Abstract
The significance of multiprotein signaling complexes in cell motility is becoming increasingly important. We have previously shown that phospholipase Cγ1 (PLCγ1) is critical for integrin-mediated cell spreading and motility (N. Jones et al., J. Cell Sci. 118:2695-2706, 2005). In the current study we show that, on a basement membrane-type matrix, PLCγ1 associates with the adaptor protein GIT1 and the Rac1/Cdc42 guanine exchange factor β-Pix; GIT1 and β-Pix form tight complexes independently of PLCγ1. The association of PLCγ1 with the complex requires both GIT1 and β-Pix and the specific array region (γSA) of PLCγ1. Mutations of PLCγ1 within the γSA region reveal that association with this complex is essential for the phosphorylation of PLCγ1 and the progression to an elongated morphology after integrin engagement. Short interfering RNA (siRNA) depletion of either β-Pix or GIT1 inhibited cell spreading in a fashion similar to that seen with siRNA against PLCγ1. Furthermore, siRNA depletion of PLCγ1, β-Pix, or GIT1 inhibited Cdc42 and Rac1 activation, while constitutively active forms of Cdc42 or Rac1, but not RhoA, were able to rescue the elongation of these cells. Signaling of the PLCγ1/GIT1/β-Pix complex to Cdc42/Rac1 was found to involve the activation of calpains, calcium-dependent proteases. Therefore, we propose that the association of PLCγ1 with complexes containing GIT1 and β-Pix is essential for its role in integrin-mediated cell spreading and motility. As a component of this complex, PLCγ1 is also involved in the activation of Cdc42 and Rac1.
Phosphoinositide-specific phospholipase C (PLC) enzymes have been established as crucial signaling molecules involved in the regulation of a variety of cellular functions (28, 54, 55). The evidence also suggests a critical involvement of members of the PLCγ family (PLCγ1 and PLCγ2) in several aspects of motility regulation (25, 27, 72). A distinct regulatory feature of PLCγ enzymes is that their activation is linked to an increase in the phosphorylation of specific tyrosine residues; multiple protein-protein interactions (mainly mediated by the SH2 and SH3 domains) also contribute to activation and have an important role in localizing PLCγ into protein complexes in different cellular compartments (4, 37, 57). As a direct substrate for a number of growth factor receptors with intrinsic tyrosine kinase activity, PLCγ1 has been shown to mediate chemotaxis towards these growth factors as a rate-limiting component (reviewed in references 26 and 70). This role of PLCγ1 has been demonstrated in different cell types (9, 33, 58), including several tumor cell lines characterized by high levels of growth factor receptors (27, 50, 66). It has also been suggested that epidermal growth factor (EGF)-driven and PLCγ1-mediated chemotaxis could have an important role in invasion in several human tumor xenograft models in vivo (68).
The possibility that PLCγ could also be an important signaling component in responses triggered directly by extracellular matrix receptors was initially suggested by observations that PLCγ1 can become phosphorylated upon integrin engagement or be recruited to integrin complexes in fibroblasts (34, 75). Further studies with highly specialized cell types (platelets and osteoclasts) supported the role of PLCγ isoforms in “outside-in” signaling, downstream of extracellular matrix receptors (22, 43, 72). Morphologically, platelets generated from mice deficient in PLCγ2 (the main PLCγ isoform in this cell type) had reduced cell spreading following the engagement of von Willebrand factor or α2β1 and αIIbβ3 integrins (22, 39, 72). Recently, we have shown that PLCγ1 has a key role in integrin-dependent cell motility, the invasion of diverse cancer cell types, and the morphogenesis of endothelial cells on basement membranes (25). Analysis of several cancer cell lines and endothelial cells where PLCγ1 was depleted using short interfering RNA (siRNA), as well as PLCγ1−/− fibroblasts, demonstrated that all cell types retain their initial rounded morphology, lacking stable cell protrusions, while the wild-type cells subsequently undergo polarization and elongation; this was particularly pronounced on some three-dimensional matrices (25). Similar observations, suggesting a role for PLCγ1 in integrin-mediated cell spreading, were also reported for PLCγ1−/− fibroblasts plated on fibronectin (69). Together, these data demonstrate that PLCγ1 is more widely involved in processes related to cell motility than originally anticipated and that, in addition to chemotaxis triggered by growth factor receptors, PLCγ1 is stimulated through integrin activation in different cell types.
Cell attachment, spreading, and motility are complex processes requiring the integration of diverse signaling networks and structural assemblies (17, 18, 26, 70, 71). Early signaling from integrin-mediated attachments is associated with pathways that lead to Rac and Cdc42 activation and actin polymerization. This then links to dynamic formation and disassembly of adhesion complexes, suggested to have a role in localizing and stabilizing actin protrusions (6, 11, 46). Although some of the key regulators have been identified, the identities of all components of these super-molecular formations and how they act together are not well understood. In the case of PLCγ1, despite considerable circumstantial evidence for the involvement of PLCγ1 in the reorganization of cytoskeletal components (26, 70), there are few observations that suggest an underlying molecular mechanism (42, 53, 64) or a link with the known regulators of actin cytoskeleton, such as small GTPases from the Rho family (1, 15). Also, most previous studies have involved the use of overexpression systems and little has been done to investigate the functioning and interactions of endogenous PLCγ1.
Here we investigated possible endogenous interaction partners for PLCγ1 and their functional implications for maintaining a motile phenotype on three-dimensional extracellular matrices. In this study, we used different cell types and combined several experimental approaches, including the depletion of specific components and direct and indirect rescue experiments using their wild-type and mutant variants. We show that, upon the engagement of β1 integrins on basement membrane, PLCγ1 associates with the adaptor protein GIT1 and the Rac1/Cdc42 guanine nucleotide exchange factor (GEF) β-Pix; for PLCγ1 to fully engage this complex, both GIT1 and β-Pix are required. The binding is mediated by the region in PLCγ1 specific to the PLCγ family (specific array [γSA]), containing critical SH2 domains, and is essential for its role in cell spreading and motility. Our data also suggest that the β-Pix/GIT1/PLCγ1 complex acts upstream of Cdc42 and Rac1 and that several signaling events, including the activation of calpains, could provide the link with Cdc42 and Rac1 activation.
MATERIALS AND METHODS
The following chemicals were from Calbiochem: PLC inhibitor U73122, Rac inhibitor NSC23766, ROCK inhibitor Y-27632, calcium chelator bis-(aminophenoxy)-ethane-N,N,N′,N′-tetraacetic acid (BAPTA-AM), src inhibitor PP2 (and PP3), and calpeptin. Mouse and rabbit horseradish peroxidase (HRPO)-conjugated secondary antibodies, lysozyme, and protease inhibitor cocktails (1 and 2) were from Sigma. Texas Red-X phalloidin and tert-butoxycarbonyl-Leu-Met-chloromethylaminocoumarin (Boc-LM-CMAC; fluorescence-linked calpain substrate) were from Molecular Probes. siRNA oligonucleotides were obtained from Dharmacon, Inc. (Smartpool: PLCγ1, GIT1, β-Pix, Rac1, Cdc42, and RhoA) or Santa Cruz (Tiam1, Crk1/2, and p130CAS). Dharmafect siRNA reagent was from Dharmacon, and Lipofectamine/Lipofectamine plus and OPTIMEM were from Invitrogen.
Anti-PLCγ1 (for immunoprecipitation), anti-Rac, anti-Src, and anti-p130CAS antibodies were from Upstate. Anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) was purchased from Research Diagnostics, Inc. Anti-β-Pix, anti-GIT1, anti-Cdc42, anti-Rac1, anti-RhoA, anti-Tiam1, anti-Crk1/2, and anti-hemagglutinin (anti-HA) antibodies (all for Western blotting) and HRPO-anti-goat secondary antibody were supplied by Santa Cruz. Anti-GIT1 antibody (for immunoprecipitation), cell recovery solution, and Matrigel were obtained from BD Bioscience. Antifilamin and anti-β-Pix antibodies (for immunoprecipitation) were from Chemicon. Anti-HA (for immunoprecipitation), antispectrin (fodrin), and anti-phospho-PLCγ1 (Y783) antibodies were from Cell Signaling Technologies. Anti-penta-His antibody was from QIAGEN, and anti-integrin b1 (4B4) (blocking antibody) was from Beckman. Protein G-agarose and glutathione-Sepharose were from Roche. The following reagents were previously described: PLCγ1−/− and PLCγ1+ (add back) mouse embryonic fibroblasts (MEFs) (24), rat PLCγ1-green fluorescent protein (GFP) construct (75), HA-tagged Src homology 2 (SH2) mutant PLCγ1 constructs (8, 23), β-Pix constructs (47), β-Pix-glutathione S-transferase (GST) construct (65), GIT1 constructs (29), GIT1 and β-Pix baculoviruses (49), Cdc42 (N17 and L61), Rac1 (N17 and V12) and RhoA (N19 and V14) constructs (56), PAK Rho family binding domain (RBD) construct (10) for Rac1 pull down, and N-WASP CRIB for Cdc42 pull down (30).
Cell culture, siRNA treatment, and transfection.
BE (colon carcinoma) and A431 (adenocarcinoma) cells and PLCγ1−/− and PLCγ1+ MEFs were maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. For all siRNA protocols, preannealed purified siRNA probes were rehydrated prior to transfection using the standard protocol. siRNA probes (200 nM) were transfected using Dharmafect reagent, using a multiple transfection strategy that involved successive siRNA transfections on 3 consecutive days. Treated cells were then either used for experiments 72 h after the first transfection or had their protein extracted for Western blotting to check protein depletion. For siRNA rescue experiments, BE cells were transfected with suitable DNA constructs (resistant to siRNA) on day 3 of the multiple transfection protocol, prior to the third and final siRNA transfection. Briefly, 1 μg of DNA was transfected into the cells for 4 h using the Lipofectamine plus transfection system. Subsequently, following extensive washing, the third and final siRNA transfection was performed in a fashion similar to the previous siRNA transfections. The effects of the rescue were then analyzed 24 h later (72 h after the first siRNA transfection). Transfection-only experiments or transfections of other cell lines (i.e., MEFs) were also performed in a similar fashion, using the Lipofectamine plus system with effects analyzed 24 h after transfection.
Preparation and processing of cell extracts.
