although the proximal site was used dominantly, mRNA isoforms produced by usage of the distal site were detectable. However, an SNP (rs3775296) at the “N” nucleotide of the distal AG (C-to-A change) could suppress the use of the distal site in wobble splicing (Fig. 7C). In addition, we analyzed the AP1GBP1 gene (NP_542117) that contains the CAGCAG tandem motif and generated two transcripts by alternative splicing at this tandem motif. However, SNP (rs12944821) at the “N” nucleotide of the distal AG (CAGCAG→CAGGAG) completely abolished the distal AG choice (Fig. 7D). These results are consistent with our above-described findings that the NAGNAG motif influences wobble splicing.
DISCUSSION
Alternative splicing of pre-mRNA involves alternative 5′ or 3′ splice site selection. Approximately 30% of human genes contain the evolutionarily conserved 3′ tandem splice site sequence NAGNAG (14). Among these genes, wobble splicing may result in one amino acid insertion or deletion in the encoded proteins. In this report, we first addressed the question of how 3′ NAGNAG wobble splicing is regulated. Our data suggested that the NAGNAG 3′ tandem splice site is crucial but not sufficient for wobble splicing. Moreover, we demonstrated that the BPS-to-NAGNAG region is likely to play a fundamental role in determining 3′ splice site usage in wobble splicing.
Infrequent use of GAG as the 3′ splice site was coincident with its rarity in the 3′ tandem splice site motif (1.4% GAGNAG and 6.1% NAGGAG among 441 cases) (see Table S3 in the supplemental material). Perhaps inefficient splicing at the GAG site has driven its loss during evolution, resulting in its scarcity in modern genomes. However, an exception observed here is the human ARID1A gene, in which GAG could compete for the use of CAG as the 3′ splice site (Fig. 6B and C). Moreover, we observed that the distal AG was preferred in the YAGAAG and the duplicate (NAGN′ AG; N = N′) tandem motifs of the human ING4 exon 4-5 boundary. In other words, C/UAG may lose its priority when located upstream of AAG, at least in human ING4 (Table 1). Interestingly, human and mouse ING4 intron 4 showed different preferences in the use of the UAGAAG tandem site, that is, AAG was more preferred in mouse than in human (Fig. 3A); such a difference is probably due to the intronic cis elements that differ between the two species. Moreover, the intron strength may also explain the use of inert GAG in 3′ wobble splicing of the minigenes with either the NAGGAG or the GAGNAG sequence (Table 1, ING4 minigene containing the AAGGAG tandem splice site and Fig. 6B, human ARID1A minigene containing the GAGCAG tandem motif). Nevertheless, future studies are required to elucidate the detailed mechanism.
Our observation suggested that the intronic sequence immediately upstream of the NAGNAG motif might mediate the selection of the 3′ tandem splice site. Akerman and Mandel-Gutfreund (1) compared constitutive and alternative splicing at the 3′-NAGNAG acceptor by using a bioinformatics approach and concluded that tandem splice site-containing genes possess a relatively conserved intronic sequence upstream of 3′-NAGNAG. This result suggests an abundance of cis elements nearby the 3′ tandem splice site. Zavolan et al. (40) suggest that the wobble splicing is the result of stochastic binding of the spliceosome at neighboring splice sites. Based on this hypothesis, Chern et al. recently developed a simple physical model that could predict whether splicing occurs only at one site or at two alternative sites at the tandem splice site (8). In our study, except for the 3′ tandem splice site and its adjacent intronic sequence governing splice site selection, we demonstrated that exonic sequences can also determine splice site selection. Thus, our results agree that wobble splice site recognition may involve thermodynamic interactions between various cis elements.
Exonic SNPs have a significant effect on protein function and impact cell physiology and disease progression. However, there is increasing evidence that both exonic and intronic SNPs affect pre-mRNA splicing, which could alter gene expression patterns and expand protein diversity (6, 25). Moreover, wobble splicing may particularly cause insertion or deletion of one amino acid in proteins. For example, an SNP in the ABCR gene (2588G→C) is frequently found in patients with Stargardt disease 1 (STGD1), resulting in an active TAGC2588AG motif, which generates two wobble splicing isoforms (26). In this study, we identified nine SNPs in the NAGNAG 3′ tandem splice site and demonstrated that SNPs interfered with wobble splicing in LAP1B and Toll-like receptor 3 (see Table S4 in the supplemental material). Using a genome-wide screen, Hiller et al. (13) recently identified 121 SNPs at the 3′ tandem splice site and found that 64 SNPs may affect alternative NAGNAG splicing, of which 18 are associated with known disease genes. Experiments are still required to confirm that these SNP sites are involved in wobble splicing regulation. Since our data indicated that the region between the BPS and the PPT is also important for wobble splicing (Fig. 5, Fig. 6, and Table 2), SNPs in this region may cause an imbalance between the wobble mRNA isoforms. Accordingly, we identified SNPs within the BPS and the PPT (8 SNPs in the BPS and 29 SNPs in the PPT) that might affect NAGNAG-based wobble splicing; this finding needs to be examined by future experiments (see Table S4 in the supplemental material).