Cells were scraped into ice-cold lysis buffer (20 mM Tris-HCl [pH 7.4], 1 mM EDTA, 50 mM KCl, 1 mM dithiothreitol [DTT], 1 mM Na3VO4, 5 mM MgCl2, 5 mM NaF, 10% [vol/vol] glycerol, 1% [vol/vol] Triton TX-100) supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail II (Sigma) and then homogenized by passing the lysate through a Hamilton syringe (22 gauge) 10 times. After standing on ice for 10 min, the lysates were clarified by centrifugation (15,800 × g/10 min) and the supernatant used as the cell extract. Extracts were used for immunoprecipitation (see below), for protein concentration determination (by the Bradford methods), or directly for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE)/blotting following the addition of SDS sample buffer. In most circumstances, cell extracts were obtained from Matrigel. Briefly, cells on Matrigel were washed and then incubated for 30 min with phosphate-buffered saline (PBS) containing phosphatase inhibitor cocktail 2 (Sigma). Following this, the cells were scraped into a suitable volume (2 ml per cm2) of ice-cold cell recovery solution (BD Biosciences) and left on ice for 1 h to digest the Matrigel. Following digestion, the cell pellet was recovered by centrifugation (160 × g for 5 min) and the cells lysed and processed in accordance with the above lysis protocol. For immunoprecipitation protocols, cell extracts were incubated first with the equilibrated protein G-agarose beads for 2 h at 4°C as a preclearing step. Following this, the supernatant was incubated overnight at 4°C with the desired primary antibody. Protein G-agarose was added to this mixture, and the incubation allowed to proceed for a further 2 h at 4°C. The beads were then washed three times with lysis buffer and combined with SDS sample buffer. Samples were separated by 10% SDS-PAGE, and the proteins transferred to polyvinylidene difluoride membrane. After being blocked, the membranes were incubated with the required primary antibody (1 in 1,000 dilution) overnight at 4°C. Following washing with 0.1% Tris-buffered saline-Tween, the membranes were incubated with suitable HRPO-conjugated secondary antibody for 2 h at 24°C. After further washing, proteins were visualized using the enhanced chemiluminescence detection method (Amersham Bioscience).
Analysis of cell morphology and migration on Matrigel.
Cell culture dishes were coated with a thin layer of Matrigel (diluted at 2 parts to 1 part Dulbecco's modified Eagle's medium). This was then allowed to set at 37°C for 90 min (approximately 200 μl of this mix was used per cm of dish area). BE cells either pretreated with siRNA or incubated in the presence or absence of inhibitors (as specified in figure legends) were plated on the Matrigel, and the ability of the cells to elongate and subsequently form connected networks (through cell movement and cell-cell contacts) was analyzed after various, specified time points (usually 4 h, as in control treatments this was found to be the time point where the majority of cells had a clearly defined elongated phenotype) using phase contrast and fluorescence microscopy (Nikon microscope camera and OpenLab software). Treatments with either siRNA or pharmacological inhibitors in most cases resulted in uniform changes in cell morphology, as shown when four fields of ∼100 cells were analyzed. For quantification experiments, the numbers of elongating cells per 100 cells counted were recorded and used to produce graphical representations of the effects of various treatments on cell spreading. When rescue experiments that involved the transfection of plasmids (specified in figure legends) were performed, the analysis was, in most instances, based on a procedure previously described for PLCγ1 plasmids used for the transfection of siRNA-pretreated BE cells (25). To distinguish cells that express proteins encoded by specific constructs with efficiencies of 40 to 70%, GFP-tagged variants were transfected and the analysis took into account the cells showing green fluorescence. Typically, 3 to 4 areas with ∼100 fluorescent cells were analyzed and the mean value calculated.
Immunofluorescence.
Cells were fixed in 3.7% formaldehyde in PBS and permeabilized in 0.2% Triton X-100 in PBS. Following being blocked in 2% bovine serum albumin in PBS for 1 h, fixed cells were incubated with Texas Red-phalloidin (1 in 1,000) for 2 h. After being washed extensively, the cells were observed by using immunofluorescence microscopy (Nikon Eclipse E600) and a Bio-Rad confocal setup plus Laser Sharp software (Bio-Rad MRC1024).
Analysis of protein-protein interactions.
For the protein-based pull-down interactions of GIT1, β-Pix, and PLCγ1SA, the proteins were purified as GST fusion proteins (GST-GIT1, GST-β-Pix, and GST-γ1SA protein were purified).
Purification of the GST-fused proteins, γ1SA (57), GIT1 (29), and β-Pix (65), was as previously described. Briefly, bacterial cell pellets expressing proteins encoded by appropriate pGEX vectors were lysed using ice-cold STE buffer (10 mM Tris [pH 7.5], 1 mM NaCl, 1 mM EDTA) supplemented with protease inhibitors (Sigma). Lysozyme (100 μg/ml) was added, and the reaction mixture left for 15 min on ice. DTT (5 mM), Tween 20 (to 0.1%), and SDS (to 0.03%) were added to the lysate, which was then centrifuged at 13,000 × g (4°C) for 40 min. The resulting supernatant was incubated for 1 h with prewashed glutathione-Sepharose at 4°C for 1 h to allow the GST proteins to bind to the beads. Following centrifugation, the beads were washed three times with STE buffer and used immediately in the pull-down assay.
For the expression and processing of baculoviruses expressing GIT1 and β-Pix proteins, methods similar to those of Premont et al. (49) were employed. Briefly, SF9 cells were infected with GIT1, β-Pix, or both GIT1 and β-Pix viruses and infection/expression allowed to proceed for 72 h. SF9 cell pellets were collected by centrifugation (2,000 × g) and frozen at −80°C until required. For processing, the pellets were resuspended in lysis buffer (20 mM Tris, [pH 7.5], 2 mM NaCl, 2 mM EDTA) and DNase I (10 μg/ml), NP-40 (0.5%), and DTT (1 mM) added. These samples were then incubated for 1 h at 4°C with constant rotation. Subsequently, the samples were centrifuged at 10,000 × g for 40 min. The resulting supernatants were collected, standardized for protein levels, and used in pull-down experiments.
To analyze protein-protein interactions, standard pull-down assays, based on immobilized GST fusion proteins, were used (57). For in vitro studies, freshly prepared (see above) γ1SA (His tagged) was incubated with either GST-GIT1 or GST-β-Pix immobilized on glutathione-Sepharose beads (30 μl) for 1 h at 4°C. Alternatively, crude SF9 cell lysates containing GIT1, β-Pix, or GIT1/β-Pix were incubated with GST fusion protein (γ1SA) immobilized on glutathione-Sepharose beads (30 μl) for 1 h at 4°C. In all cases, the beads were subsequently washed three times in lysis buffer and then processed for SDS-PAGE and Western blotting. Processed samples were subjected to SDS-PAGE, transferred, and incubated with β-Pix, GIT1, or His antibodies, as indicated for individual experiments.
For cell-based studies, freshly prepared BE cell extracts were standardized for protein amount and equal amounts of cell extracts (in a final volume of 150 μl) were incubated with GST fusion protein (γ1SA) immobilized on glutathione-Sepharose beads (30 μl) for 1 h at 4°C. The beads were subsequently washed three times in lysis buffer and then processed for SDS-PAGE and Western blotting. Processed samples were subjected to SDS-PAGE, transferred, and incubated with β-Pix or GIT1 antibodies, as indicated for individual experiments. Samples of total cell extracts and the pull-down supernatant were also analyzed by Western blotting.
Assays for Cdc42/Rac1 activity.
The amount of active (GTP-bound) Cdc42 or Rac1 was analyzed using binding domains from proteins that only bind to active forms of Cdc42 or Rac1; the Cdc42 or Rac1 in the resulting complexes was subsequently visualized by Western blotting. For Rac1, the CRIB domain of p21PAK (PAK-RBD) is commonly used (10), and for Cdc42, the GTPase binding domain of N-WASP (WASP-CRIB [RBD]) is considered more suitable (30). These domains were expressed as GST fusion proteins and processed as described above for the other GST fusion proteins. After purification, the GST fusion proteins were bound to glutathione-Sepharose beads as described above and used immediately in pull-down studies of cell extracts. Briefly, freshly prepared BE cell extracts were standardized for protein concentration (in a final volume of 150 μl) and incubated with GST-fused proteins (PAK- or WASP-RBD) immobilized to glutathione-Sepharose beads (30 ul) for 1 h at 4°C. The beads were subsequently washed three times in lysis buffer and subjected to Western blotting using antibody to either Rac1 (PAK-RBD pull down) or Cdc42 (WASP-RBD pull down). Total Rac1 or Cdc42 was also analyzed by Western blotting of cell extracts, and the ratios of active GTPase to total cellular GTPase were calculated by densitometry measurements (using Western blot autoradiographs and NIH image software) and corrected for protein levels based on the amounts of Src and GAPDH in cell extracts and pull-down supernatant samples. The resulting activity measurements from several experiments for each GTPase were expressed as percentages of the GTPase activity of control cells and presented in graphical form.
Assay for calpain activity.
To assay calpain activity, methods similar to those of Satish et al. were used (62). These involved using Boc-LM-CMAC, a nonfluorescent calpain substrate which, when cleaved by calpain, generates a fluorescent cleavage product that can be detected by fluorescence microscopy. Briefly, BE cells were pretreated with inhibitors (30 min) or siRNA prior to the addition of the calpain substrate Boc-LM-CMAC. Boc-LM-CMAC (50 μm) was then added to the cells for 30 min prior to trypsinization to allow for cellular uptake of the substrate. The cells were then plated on Matrigel and allowed to elongate for 4 h. After this, the cells were fixed and the presence of the fluorescent cleavage product of the Boc-LM-CMAC was assessed by fluorescence microscopy (Nikon microscope and Simple PCI software). Several images were taken for each treatment, and the amount of fluorescence in each cell was quantified using densitometry (Image J software) to establish the relative calpain activity.
RESULTS
PLCγ1 complexes with GIT1 and β-Pix.
We have previously shown that in BE cells, PLCγ1 becomes phosphorylated on the critical tyrosine residue Y738 upon the engagement of β1 integrins by components of basement membrane matrices (Matrigel). The involvement of Src kinase in PLCγ phosphorylation was further documented by using Src kinase inhibitors, by the ability of this kinase to directly phosphorylate the PLCγ1 Y738 residue, and by the presence of Src kinase in complexes with PLCγ1 isolated by immunoprecipitation of PLCγ1 from motile BE cells (25). We have also detected the adapter protein GIT1 in these complexes (25). Since in some cellular systems, GIT1 forms complexes with the Rac/Cdc42 GEF β-Pix (38, 49, 76), we extended our studies to include several cell types (BE [colorectal], A431 [squamous cell carcinoma], and PLCγ1− and PLCγ1+ MEFs) and monitored the endogenous expression and coimmunoprecipitation of PLCγ1, β-Pix, and GIT1 in these cell types (Fig. 1).
FIG. 1.