The 3′ wobble splicing at the NAGNAG tandem motif occurs with a higher frequency than 5′ GTNGT splicing, since the 3′ end of introns has a more intricate set of regulatory elements. However, the exact mechanism of the 3′ NAGNAG-based wobble splicing is not well understood. Using a prototypical example of 3′ wobble splicing, ING4, we have illustrated the important region between the BPS and the tandem motif. In mammalian introns, both the BPS and PPT are essential for splicing (2, 39). Mutations or SNPs within these elements disturb splicing or induce alternative splice site utilization (21, 29, 31). A recent genome-wide study shows that mutations in putative BPSs cause a shift from constitutive to alternative splicing and alter the exon inclusion/skipping ratio (19). Our present study shows that mutations at a putative branch point of ARID1A, SIPA1L1, and RAGE had no apparent effect on splicing efficiency (data not shown) but affected wobble splicing (Fig. 6C and D), suggesting that the BPS may also play a role in determining 3′ splice site selection. Selection of the 3′ splice site depends on both the BPS-to-AG distance and the distance between the two adjacent AGs (9). Therefore, the BPS mutations may activate an aberrant branch point and thus alter the BPS-to-AG distance, changing the wobble splicing pattern. However, the minor change of distance (4 bp) between the new BPS (ACTTAAC) and the NAGNAG sequence did not alter AG selection. Therefore, our results suggest that the PPT sequence is more important in determining the 3′ splice site usage of ING4 than the BPS-to-NAGNAG distance (Fig. 5 and Fig. 6D). We considered that the BPS-to-NAGNAG distance and PPT strength may serve as major factors to determine 3′ NAGNAG wobble splicing. However, this hypothesis needs to be examined further.
In summary, this study provides experimental evidence showing that intronic cis elements, particularly in the region between the BPS and the NAGNAG 3′ splice site, play an important role in the 3′ tandem splice site selection. Moreover, SNPs within these regions may directly affect 3′ splice site selection and thus alter the mRNA isoform pattern generated by wobble splicing.
Supplementary Material
Acknowledgments
We thank Chun-Hung Lai and Shiao-Wei Kao for excellent technical assistance.
This study was supported in part by grants from the Academia Sinica and the National Science Council, Taiwan, Republic of China (94-2311-B001-033 and 95-2311-B-001-012, respectively).
Footnotes
Published ahead of print on 11 June 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akerman, M., and Y. Mandel-Gutfreund. 2006. Alternative splicing regulation at tandem 3′ splice sites. Nucleic Acids Res. 34:23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berglund, J. A., N. Abovich, and M. Rosbash. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 4.Brett, D., J. Hanke, G. Lehmann, S. Haase, S. Delbruck, S. Krueger, J. Reich, and P. Bork. 2000. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 474:83-86. [DOI] [PubMed] [Google Scholar]
- 5.Brow, D. A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36:333-360. [DOI] [PubMed] [Google Scholar]
- 6.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3:285-298. [DOI] [PubMed] [Google Scholar]
- 7.Chen, S., K. Anderson, and M. J. Moore. 2000. Evidence for a linear search in bimolecular 3′ splice site AG selection. Proc. Natl. Acad. Sci. USA 97:593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chern, T. M., E. van Nimwegen, C. Kai, J. Kawai, P. Carninci, Y. Hayashizaki, and M. Zavolan. 2006. A simple physical model predicts small exon length variations. PLoS Genet. 2:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chua, K., and R. Reed. 2001. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell. Biol. 21:1509-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condorelli, G., R. Bueno, and R. J. Smith. 1994. Two alternatively spliced forms of the human insulin-like growth factor I receptor have distinct biological activities and internalization kinetics. J. Biol. Chem. 269:8510-8516. [PubMed] [Google Scholar]
- 11.Englert, C., M. Vidal, S. Maheswaran, Y. Ge, R. M. Ezzell, K. J. Isselbacher, and D. A. Haber. 1995. Truncated WT1 mutants alter the subnuclear localization of the wild-type protein. Proc. Natl. Acad. Sci. USA 92:11960-11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox-Walsh, K. L., Y. Dou, B. J. Lam, S. P. Hung, P. F. Baldi, and K. J. Hertel. 2005. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl. Acad. Sci. USA 102:16176-16181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiller, M., K. Huse, K. Szafranski, N. Jahn, J. Hampe, S. Schreiber, R. Backofen, and M. Platzer. 2006. Single-nucleotide polymorphisms in NAGNAG acceptors are highly predictive for variations of alternative splicing. Am. J. Hum. Genet. 78:291-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiller, M., K. Huse, K. Szafranski, N. Jahn, J. Hampe, S. Schreiber, R. Backofen, and M. Platzer. 2004. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet. 36:1255-1257. [DOI] [PubMed] [Google Scholar]