PLCγ1, β-Pix, and GIT1 form interaction complexes in various cell lines. (A) Expression of PLCγ1, β-Pix, and GIT1 in BE (colorectal) and A431 (squamous carcinoma) cells and PLCγ1− and PLCγ1+ MEFs analyzed by Western blotting. (A faint nonspecific band is observed in the PLCγ1− MEF with the Sigma PLCγ1 antibody but not with the other antibodies used in this study; further-more, these cells have been well characterized as being deficient in PLCγ1 [24]). (B) PLCγ1, β-Pix, and GIT1 coimmunoprecipitate. BE or A431 cells or MEFs were plated on Matrigel for 4 h and then recovered from Matrigel and lysed, and immunoprecipitation (IP) was performed with PLCγ1, β-Pix, or GIT1 antibodies as indicated below the panels. Subsequent Western blotting of these samples was performed using PLCγ1, β-Pix, or GIT1 antibodies as indicated. (C) Complex formation between PLCγ1 and GIT1/β-Pix requires both GIT1 and β-Pix. Western blots showing the effect of the indicated siRNA depletion of PLCγ1, β-Pix, or GIT1 protein in BE cells are shown in the left panel. For IP experiments (right panel), BE cells previously treated with the indicated siRNAs (Scr, scrambled; PLCγ1, smart pool to PLCγ1; β-Pix, smart pool to β-Pix; and GIT1, smart pool to GIT1) were plated on Matrigel for 4 h and then recovered from Matrigel, lysed, and subjected to IP using anti-PLCγ1 antibody. Subsequent Western blotting of these samples was performed using PLCγ1, β-Pix, or GIT1 antibodies as indicated; Western blots for GAPDH levels in total cell lysate (TCL) were used as a loading control (bottom panels). (D) PLCγ1, β-Pix, and GIT1 siRNAs inhibit cell elongation on Matrigel. Pretreated (with the indicated siRNA, as described for panel C) BE cells were plated on Matrigel, and the level of elongation analyzed by phase-contrast microscopy (large panels). Cells were also stained with phalloidin and analyzed for individual cell spreading (insert panels). (E) Graphical representation of the effects of PLCγ1, β-Pix, and GIT1 siRNAs (as described for panel C) on BE cell elongation on Matrigel. The number of elongated cells (per 100 cells) were counted and expressed as a percentage of the total cell number (n = 4 fields analyzed). Error bars show standard deviations.
BE and A431 cells and PLCγ1+ MEFs were found to contain appreciable levels of PLCγ1, β-Pix, and GIT1, whereas PLCγ1− MEFs, as expected, only contained GIT1 and β-Pix (Fig. 1A). Immunoprecipitation experiments revealed that GIT1, β-Pix, and PLCγ1 coimmunoprecipitate from BE and A431 cells (Fig. 1B, top panels). While immunoprecipitation with PLCγ1 pulled down a substantial quantity of not only PLCγ1, but also β-Pix and GIT1, immunoprecipitation with GIT1 or β-Pix antibodies only pulled down a fraction of the total cellular PLCγ1. In PLCγ1+ MEFs, immunoprecipitation with PLCγ1 or GIT1 antibodies again showed PLCγ1, β-Pix, and GIT1 to coimmunoprecipitate, whereas in PLCγ1− cells, the PLCγ1 antibody did not nonspecifically pull down any of the components; in PLCγ1− MEFs, GIT1 antibody pulled down GIT1 and β-Pix (Fig. 1B, bottom panels).
Based on observations from PLCγ1− MEFs that β-Pix and GIT1 are tightly complexed irrespective of the presence of PLCγ1 (Fig. 1B, bottom panels), we analyzed further the interaction between PLCγ1 and the β-Pix/GIT1 complex in BE cells. siRNA treatments were used to deplete the cells of PLCγ1, GIT1, or β-Pix, respectively (Fig. 1C, left panel), and the interactions analyzed postdepletion by immunoprecipitation and Western blotting (Fig. 1C, right panel). Depletion of PLCγ1 prevented the pull down of GIT1 or β-Pix by the PLCγ1 antibody. However, siRNA depletion of either β-Pix or GIT1 prevented any coimmunoprecipitation of both GIT1 and β-Pix by PLCγ1 (Fig. 1C, right panel). This suggests that both β-Pix and GIT1 are necessary for the complete interaction of the β-Pix/GIT1 complex with PLCγ1 and supports the notion of a tight, semipermanent β-Pix/GIT1 complex in cells.
We also investigated the possibility that some other proteins previously suggested to interact and form complexes with PLCγ, namely, Tiam1 (15), p130CAS, and its binding partner Crk1/2 (44, 67), were present in our PLCγ1/GIT1/β-Pix complex in BE cells following plating on Matrigel. Immunoprecipitation with PLCγ1 antibody did not pull down appreciable levels of Tiam1, p130CAS, or Crk1/2 (compared to the levels of GIT1 and β-Pix which coimmunoprecipitate with PLCγ1), although readily detectable levels of Tiam1, P130CAS, and Crk1/2 were present in the cells (Fig. 2A). This suggests that in cell spreading on Matrigel, the system analyzed here, the main binding partners for PLCγ1 are GIT1 and β-Pix.
FIG. 2.
Tiam1, p130CAS, and Crk1/2 do not interact with PLCγ1 or affect the elongation of BE cells on Matrigel. (A) PLCγ1 does not complex with Tiam1, p130CAS, or Crk1/2. BE cells were extracted from Matrigel 4 h after plating, lysed, and subjected to immunoprecipitation with PLCγ1 antibody. Both the supernatants (SN) and immunoprecipitates (IP) were subjected to Western blotting using PLCγ1, GIT1, Tiam1, p130CAS, and Crk1/2 antibodies. (B) siRNA treatment against Tiam1, p130CAS, or Crk1/2 does not inhibit cell spreading. BE cells were pretreated with the indicated siRNA probes (scrambled [Scr] and against PLCγ1, Tiam1, p130CAS or Crk1/2) and plated on Matrigel for 4 h. Representative Western blots for Tiam1, p130CAS, and Crk1/2 siRNA treatments are shown in upper panels. The level of elongation was analyzed by phase-contrast microscopy (lower panels). Cells were also stained with phalloidin and analyzed for individual cell spreading (insert panels). (C) Graphical representation of the effects of PLCγ1, Tiam1, p130CAS, and Crk1/2 siRNAs (as described for panel B) on BE cell elongation on Matrigel. The number of elongated cells (per 100 cells) was determined and expressed as a percentage of the total cell number (n = 4 fields analyzed). Error bars show standard deviations.
Our previous studies that established the requirement for PLCγ1 for cell spreading focused on early changes in BE cell morphology. In addition to early morphological differences (marked by the presence of cell protrusions), the wild-type BE cells subsequently undergo marked elongation and network formation on Matrigel. This helps to easily distinguish the wild-type cells from PLCγ1-deficient cells, which remain rounded, with few cell-cell contacts (25). We used the same conditions to analyze BE cells after the depletion of β-Pix or GIT1 using siRNA. The depletion of either GIT1 or β-Pix yielded an inhibition of cell elongation similar to that seen with PLCγ1 knockdown, with the cells retaining the initial rounded-cell phenotype (Fig. 1D and E). The inhibition of cell spreading was clearly visible after 4 h (as shown in Fig. 1D and E), a time point where control cells were clearly elongated; however, it was also observed that the inhibited cells retained the rounded-cell phenotype 20 h after plating on Matrigel (24; data not shown). We also explored the effects of depletion of the Tiam1, p130CAS, and Crk1/2 proteins. BE cells pretreated with these siRNAs spread normally, and these data further support the hypothesis that in this system, these proteins are not linked to PLCγ1 and its role in establishing the elongated-cell phenotype (Fig. 2B and C).
Association of PLCγ1 with GIT1/β-Pix complexes is essential for its function in cell spreading.
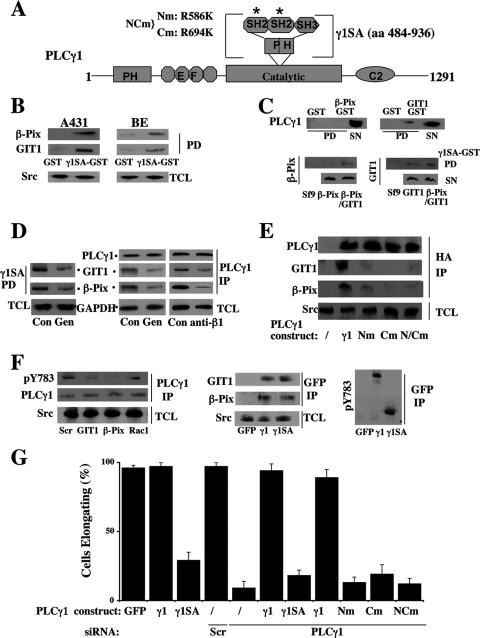
All three components, β-Pix, GIT1, and PLCγ1, are multidomain proteins that could be involved in a number of protein-protein interactions (21, 54, 60). PLCγSA contains two SH2 domains, an SH3 domain, and an internal PH domain (Fig. 3A) and has been implicated in various protein-protein interactions (55). To analyze the importance of γSA in our system, we first demonstrated that purified, immobilized γSA protein (amino acids [aa] 484 to 936) can pull down considerable amounts of the β-Pix and GIT 1 present in the cell lysate of BE cells plated on matrigel (Fig. 3B). However, interactions between purified β-Pix and γSA could not be detected, while the binding of purified GIT1 to γSA was very weak; interestingly, coexpression of β-Pix and GIT1 in SF9 cells to some degree increased the binding of both to γSA protein when compared to the binding of each protein expressed individually in the same system (Fig. 3C). Previous attempts to express GIT1 and β-Pix suggested that protein stability and the ability to form GIT1/β-Pix complexes were required for their coexpression (49). This could also be one of the reasons for our observations, but could not fully explain the low levels of complex formation with γSA even when β-Pix and GIT1 are coexpressed. Another important factor for GIT1 and β-Pix binding to γSA could be the need for further posttranslational modifications that are difficult to reproduce correctly in insect cells. Consistent with this possibility, our data show a marked reduction in complex formation in BE cells in the presence of genistein, an inhibitor of tyrosine phosphorylation (Fig. 3D, right panel). Furthermore, the association of GIT1 and β-Pix present in cell lysates from genistein-treated cells with the purified, unmodified γSA protein was greatly reduced (Fig. 3D, left panel). It is therefore likely that tyrosine phosphorylation of the β-Pix/GIT1 complex contributes largely to the interaction with γSA.
FIG. 3.
Importance of PLCγ1SA domain in interactions with GIT1 and β-Pix. (A) Domain organization of PLCγ1, showing the SA region (aa 484 to 936) and positions of mutations in SH2 domains (*) in PLCγ1. Nm, R586K mutation in N-terminal SH2 domain; Cm, R694K mutation in C-terminal SH2; NCm, R586K and R694K mutations. (B) Cellular β-Pix and GIT1 interact with the γSA domain of PLCγ1. A431 or BE cells were lysed, and the cell extract incubated with GST-PLCγ1SA or GST alone bound to glutathione beads. The resulting pull-down complex was analyzed by Western blotting; Src present in the initial total cell lysate (TLC) was included as a loading control. (C) Purified GIT1 and β-Pix weakly interact with the γSA domain of PLCγ1. The purified γSA domain of PLCγ1 containing a histidine tag was incubated with either GST-GIT1 (upper right panel) or GST-β-Pix (upper left panel) or GST alone bound to glutathione beads (upper panels). Sf9 lysates containing GIT1, β-Pix, or GIT1/β-Pix were incubated with GST-γ1SA bound to glutathione beads (lower panels). The resulting pull-down complexes were analyzed by Western blotting using the antibodies indicated. (D) Cellular tyrosine phosphorylation is important for interactions between PLCγ1 and GIT1/β-Pix. BE cells pretreated with genistein (50 μM) were plated on Matrigel for 4 h and lysed, and the cell extract incubated with GST-γ1SA. The resulting pull-down complex was analyzed by Western blotting (left panel). BE cells pretreated (1 h) with genistein (50 μM) or β1 integrin-blocking antibody (anti-β1, 10 μg/ml), were plated on Matrigel for 4 h, lysed, and subjected to immunoprecipitation (IP) using anti-PLCγ1 antibody. Subsequent Western blotting of these samples was performed using PLCγ1, β-Pix, or GIT1 antibodies as indicated; Western blots for GAPDH levels in total cell lysate (TCL) were used as a loading control (right panel). (E) SH2 domains of PLCγ1 are important for PLCγ1 interactions with GIT1/β-Pix. BE cells transfected with HA-tagged full-length PLCγ1 (γ1) or full-length PLCγ1 with point mutations in either N-SH2 (Nm), C-SH2 (Cm) or N- and C-SH2 (NCm) were extracted from Matrigel 4 h after plating, lysed, and subjected to IP with HA antibody. Subsequent Western blotting of these samples was performed using antibodies to PLCγ1, β-Pix, or GIT1 or, as a loading control in TCL, using antibodies to Src. (F) The interaction of GIT1 and β-Pix with PLCγ1SA facilitates the phosphorylation of PLCγ1SA on the activating tyrosine 783 site. BE cells treated with siRNAs (scrambled [Scr], GIT1, β-Pix, or Rac1 [for effectiveness of GIT1 and β-Pix siRNAs, see Fig. 1C, and for Rac1 siRNA, see Fig. 6A]) were extracted from Matrigel 4 h after plating, lysed, and subjected to IP with PLCγ1 antibody. Subsequent Western blotting of these samples was performed using either phospho-PLCγ1 (pY783) or PLCγ1 antibodies as indicated; Western blots for Src levels were used as a loading control (left panels). BE cells, transfected with GFP alone (GFP), GFP-tagged full-length PLCγ1 (γ1), or GFP-tagged PLCγ1SA (γ1SA), were extracted from Matrigel 4 h after plating, lysed, and subjected to IP with GFP antibody, and the Western blots analyzed using GIT1, β-Pix (middle panels), or pY783 (right panel) antibodies. (G) Graphical representation of the effects of PLCγ1 constructs on BE cell elongation and the ability of PLCγ1 constructs to rescue cells treated with PLCγ1 siRNA. The numbers of elongating BE cells on Matrigel after transfection with GFP (GFP) and GFP-tagged PLCγ1 constructs (γ1 and γ1SA) were analyzed in untreated cells (bars 1 to 3). The numbers of elongating BE cells on Matrigel after siRNA treatments and subsequent transfection with the indicated PLCγ1 constructs are shown in bar 4 (Scr, scrambled siRNA) and bars 5 to 11 (PLCγ1 siRNA). The number of elongated cells was determined as described in Materials and Methods and expressed as a percentage of the total cell number selected for the analysis. Error bars show standard deviations.
The experiments carried out with BE cells using different PLCγ1 variants further supported the requirement of γSA for the complex formation and, more importantly, suggested that this complex formation is critical for the activation of PLCγ1 and its role in cell elongation. As shown in Fig. 3E, we found that point mutations in the SH2 domains (known to abolish protein-protein interactions (8, 23) introduced into the full-length PLCγ1 prevented the association of PLCγ1 with the GIT1/β-Pix complex in transfected BE cells. This reduction was even more striking than that observed with the treatment with genistein. Furthermore, the association of PLCγ1 with the GIT1/β-Pix complex was also found to be necessary for the phosphorylation of PLCγ1 on the tyrosine 783 site associated with PLCγ1 activation. On Matrigel, BE cells depleted of either GIT1 or β-Pix were found to have a dramatically reduced phosphorylation of PLCγ1 Y783 (Fig. 3F, left panel). In transfected cells, both PLCγ1 and γ1SA are incorporated in GIT1/β-Pix complexes, providing further evidence that γ1SA is the main region of PLCγ1 involved in the association with the GIT1- and β-Pix-containing complexes (Fig. 3F, middle panel). We have also established that, like the full-length PLCγ1, isolated γ1SA becomes phosphorylated on the critical Y783 (Fig. 3F, right panel).
Subsequently, we tested different PLCγ1 constructs for the ability to restore the elongated morphology of PLCγ1-depleted cells, demonstrating that for PLCγ1 variants incorporating mutations in SH2 domains, this ability correlates with the ability to form complexes with GIT1/β-Pix (Fig. 3G). Interestingly, γ1SA (which does not contain the catalytic activity of PLCγ1) acted as a dominant negative construct in respect to cell elongation (Fig. 3G) and, despite the ability to be incorporated in the complex and phosphorylated (Fig. 3F, middle and right panels), did not restore the elongated morphology in BE cells depleted of endogenous PLCγ1 (Fig. 3G). This is consistent with previous findings and the data shown here using catalytically inactive variants and pharmacological inhibitors of PLC activity (25) (see Fig. 9A, lower panel).
FIG. 9.
PLCγ1 signaling pathways utilize calpain activity as one route by which they signal to Cdc42/Rac1. (A) Calpain activity and calcium facilitate Cdc42 activation. Cells were plated on Matrigel and either left untreated (Con) or incubated for 4 h in the presence of 2 μM PLC inhibitor U73122 (U73122), 10 μM calcium chelator BAPTA-AM (BAPTA), or 75 μg/ml calpain inhibitor calpeptin (Calp). BE cells were either analyzed for active Cdc42 (upper panel) or fixed, stained with phalloidin, and visualized by fluorescence microscopy (lower panel). (B) On Matrigel, the PLCγ1/GIT1/β-Pix complex signals to calpains. BE cells pretreated with calpeptin (75 μg/ml) (upper panel), siRNA (middle panel), or inhibitor (anti-β1 integrin antibody [anti-β1; 10 μg/ml], Src inhibitor [PP2; 10 μM], U73122 [2 μM], BAPTA [10 μM], Rac inhibitor [NCS23766 10 μM], or ROCK inhibitor [Y27632; 10 μM]) (lower panel) were incubated with the calpain substrate Boc-LM-CMAC (50 μM) prior to plating on Matrigel for 4 h. Cells were fixed and visualized by fluorescence microscopy for the presence of the fluorescent cleavage product that signifies calpain activity. For control and calpeptin treatments in top panels, representative fluorescent images (top left) are shown along with bright field images (top right) to show typical fields of view. Scram, scrambled siRNA. (C) Graphical representation of quantification of fluorescence in images from calpain activity assay. From the images from several independent experiments, individual cells were selected and fluorescence intensity quantified (Image-J). Calpain activity is expressed as a percentage of activity in the control (Con) (n = 60 cells analyzed). BE cells pretreated with calpain inhibitor, Calpeptin, or siRNA (upper panel); BE cells pretreated with PLCγ1 pathway inhibitor (lower panel). Scr, scrambled siRNA. (D) Western blot analysis of possible calpain substrates. BE cells pretreated with calpeptin (75 μg/ml), siRNA (scrambled [Scr] or PLCγ1), or U73122 (2 μM) were plated on Matrigel for 4 h, and the cells then extracted, processed for Western blotting, and probed with the antibodies to spectrin (fodrin) (full length, 240 kDa; cleavage product, 150 kDa) (upper left panel), filamin (full length, 280 kDa; cleavage product, 90 kDa) (upper right panel), and RhoA (full length, 23 kDa; cleavage product, 20 kDa) (lower right panel). Error bars show standard deviations. Con, control.
The role of β-Pix in the GIT1/β-Pix complex interaction with PLCγ1 was further investigated in transfected cells by using a series of β-Pix domain deletion mutants (Fig. 4A). Immunoprecipitation with either anti-GFP (to pull down the β-Pix constructs) or PLCγ1 revealed that the SH3 domain of β-Pix (aa 1 to 60) could facilitate, directly or indirectly, the interaction of β-Pix with PLCγ1 (Fig. 4A). The SH3 domain of β-Pix has previously been suggested to bind other proteins (41, 65), so it could be that binding of other factors to this region stabilizes the complex or facilitates the binding of PLCγ1. Deletion of the SH3 domain of β-Pix did not appear to affect its binding to GIT1, which is reported to associate with the C-terminal regions of β-Pix (aa 416 to 646) (49, 76). The DH and PH domains did not appear to be required in the interaction of β-Pix with PLCγ1. Interestingly, in a functional assay, a β-Pix mutant with the DH domain deleted was not able to restore the elongated phenotype in β-Pix-depleted cells (Fig. 4B), suggesting that other properties of this domain (such as its GEF activity [7]) could be important for the function of β-Pix in promoting cell elongation.
FIG. 4.
Importance of GIT1 and β-Pix in forming complexes with PLCγ1. (A) Interaction between PLCγ1 and β-Pix in BE cells. Domain organization of β-Pix: the wild type (WT) and various deletion variants (ΔSH3, ΔPH, and ΔDH) of β-Pix are shown (upper panel). The expression of the indicated GFP-tagged β-Pix deletion variants was analyzed by Western blotting using anti-GFP antibody (middle panel). Results of immunoprecipitation of BE cells transfected with β-Pix constructs and subsequent Western blotting are shown in lower panels; immunoprecipitation (IP) was either with GFP antibody (lower left panel) or PLCγ1 antibody (lower right panel). Src (present in total cell lysates [TCL]) was included as a loading control in both cases. (B) Graphical representation of the ability of GFP-tagged GIT1 or β-Pix variants to rescue the elongation on Matrigel of BE cells treated with either GIT1 or β-Pix siRNA. The following GFP-tagged GIT1 variants were used in cells treated with GIT1 siRNA (bars 2 to 5): GFP alone (GFP), wild-type GIT1 (GIT), GIT1 GAP region (aa 1 to 374) (GAP), and GIT1 SP region (aa 486 to 645) (SP). β-Pix siRNA-treated cells (bars 6 to 10) were transfected with GFP only or the following GFP-tagged variants of β-Pix: wild-type β-Pix (Pix), the SH3 domain deletion variant (ΔSH3), the PH domain deletion variant (ΔPH), and the DH domain deletion variant (ΔDH). The number of elongated cells was determined as described in Materials and Methods and expressed as a percentage of the total cell number selected for the analysis. Error bars show standard deviations. Scr, scrambled siRNA.