- 15.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 16.Kan, Z., E. C. Rouchka, W. R. Gish, and D. J. States. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11:889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao, H. W., H. C. Chen, C. W. Wu, and W. C. Lin. 2003. Tyrosine-kinase expression profiles in human gastric cancer cell lines and their modulations with retinoic acids. Br. J. Cancer 88:1058-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kay, P. H., and M. R. Ziman. 1999. Alternate Pax7 paired box transcripts which include a trinucleotide or a hexanucleotide are generated by use of alternate 3′ intronic splice sites which are not utilized in the ancestral homologue. Gene 230:55-60. [DOI] [PubMed] [Google Scholar]
- 19.Kol, G., G. Lev-Maor, and G. Ast. 2005. Human-mouse comparative analysis reveals that branch-site plasticity contributes to splicing regulation. Hum. Mol. Genet. 14:1559-1568. [DOI] [PubMed] [Google Scholar]
- 20.Kralovicova, J., S. Houngninou-Molango, A. Kramer, and I. Vorechovsky. 2004. Branch site haplotypes that control alternative splicing. Hum. Mol. Genet. 13:3189-3202. [DOI] [PubMed] [Google Scholar]
- 21.Kralovicova, J., H. Lei, and I. Vorechovsky. 2006. Phenotypic consequences of branch point substitutions. Hum. Mutat. 27:803-813. [DOI] [PubMed] [Google Scholar]
- 22.Lai, C. H., L. Y. Hu, and W. C. Lin. 2006. Single amino-acid InDel variants generated by alternative tandem splice-donor and -acceptor selection. Biochem. Biophys. Res. Commun. 342:197-205. [DOI] [PubMed] [Google Scholar]
- 23.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez, A. J. 1998. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32:279-305. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Bigas, N., B. Audit, C. Ouzounis, G. Parra, and R. Guigo. 2005. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 579:1900-1903. [DOI] [PubMed] [Google Scholar]
- 26.Maugeri, A., M. A. van Driel, D. J. van de Pol, B. J. Klevering, F. J. van Haren, N. Tijmes, A. A. Bergen, K. Rohrschneider, A. Blankenagel, A. J. Pinckers, N. Dahl, H. G. Brunner, A. F. Deutman, C. B. Hoyng, and F. P. Cremers. 1999. The 2588G→C mutation in the ABCR gene is a mild frequent founder mutation in the Western European population and allows the classification of ABCR mutations in patients with Stargardt disease. Am. J. Hum. Genet. 64:1024-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rappsilber, J., U. Ryder, A. I. Lamond, and M. Mann. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12:1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340-345. [DOI] [PubMed] [Google Scholar]
- 29.Reed, R., and T. Maniatis. 1988. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 2:1268-1276. [DOI] [PubMed] [Google Scholar]
- 30.Rio, D. C. 1993. Splicing of pre-mRNA: mechanism, regulation and role in development. Curr. Opin. Genet. Dev. 3:574-584. [DOI] [PubMed] [Google Scholar]
- 31.Ruskin, B., J. M. Greene, and M. R. Green. 1985. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell 41:833-844. [DOI] [PubMed] [Google Scholar]
- 32.Smith, C. W., T. T. Chu, and B. Nadal-Ginard. 1993. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol. Cell. Biol. 13:4939-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, C. W., E. B. Porro, J. G. Patton, and B. Nadal-Ginard. 1989. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature 342:243-247. [DOI] [PubMed] [Google Scholar]
- 34.Tadokoro, K., M. Yamazaki-Inoue, M. Tachibana, M. Fujishiro, K. Nagao, M. Toyoda, M. Ozaki, M. Ono, N. Miki, T. Miyashita, and M. Yamada. 2005. Frequent occurrence of protein isoforms with or without a single amino acid residue by subtle alternative splicing: the case of Gln in DRPLA affects subcellular localization of the products. J. Hum. Genet. 50:382-394. [DOI] [PubMed] [Google Scholar]
- 35.Tsai, K. W., and W. C. Lin. 2006. Quantitative analysis of wobble splicing indicates that it is not tissue specific. Genomics 88:855-864. [DOI] [PubMed] [Google Scholar]
- 36.Unoki, M., J. C. Shen, Z. M. Zheng, and C. C. Harris. 2006. Novel splice variants of ING4 and their possible roles in the regulation of cell growth and motility. J. Biol. Chem. 281:34677-34686. [DOI] [PubMed] [Google Scholar]
- 37.Vogan, K. J., D. A. Underhill, and P. Gros. 1996. An alternative splicing event in the Pax-3 paired domain identifies the linker region as a key determinant of paired domain DNA-binding activity. Mol. Cell. Biol. 16:6677-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen, F., F. Li, H. Xia, X. Lu, X. Zhang, and Y. Li. 2004. The impact of very short alternative splicing on protein structures and functions in the human genome. Trends Genet. 20:232-236. [DOI] [PubMed] [Google Scholar]
- 39.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832-835. [DOI] [PubMed] [Google Scholar]
- 40.Zavolan, M., S. Kondo, C. Schonbach, J. Adachi, D. A. Hume, Y. Hayashizaki, and T. Gaasterland. 2003. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 13:1290-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.