In similar functional studies of cell elongation, only full-length GIT1 (aa 1 to 770), but not variants expressing just the GAP domain (aa 1 to 374) or the SP region (aa 486 to 645) of GIT1, was able to rescue the elongation of BE cells depleted of GIT1, again emphasizing the need for functional components of the PLCγ1/GIT1/β-Pix complex (Fig. 4B).
Complexes containing GIT1, β-Pix, and PLCγ1 are required for activation of Cdc42 and Rac1.
To further analyze the functional relationships among PLCγ1, GIT1, and β-Pix, we performed rescue experiments with different knockdown BE cells (Fig. 5A and C, upper panel). The addition of a PLCγ1 expression vector was unable to rescue cells from either a GIT1 or β-Pix siRNA-induced rounded phenotype, although it could rescue PLCγ1 siRNA depletion. Similarly, the addition of a GIT1 expression vector could only rescue the elongation of cells depleted of GIT1, but not that of cells depleted of PLCγ1 or β-Pix. The addition of a β-Pix expression vector had no effect on the rounded-cell phenotype in cells with PLCγ1 or GIT1 depleted, but could restore elongation in cells treated with β-Pix siRNA. This suggests that all three components (PLCγ1, GIT1, and β-Pix) of the complex are necessary and that overexpression of individual components is not sufficient to replace deficiencies in another component of the PLCγ1/GIT1/β-Pix complex.
FIG. 5.
PLCγ1, GIT1, and β-Pix are all required for cell spreading on Matrigel. (A) PLCγ1, β-Pix, and GIT1 siRNAs inhibit the elongation of BE cells on Matrigel, and rescue of cell spreading is specific to restoration of expression of the siRNA's targeted protein. BE cells pretreated with siRNA were transfected with specific PLCγ1, GIT1, or β-Pix plasmids (GFP-tagged variants that are not targeted by PLCγ1, GIT1, or β-Pix siRNA, respectively) or scrambled siRNA (Scr). Cells were stained with phalloidin, and individual cell spreading analyzed. (B) Cdc42 and Rac1, but not RhoA, GTPases can rescue PLCγ1, GIT1, or β-Pix knockout effects on cell spreading. BE cells were transfected with constitutively active Rho family GTPases (Rac1 V12, Cdc42 L61, or RhoA V14) after prior treatment with PLCγ1, GIT1, or β-Pix siRNA and plated on Matrigel. Cells were stained with phalloidin, and individual cells analyzed by fluorescence microscopy. (C) Graphical representation of the effects of PLCγ1, GIT1, and β-Pix rescue constructs (upper panel) and constitutively active (CA) Rac1, Cdc42, and RhoA constructs (lower panel) on the elongation on Matrigel of BE cells pretreated with either PLCγ1, β-Pix, or GIT1 siRNA. For the experiments shown in the upper panel, conditions were as described for panel A. BE cells were treated with scrambled siRNA (bar 1), PLCγ1 siRNA (bars 2 to 5), GIT1 siRNA (bars 6 to 9), or β-Pix siRNA (bars 10 to 13). The transfection was with GFP-tagged constructs of PLCγ1, GIT1, or β-Pix as indicated. Lower panel shows quantitative analysis of data using conditions as described for panel B. BE cells were treated with siRNA as described for upper panel and then transfected with constitutively active Cdc42L61 (Cdc), Rac1V12 (Rac), or RhoAV14 (Rho) construct as indicated. In both cases, the number of elongated cells was determined and expressed as a percentage of the total cell number selected for the analysis. Error bars show standard deviations.
Numerous studies have implicated Rho family GTPases as being critical in integrin signaling leading to early actin remodeling, cell spreading, and motility (6, 11, 13, 20, 46, 52). Therefore, we tested the abilities of the constitutively active variants of Rac1, Cdc42, and RhoA to rescue BE cells depleted of PLCγ1, GIT1, or β-Pix by the specific siRNA (Fig. 5B and C, lower panel). In these experiments, active Rac1 and Cdc42 were able to rescue the cell elongation phenotype in cells depleted of PLCγ1, GIT1, or β-Pix. This shows that Cdc42 and Rac1 signaling are sufficient to establish BE cell spreading on Matrigel and suggests that the major signaling role of the complex is to activate the Cdc42/Rac1 cascade. Active RhoA was unable to rescue the spreading phenotype in BE cells treated with PLCγ1, GIT1, or β-Pix siRNA, emphasizing that RhoA is not involved in the suggested signaling cascade. Essentially the same conclusion was obtained when using PLCγ1− MEFs (Fig. 6C, lower panel, and D, lower panel).
FIG. 6.
Cdc42 and Rac1, but not RhoA, are essential for cell spreading on Matrigel. (A) Cdc42 or Rac1 siRNA treatment of BE cells prevents cell elongation. BE cells pretreated with the indicated siRNA were plated on Matrigel, and cell morphology analyzed by phase-contrast microscopy (upper panel). Cells were also labeled with phalloidin, and individual cells analyzed by fluorescence microscopy (lower panel). Western blot of BE cells from Matrigel (right panels) shows the effectiveness of the Cdc42, Rac1, and RhoA siRNA (smart pool) probes. (B) Dominant negative (DN) variants or inhibitors of Cdc42 or Rac1, but not RhoA, inhibit cell spreading. BE cells were treated with inhibitors of Rac (10 μM NCS23766) or a Rho target, ROCK (10 μM Y27632), or transfected with dominant negative GTPase constructs (Rac1 N17, Cdc42 N17, or RhoA N19) and then plated on Matrigel. Cells were stained with phalloidin, and individual cells analyzed by fluorescence microscopy. (C) Cdc42 or Rac1 can signal downstream of PLCγ1 in MEFs. PLCγ+ MEFs were transfected with the indicated dominant negative variants of these GTPases (upper panel). PLCγ− MEFs were transfected with plasmids encoding constitutively active forms of Rac1, Cdc42, or RhoA prior to plating on Matrigel (lower panel). In both cases, the cells were plated on Matrigel, stained with phalloidin, and analyzed by fluorescence microscopy. (D) Graphical representation of the effects of Rac1, Cdc42, and RhoA siRNAs, Rho family constructs, and pharmacological inhibitors on cell elongation on Matrigel. BE cells treated with the indicated siRNA, dominant negative construct, or inhibitor using conditions as described for panels plated on Matrigel, and the number of elongating cells determined 4 h after plating (upper panel). For siRNA treatment (bars 1 to 4), the following probes were used: scrambled siRNA (Scr), Cdc42 siRNA (Cdc), Rac1 siRNA (Rac), or RhoA siRNA (Rho). BE cells were also transfected with dominant negative construct (bars 6 to 9) Cdc42 N17 (Cdc), Rac1 N17 (Rac), or RhoAN19 (Rho) or treated with pharmacological inhibitor (bars 9 and 10) using 10 μM Rac inhibitor NCS23766 (*) and 10 μM Rock inhibitor Y27632 (**). Conditions for quantitative analysis of MEF elongation (lower panel) were as described for panel C. Cells transfected with the indicated dominant negative (PLCγ+ MEFs, bars 2 to 4) or constitutively active (CA) (PLCγ− MEFs, bars 5 to 8) constructs were plated on Matrigel and the number of elongating cells determined 4 h after plating. Dominant negative constructs, bars 2 to 4: Cdc42 N17 (Cdc), Rac1 N17 (Rac), and RhoA N19 (Rho). Constitutively active constructs, bars 6 to 8: Cdc42 L61 (Cdc), Rac1 V12 (Rac), and RhoA V14 (Rho).
In further studies of Rho family GTPases in our system, we used a combination of established reagents to confirm their critical role in the spreading and motility of BE cells. The use of an siRNA strategy against RhoA, Rac1, or Cdc42 revealed that, while depletion of either Cdc42 or Rac1 abolished the cell elongation phenotype, depletion of RhoA had no effect on spreading and the cells elongated normally (Fig. 6A and D, upper panel). Further analysis revealed that dominant negative variants of Rac1 or Cdc42 or chemical inhibitors in either BE (Fig. 6B and D, upper panel) cells or PLCγ1+ MEFs (Fig. 6C, upper panel, and D, lower panel) yielded the rounded-cell phenotype indicative of an inhibition of cell spreading.
Since we confirmed that Cdc42 and Rac1 were important in establishing cell spreading, we decided to further investigate signaling to these GTPases. The RBD domain of p21PAK (10) was used to pull down active (GTP bound) Rac1 from cells on Matrigel. Similarly, the CRIB domain of WASP (30) was used to pull down the active form of Cdc42. A time course for Cdc42 activation after plating on Matrigel was performed (Fig. 7A), and this established 4 h to be the optimum time point, as Cdc42 activity was high at this point and correlated with the observed cell-spreading phenotype. Initial studies with BE cells treated with U73122 or the comparison of PLCγ1+ with PLCγ1− MEFs revealed that PLCγ1 activity is required to generate active Rac1 and Cdc42 (Fig. 7B). This correlated with the cell-spreading profiles of these cells/treatments on Matrigel and suggested that PLCγ1 may lie upstream of Rac1 and Cdc42. In further experiments, siRNA against PLCγ1, GIT1, or β-Pix was found to inhibit the activation of Rac1, as monitored by a PAK pull-down assay (Fig. 8A). The siRNA probes against PLCγ1, GIT1, and β-Pix were also found to inhibit Cdc42 activation (Fig. 9B).
FIG. 7.
Requirements for activation of Cdc42 and Rac1. (A) Time course for Cdc42 activation on Matrigel. Serum-starved (24 h) BE cells were plated on Matrigel for the times shown (pre: preplating). Cell extracts were then analyzed in the N-WASP RBD pull down (PD) to isolate active Cdc42 and processed as described for panel B. Results from several experiments were analyzed, and the results expressed graphically as relative Cdc42 activity. A representative Western blot is also shown. TCL, total cell lysates. (B) BE cells, control and treated with U73122 (2 μM) (U731), or MEFs (PLCγ+ and PLCγ−) were plated on Matrigel for 4 h. The cell extracts were subsequently analyzed by PAK RBD pull-down assay to isolate active Rac1 (left panel) or subjected to N-WASP RBD pull down to isolate active Cdc42 (right panel). Western blots using anti-Rac1 and anti-Cdc42 antibodies were performed on the pull-down precipitates and on total cell lysates to assess both the amount of GTP-bound Rac1 or Cdc42 (in pull downs) and total Rac1 or Cdc42 (in total cell lysates). The samples of total cell lysates were also subjected to Western blotting using anti-Src antibody to assess total protein levels (loading control). All blots were quantified by densitometry, and the Cdc42 or Rac1 activity calculated based on the ratios of active Rac1 or Cdc42 to total Rac1 or Cdc42 when corrected for protein loading. Results from several experiments were analyzed and were expressed graphically as a percentage of the Cdc42 or Rac1 activity in control, untreated BE cells (n = 4 experiments). Representative Western blots are also shown. (C) Control BE cells (Con) and BE cells transfected with the indicated constructs encoding dominant negative Rho family GTPases (Rac1 N17, Cdc42 N17, and RhoA N19) were plated on Matrigel for 4 h, and GTPase activity assessed by pull-down assays. Rac1 activity (left panel) and Cdc42 activity (right panel) were analyzed and are presented as described for panel B. Error bars show standard deviations.
FIG. 8.
PLCγ1/GIT1/β-Pix signal to activate Cdc42 and Rac1. (A) siRNA knockdown of PLCγ1, GIT1, β-Pix, Cdc42, Rac1, and RhoA in BE cells was performed as described in the Fig. 1 and 6 legends. Cells treated with siRNA were plated on Matrigel for 4 h, and subsequently, the cell extracts subjected to PAK RBD pull-down assay to isolate active Rac; the resulting precipitates and the total cell extracts were analyzed by Western blotting using antibodies to Rac1 and Src, for a loading control. After quantification of selected areas on Western blots, the data were normalized for loading and the Rac activity expressed as the percentage of Rac activity in control, untreated BE cells (n = 4 independent experiments). Representative Western blots are also shown. (B) BE cells were processed as described for panel A, except that N-WASP RBD was used to isolate active Cdc42 that was visualized using anti-Cdc42 antibody. A graphical representation of normal Cdc42 activities relative to the activity of the control (expressed as percentages) is shown, as well as representative Western blots. PD, pull down; TCL, total cell lysates; Scr, scrambled siRNA. Error bars show standard deviations.
In initial experiments to establish an assay for the detection of activated Rac and Cdc42 in our system, we used control conditions where dominant negative variants of Rho GTPases were introduced into cells (Fig. 7C). The dominant negative RhoA variant had no effect on either Cdc42 or Rac1 activity, and the Rac1 mutant only affected Rac1 activity. However, the Cdc42 mutant not only inhibited Cdc42 activity, but also inhibited Rac1 activity to a degree, suggesting that Cdc42 may lie upstream of Rac1 and play a role, along with other components, in activating Rac1 (Fig. 7C, right panel). Use of the siRNAs against Rac1, Cdc42, and RhoA further supported these observations (Fig. 8A and B).
Calpains are involved downstream of the PLCγ1 complex in signaling to Cdc42/Rac.
In some cellular systems, the action of calcium-activated proteases and calpains is required for the activation of Cdc42 and Rac (3, 32, 51, 59, 62). Calpains have also been previously implicated in cell motility events on Matrigel (5). Since we identified PLCγ1-dependent increases in intracellular calcium as being critical for cell elongation, rather than the activation of protein kinase C (25), we tested whether the increase in calcium concentration generated by PLCγ1 could contribute to Cdc42 and Rac1 activation through the action of calpains (Fig. 9A, upper panel). Chemical inhibition of calpains or chelation of intracellular calcium dramatically inhibited the activation of Cdc42. Chemical inhibition of calpains was also found to inhibit BE cell spreading (Fig. 9A, lower panel). This suggests that calcium release is important in Cdc42/Rac1 activation and that calcium-regulated calpains could have a role in the activation of Cdc42/Rac1 and in establishing the cell elongation phenotype. The activation of calpains in the cell-spreading phenotype was then investigated by using a fluorescence-linked calpain substrate (62). Calpains were activated in elongating BE cells on Matrigel (Fig. 9B and C), and siRNA depletion of PLCγ1, GIT1, or β-Pix dramatically reduced calpain activity. The depletion of Cdc42 or Rac1 by siRNA or chemical inhibition of Rac1 activity, while preventing cell spreading, did not affect calpain activity, supporting the notion that calpain activity lies upstream of Cdc42 and Rac1 but downstream of the PLCγ1 complex (Fig. 9B and C). The use of chemical inhibitors of the PLCγ1 pathways previously implicated in cell spreading on Matrigel (25) also suggested a requirement for PLCγ1 signaling upstream of calpain signaling (Fig. 9B and C, lower panel). The depletion of RhoA by siRNA or chemical inhibition of ROCK was found not to affect calpain activity.
We then looked at possible calpain substrates, which could have a role to play in calpain signaling to Cdc42/Rac1. The calpain-dependent cleavage of the cytoskeleton-linked protein spectrin (fodrin) to yield an active 150-kDa product has previously been implicated upstream of Rac1 activation (3). In our cell system, spectrin was cleaved in a calpain-dependent manner in BE cells spreading on Matrigel (Fig. 9D, upper left panel). Spectrin cleavage was also greatly reduced in cells where PLCγ1 was either depleted by siRNA or inhibited by U73122 (Fig. 9D, upper left panel). This implicates the cleavage of spectrin as one route by which the PLCγ1 complex may utilize calpain activation to facilitate the activation of Cdc42 and Rac1.
The cytoskeleton-linked protein filamin was also found to be cleaved in BE cells plated on Matrigel; however, the inhibition of calpain or PLCγ1 activity did not prevent this cleavage, suggesting that filamin cleavage is not important in our system (Fig. 9D, right panel). The calpain cleavage of RhoA to yield a dominant negative fragment of RhoA has also been suggested to be an important motility event in some cell types (31). In BE cells plated on Matrigel, we were unable to detect any RhoA cleavage products. This suggests that the inhibition of RhoA, which could facilitate Cdc42- and Rac1-mediated responses, is not part of the regulatory mechanism in this cellular system. As both calcium and calpains have many downstream targets, it is possible that, in addition to the links described above, their other effects that are related to or independent of Cdc42/Rac1 activation could be involved in the cell spreading observed in this cellular system.
DISCUSSION
A number of recent observations suggest a critical involvement of members of the PLCγ family in several aspects of motility regulation, including chemotaxis triggered by growth factor receptors and cell spreading and movement regulated by the engagement of integrins by the components of the extracellular matrix (25, 27, 72). However, the underlying molecular mechanisms and signaling links with the known regulators of cell motility have not been clarified. Here we show that in BE cells plated on a basement membrane-type matrix, the function of PLCγ1 in cell spreading and motility requires the association of PLCγ1 with β-Pix/GIT1-containing complexes, leading to the activation of the Rho family GTPases Cdc42 and Rac1. The type of cell elongation observed here is similar to cell spreading and motility phenotypes we previously described for several cell lines (BE, DU145, and HUVEC) (25) and to the well-characterized elongated motility (61).
Our data (Fig. 1) suggest that the association of endogenous PLCγ1 with the β-Pix and GIT1 proteins requires both components, as we found that siRNA depletion of either β-Pix or GIT1 abolishes interaction with PLCγ1. This supports a model where GIT1 and β-Pix form a tight multimeric complex to which other proteins can bind (49). It is known that the β-Pix and GIT1 proteins can homodimerize and that subsequent complexes assembled in cells by β-Pix and GIT1 interactions are likely to be very large (1 to 2 MDa) (49). The other components often present in GIT1/β-Pix complexes, depending on cellular localization, are PAK (36, 41) and paxillin (38, 67). Several other signaling proteins linked to this complex have also been identified, including Rac1/Cdc42 (21, 59, 65), Cbl (12, 14), and calpain (59). Since a large number of GIT1 and β-Pix molecules are present within this extended scaffold, the same domain in different GIT1 or β-Pix molecules could recruit different binding proteins at the same time and/or multiple copies of each binding protein (49).
Our studies of the structural requirements for complex formation (Fig. 3 and 4) suggest that multiple protein-protein interactions are likely to be involved in the assembly and maintenance of PLCγ1 within complexes based on the GIT1/β-Pix scaffold. Our observations suggest that the γSA region of PLCγ1 provides sites for these multiple interactions; this is consistent with the results of previous initial, separate studies of the interactions of GIT1 and β-Pix proteins with potential binding partners that suggest the involvement of PLCγ isoforms (1, 19, 63). Furthermore, our studies provide the evidence that the association of PLCγ1 with GIT1/β-Pix complexes is critical for tyrosine phosphorylation (associated with the active state) and the function of PLCγ1 in cell spreading (Fig. 3). There are several similarities between our observations in BE cells and another cellular system where PLCγ1 is a key component of large protein complexes, namely, the activation of T cells through T-cell receptors. In T cells, intricate multiprotein scaffolds are observed, where the LAT-Gads-Slp76 complex creates a platform for the recruitment of multiple signaling molecules, including PLCγ1, Grb2, Nck, Vav, cCbl, and Itk (4, 48). Similar to our observations in BE cells (Fig. 3 and 4), several protein-protein interactions, mediated by γSA, are involved in PLCγ1 recruitment into these T-cell complexes in response to the stimulation of T-cell receptors (4). In BE cells, PLCγ1, as suggested for some other proteins (49, 59), could be recruited and released from the β-Pix/GIT1 platform in a regulated manner triggered by integrins.
How PLCγ1 recruitment to the GIT1/β-Pix complex is related to the cellular functions of the complex is a complicated issue, since the β-Pix and GIT1 proteins could have multiple roles (21, 60). Both positive (73, 74) and negative roles (16) for GIT in cell motility events have been previously reported, depending on the cell type, the stimulus employed, and isoform preference (GIT1 or GIT2). Nevertheless, considering our observed cell spreading/motility defect in cells depleted of PLCγ1, β-Pix, or GIT1 (Fig. 1 and 5), PLCγ1 is likely to be linked to the paxillin-GIT1-β-Pix-PAK module implicated in the adhesion turnover and localized actin protrusion formation involved in cell spreading and migration (13, 35, 45). The function of this module is also closely linked to the Rho family GTPases Rac and Cdc42 (59). Based on our studies using specific-siRNA depletion and rescue experiments, pharmacological inhibitors, and constitutively active and dominant negative variants of different signaling components (Fig. 7 and 8), the complex formation that includes GIT1/β-Pix and PLCγ1 seems to be upstream and required for the activation of both Cdc42 and Rac1. We also observed a correlation between the involvement of these proteins in integrin signaling and the spreading/motility phenotypes of BE cells and MEFs on basement membrane matrices (Fig. 5 and 6). Our data (Fig. 7 and 8) also indicate that Cdc42 could be required for the activation of Rac1, as observed in some other cellular systems (6, 52, 61).
It is possible that several signaling events could provide a link between complexes that include β-Pix/GIT1 and PLCγ1 and the activation of Cdc42 and Rac1. Although β-Pix belongs to a family of Rho GEFs characterized by the presence of the DH domain, there are reports that Pix proteins could lead to the activation of Cdc42 and Rac1 by both GEF activity-dependent and -independent events (59). The evidence that β-Pix acts as a Rho family GEF is based on several observations (2, 7, 12). Most notably, in the case of Rac1, direct binding to β-Pix and recruitment to the plasma membrane and focal adhesions at the leading edge were proposed and shown to have a role in cell spreading; furthermore, in the same system, β-Pix was shown to be able to act as a GEF to bound Rac1 (65). In our system, a deletion mutant of β-Pix lacking the GEF activity-containing DH domain was unable to rescue the spreading of cells depleted of endogenous β-Pix (Fig. 4C), suggesting that the GEF activity of β-Pix may be important in cell spreading events. However, we cannot completely exclude the possibility that β-Pix has primarily a scaffolding role or that, in addition, other GEFs are involved and required for the activation of either Cdc42 or Rac1.
Another signaling link between complexes that include β-Pix/GIT1 and PLCγ1 and the activation of Cdc42 and Rac1 could be provided by PLCγ1-dependent calcium release. Our previous observations have suggested a key role for the catalytic activity of PLCγ1 and, in particular, increases in intracellular calcium in cell spreading on basement membranes (25). Although there are numerous potential downstream targets of calcium, the data presented here (Fig. 9) suggest that calcium-activated proteases, calpains, could be among the key mediators. This is consistent with several previous observations, including findings that members of the PLC family can regulate calpain activity (62), that calpains associate with Pix proteins (59), and that protein cleavage by calpains is, in some cellular systems, an early event required for Rac activation, although the relevant substrate and mechanism are not clear (3, 32, 59). As proposed in a model depicted in Fig. 10, the role of PLCγ1 in the complex could be to provide an increase in local calcium concentrations, leading to calpain activation that acts together with β-Pix or other GEFs to activate Rac and/or Cdc42.
FIG. 10.

Model of PLCγ1/GIT/β-Pix signaling from integrins. Engagement of components of extracellular matrix to β1 integrins leads to phosphorylation of PLCγ1 and association of PLCγ1 with large complexes containing Pix/GIT scaffold. Subsequent activation of Rac1 and Cdc42 could be mediated by several signaling events, including stimulation of GEF activity of Pix and activation of calpains by PLC-mediated increase in cellular calcium. PIP2, phosphatidylinositol 4,5 -biphosphate; DAG, diacylglycerol; IP3, inositol 1,3,5-triphosphate.
In our studies, we focused on basement membrane-like extracellular matrices and linked PLCγ1-dependent spreading phenotypes to the activation of β1 integrin in several cell types (25). However, PLCγ1 may not be involved in signaling from other integrins activated by different extracellular matrices; one example of such integrin dependence shows that protein kinase Cα is required for α5β1 but not α4β1 integrin signaling (40). Nevertheless, the data obtained by using the cellular systems described in this study underline the importance of dynamic multiprotein complexes in signaling from adhesion receptors towards a cell spreading/motile phenotype and propose PLCγ1 as an important component in the GIT1/β-Pix complex which signals to Rac1/Cdc42 and facilitates integrin-mediated cell motility events.
Acknowledgments
We thank Graham Carpenter, Eunjoon Kim, Peter Hordijk, Richard Premont, Chris Marshall, and David Sacks for their kind gift of PLCγ1, GIT1, β-Pix, Cdc42, rac PAK-RBD, and WASP_CRIB constructs.
This work was funded by a grant from Cancer Research UK.
Footnotes
Published ahead of print on 11 June 2007.
REFERENCES
- 1.Bae, J. Y., S. J. Ahn, J. E. Lee, J. E. Kim, M. R. Han, W. Han, S. W. Kim, H. J. Shin, S. J. Lee, D. Park, and D. Y. Noh. 2005. BetaPix-a enhances the activity of phospholipase Cgamma1 by binding SH3 domain in breast cancer. J. Cell. Biochem. 94:1010-1016. [DOI] [PubMed] [Google Scholar]
- 2.Baird, D., Q. Feng, and R. A. Cerione. 2005. The Cool-2/alpha-Pix protein mediates a Cdc42-Rac signaling cascade. Curr. Biol. 15:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Bialkowska, K., S. Kulkarni, X. Du, D. E. Goll, T. C. Saido, and J. E. Fox. 2000. Evidence that beta3 integrin-induced Rac activation involves the calpain-dependent formation of integrin clusters that are distinct from the focal complexes and focal adhesions that form as Rac and RhoA become active. J. Cell Biol. 151:685-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braiman, A., M. Barda-Saad, C. L. Sommers, and L. E. Samelson. 2006. Recruitment and activation of PLCgamma1 in T cells: a new insight into old domains. EMBO J. 25:774-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carragher, N. O., S. M. Walker, L. A. Scott Carragher, F. Harris, T. K. Sawyer, V. G. Brunton, B. W. Ozanne, and M. C. Frame. 2006. Calpain 2 and Src dependence distinguishes mesenchymal and amoeboid modes of tumour cell invasion: a link to integrin function. Oncogene 25:5726-5740. [DOI] [PubMed] [Google Scholar]
- 6.Cau, J., and A. Hall. 2005. Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. J. Cell Sci. 118:2579-2587. [DOI] [PubMed] [Google Scholar]
- 7.Chahdi, A., B. Miller, and A. Sorokin. 2005. Endothelin 1 induces beta 1Pix translocation and Cdc42 activation via protein kinase A-dependent pathway. J. Biol. Chem. 280:578-584. [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhyay, A., M. Vecchi, Q. Ji, R. Mernaugh, and G. Carpenter. 1999. The role of individual SH2 domains in mediating association of phospholipase C-gamma1 with the activated EGF receptor. J. Biol. Chem. 274:26091-26097. [DOI] [PubMed] [Google Scholar]
- 9.Chen, P., H. Xie, M. C. Sekar, K. Gupta, and A. Wells. 1994. Epidermal growth factor receptor-mediated cell motility: phospholipase C activity is required, but mitogen-activated protein kinase activity is not sufficient for induced cell movement. J. Cell Biol. 127:847-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clerk, A., F. H. Pham, S. J. Fuller, E. Sahai, K. Aktories, R. Marais, C. Marshall, and P. H. Sugden. 2001. Regulation of mitogen-activated protein kinases in cardiac myocytes through the small G protein Rac1. Mol. Cell. Biol. 21:1173-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeMali, K. A., K. Wennerberg, and K. Burridge. 2003. Integrin signaling to the actin cytoskeleton. Curr. Opin. Cell Biol. 15:572-582. [DOI] [PubMed] [Google Scholar]
- 12.Feng, Q., D. Baird, and R. A. Cerione. 2004. Novel regulatory mechanisms for the Dbl family guanine nucleotide exchange factor Cool-2/alpha-Pix. EMBO J. 23:3492-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipenko, N. R., S. Attwell, C. Roskelley, and S. Dedhar. 2005. Integrin-linked kinase activity regulates Rac- and Cdc42-mediated actin cytoskeleton reorganization via alpha-PIX. Oncogene 24:5837-5849. [DOI] [PubMed] [Google Scholar]
- 14.Flanders, J. A., Q. Feng, S. Bagrodia, M. T. Laux, A. Singavarapu, and R. A. Cerione. 2003. The Cbl proteins are binding partners for the Cool/Pix family of p21-activated kinase-binding proteins. FEBS Lett. 550:119-123. [DOI] [PubMed] [Google Scholar]
- 15.Fleming, I. N., C. M. Elliott, and J. H. Exton. 1998. Phospholipase C-gamma, protein kinase C and Ca2+/calmodulin-dependent protein kinase II are involved in platelet-derived growth factor-induced phosphorylation of Tiam1. FEBS Lett. 249:229-233. [DOI] [PubMed] [Google Scholar]
- 16.Frank, S. R., M. R. Adelstein, and S. H. Hansen. 2006. GIT2 represses Crk- and Rac1-regulated cell spreading and Cdc42-mediated focal adhesion turnover. EMBO J. 25:1848-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedl, P., and E. B. Brocker. 2000. The biology of cell locomotion within three-dimensional extracellular matrix. Cell. Mol. Life Sci. 57:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedl, P., and K. Wolf. 2003. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 3:362-374. [DOI] [PubMed] [Google Scholar]
- 19.Haendeler, J., G. Yin, Y. Hojo, Y. Saito, M. Melaragno, C. Yan, V. K. Sharma, M. Heller, R. Aebersold, and B. C. Berk. 2003. GIT1 mediates Src-dependent activation of phospholipase Cgamma by angiotensin II and epidermal growth factor. J. Biol. Chem. 278:49936-49944. [DOI] [PubMed] [Google Scholar]
- 20.Hall, A. 2005. Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33:891-895. [DOI] [PubMed] [Google Scholar]
- 21.Hoefen, R. J., and B. C. Berk. 2006. The multifunctional GIT family of proteins. J. Cell Sci. 119:1469-1475. [DOI] [PubMed] [Google Scholar]
- 22.Inoue, O., K. Suzuki-Inoue, W. L. Dean, J. Frampton, and S. P. Watson. 2003. Integrin alpha2beta1 mediates outside-in regulation of platelet spreading on collagen through activation of Src kinases and PLCgamma2. J. Cell Biol. 160:769-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji, Q. S., A. Chattopadhyay, M. Vecchi, and G. Carpenter. 1999. Physiological requirement for both SH2 domains for phospholipase C-γ1 function and interaction with platelet-derived growth factor receptors. Mol. Cell. Biol. 19:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji, Q. S., S. Ermini, J. Baulida, F. L. Sun, and G. Carpenter. 1998. Epidermal growth factor signaling and mitogenesis in Plcg1 null mouse embryonic fibroblasts. Mol. Biol. Cell 9:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, N. P., J. Peak, S. Brader, S. A. Eccles, and M. Katan. 2005. PLCgamma1 is essential for early events in integrin signalling required for cell motility. J. Cell Sci. 118:2695-2706. [DOI] [PubMed] [Google Scholar]
- 26.Kassis, J., D. A. Lauffenburger, T. Turner, and A. Wells. 2001. Tumor invasion as dysregulated cell motility. Semin. Cancer Biol. 11:105-117. [DOI] [PubMed] [Google Scholar]
- 27.Kassis, J., J. Moellinger, H. Lo, N. M. Greenberg, H. G. Kim, and A. Wells. 1999. A role for phospholipase C-gamma-mediated signaling in tumor cell invasion. Clin. Cancer Res. 5:2251-2260. [PubMed] [Google Scholar]
- 28.Katan, M. 2005. New insights into the families of PLC enzymes: looking back and going forward. Biochem. J. 391:e7-e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, S., J. Ko, H. Shin, J. R. Lee, C. Lim, J. H. Han, W. D. Altrock, C. C. Garner, E. D. Gundelfinger, R. T. Premont, B. K. Kaang, and E. Kim. 2003. The GIT family of proteins forms multimers and associates with the presynaptic cytomatrix protein Piccolo. J. Biol. Chem. 278:6291-6300. [DOI] [PubMed] [Google Scholar]
- 30.Kim, S. H., Z. Li, and D. B. Sacks. 2000. E-cadherin-mediated cell-cell attachment activates Cdc42. J. Biol. Chem. 275:36999-37005. [DOI] [PubMed] [Google Scholar]
- 31.Kulkarni, S., D. E. Goll, and J. E. Fox. 2002. Calpain cleaves RhoA generating a dominant-negative form that inhibits integrin-induced actin filament assembly and cell spreading. J. Biol. Chem. 277:24435-24441. [DOI] [PubMed] [Google Scholar]
- 32.Kulkarni, S., T. C. Saido, K. Suzuki, and J. E. Fox. 1999. Calpain mediates integrin-induced signaling at a point upstream of Rho family members. J. Biol. Chem. 274:21265-21275. [DOI] [PubMed] [Google Scholar]
- 33.Kundra, V., J. A. Escobedo, A. Kazlauskas, H. K. Kim, S. G. Rhee, L. T. Williams, and B. R. Zetter. 1994. Regulation of chemotaxis by the platelet-derived growth factor receptor-beta. Nature 367:474-476. [DOI] [PubMed] [Google Scholar]
- 34.Langholz, O., D. Roeckel, D. Petersohn, E. Broermann, B. Eckes, and T. Krieg. 1997. Cell-matrix interactions induce tyrosine phosphorylation of MAP kinases ERK1 and ERK2 and PLCgamma-1 in two-dimensional and three-dimensional cultures of human fibroblasts. Exp. Cell Res. 235:22-27. [DOI] [PubMed] [Google Scholar]
- 35.Lee, J., I. D. Jung, W. K. Chang, C. G. Park, D. Y. Cho, E. Y. Shin, D. W. Seo, Y. K. Kim, H. W. Lee, J. W. Han, and H. Y. Lee. 2005. p85 beta-PIX is required for cell motility through phosphorylations of focal adhesion kinase and p38 MAP kinase. Exp. Cell Res. 307:315-328. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. H., M. Eom, S. J. Lee, S. Kim, H. J. Park, and D. Park. 2001. BetaPix-enhanced p38 activation by Cdc42/Rac/PAK/MKK3/6-mediated pathway. Implication in the regulation of membrane ruffling. J. Biol. Chem. 276:25066-25072. [DOI] [PubMed] [Google Scholar]
- 37.Liao, F., H. S. Shin, and S. G. Rhee. 1993. In vitro tyrosine phosphorylation of PLC-gamma 1 and PLC-gamma 2 by src-family protein tyrosine kinases. Biochem. Biophys. Res. Commun. 191:1028-1033. [DOI] [PubMed] [Google Scholar]
- 38.Manabe, R., M. Kovalenko, D. J. Webb, and A. R. Horwitz. 2002. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J. Cell Sci. 115:1497-1510. [DOI] [PubMed] [Google Scholar]
- 39.Mangin, P., Y. Yuan, I. Goncalves, A. Eckly, M. Freund, J. P. Cazenave, C. Gachet, S. P. Jackson, and F. Lanza. 2003. Signaling role for phospholipase C gamma 2 in platelet glycoprotein Ib alpha calcium flux and cytoskeletal reorganization. Involvement of a pathway distinct from FcR gamma chain and Fc gamma RIIA. J. Biol. Chem. 278:32880-32891. [DOI] [PubMed] [Google Scholar]
- 40.Mostafavi-Pour, Z., J. A. Askari, S. J. Parkinson, P. J. Parker, T. T. Ng, and M. J. Humphries. 2003. Integrin-specific signaling pathways controlling focal adhesion formation and cell migration. J. Cell Biol. 161:155-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mott, H. R., D. Nietlispach, K. A. Evetts, and D. Owen. 2005. Structural analysis of the SH3 domain of beta-PIX and its interaction with alpha-p21 activated kinase (PAK). Biochemistry 44:10977-10983. [DOI] [PubMed] [Google Scholar]
- 42.Mouneimne, G., L. Soon, V. DesMarais, M. Sidani, X. Song, S. C. Yip, M. Ghosh, R. Eddy, J. M. Backer, and J. Condeelis. 2004. Phospholipase C and cofilin are required for carcinoma cell directionality in response to EGF stimulation. J. Cell Biol. 166:697-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura, I., L. Lipfert, G. A. Rodan, and L. T. Duong. 2001. Convergence of alpha(v)beta(3) integrin- and macrophage colony stimulating factor-mediated signals on phospholipase Cgamma in prefusion osteoclasts. J. Cell Biol. 152:361-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natarajan, K., G. Yin, and B. C. Berk. 2004. Scaffolds direct Src-specific signaling in response to angiotensin II: new roles for Cas and GIT1. Mol. Pharmacol. 65:822-825. [DOI] [PubMed] [Google Scholar]
- 45.Nayal, A., D. J. Webb, C. M. Brown, E. M. Schaefer, M. Vicente-Manzanares, and A. R. Horwitz. 2006. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 173:587-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nobes, C. D., and A. Hall. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81:53-62. [DOI] [PubMed] [Google Scholar]
- 47.Park, E., M. Na, J. Choi, S. Kim, J. R. Lee, J. Yoon, D. Park, M. Sheng, and E. Kim. 2003. The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. J. Biol. Chem. 278:19220-19229. [DOI] [PubMed] [Google Scholar]
- 48.Phee, H., R. T. Abraham, and A. Weiss. 2005. Dynamic recruitment of PAK1 to the immunological synapse is mediated by PIX independently of SLP-76 and Vav1. Nat. Immunol. 6:608-617. [DOI] [PubMed] [Google Scholar]
- 49.Premont, R. T., S. J. Perry, R. Schmalzigaug, J. T. Roseman, Y. Xing, and A. Claing. 2004. The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell. Signal. 16:1001-1011. [DOI] [PubMed] [Google Scholar]
- 50.Price, J. T., T. Tiganis, A. Agarwal, D. Djakiew, and E. W. Thompson. 1999. Epidermal growth factor promotes MDA-MB-231 breast cancer cell migration through a phosphatidylinositol 3′-kinase and phospholipase C-dependent mechanism. Cancer Res. 59:5475-5478. [PubMed] [Google Scholar]
- 51.Price, L. S., M. Langeslag, J. P. ten Klooster, P. L. Hordijk, K. Jalink, and J. G. Collard. 2003. Calcium signaling regulates translocation and activation of Rac. J. Biol. Chem. 278:39413-39421. [DOI] [PubMed] [Google Scholar]
- 52.Price, L. S., J. Leng, M. A. Schwartz, and G. M. Bokoch. 1998. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol. Biol. Cell 9:1863-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raucher, D., T. Stauffer, W. Chen, K. Shen, S. Guo, J. D. York, M. P. Sheetz, and T. Meyer. 2000. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell 100:221-228. [DOI] [PubMed] [Google Scholar]
- 54.Rebecchi, M. J., and S. N. Pentyala. 2000. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol. Rev. 80:1291-1335.11015615 [Google Scholar]
- 55.Rhee, S.-G. 2001. Regulation of phophoinositide-specific phospholipase C. Annu. Rev. Biochem. 70:281-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ridley, A. J., A. J. Self, F. Kasmi, H. F. Paterson, A. Hall, C. J. Marshall, and C. Ellis. 1993. Rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J. 12:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez, R., M. Matsuda, O. Perisic, J. Bravo, A. Paul, N. P. Jones, Y. Light, K. Swann, R. L. Williams, and M. Katan. 2001. Tyrosine residues in phospholipase Cgamma 2 essential for the enzyme function in B-cell signaling. J. Biol. Chem. 276:47982-47992. [DOI] [PubMed] [Google Scholar]
- 58.Ronnstrand, L., A. Siegbahn, C. Rorsman, M. Johnell, K. Hansen, and C. H. Heldin. 1999. Overactivation of phospholipase C-gamma1 renders platelet-derived growth factor beta-receptor-expressing cells independent of the phosphatidylinositol 3-kinase pathway for chemotaxis. J. Biol. Chem. 274:22089-22094. [DOI] [PubMed] [Google Scholar]
- 59.Rosenberger, G., A. Gal, and K. Kutsche. 2005. AlphaPIX associates with calpain 4, the small subunit of calpain, and has a dual role in integrin-mediated cell spreading. J. Biol. Chem. 280:6879-6889. [DOI] [PubMed] [Google Scholar]
- 60.Rosenberger, G., and K. Kutsche. 2006. AlphaPIX and betaPIX and their role in focal adhesion formation. Eur. J. Cell Biol. 85:265-274. [DOI] [PubMed] [Google Scholar]
- 61.Sahai, E., and C. J. Marshall. 2003. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat. Cell Biol. 5:711-719. [DOI] [PubMed] [Google Scholar]
- 62.Satish, L., H. C. Blair, A. Glading, and A. Wells. 2005. Interferon-inducible protein 9 (CXCL11)-induced cell motility in keratinocytes requires calcium flux-dependent activation of μ-calpain. Mol. Cell. Biol. 25:1922-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitz, U., M. Ishida, and B. C. Berk. 1997. Angiotensin II stimulates tyrosine phosphorylation of phospholipase C-gamma-associated proteins. Characterization of a c-Src-dependent 97-kD protein in vascular smooth muscle cells. Circ. Res. 81:550-557. [DOI] [PubMed] [Google Scholar]
- 64.Scott, C. C., W. Dobson, R. J. Botelho, N. Coady-Osberg, P. Chavrier, D. A. Knecht, C. Heath, P. Stahl, and S. Grinstein. 2005. Phosphatidylinositol-4,5-bisphosphate hydrolysis directs actin remodeling during phagocytosis. J. Cell Biol. 169:139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.ten Klooster, J. P., Z. M. Jaffer, J. Chernoff, and P. L. Hordijk. 2006. Targeting and activation of Rac1 are mediated by the exchange factor beta-Pix. J. Cell Biol. 172:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomas, S. M., F. M. Coppelli, A. Wells, W. E. Gooding, J. Song, J. Kassis, S. D. Drenning, and J. R. Grandis. 2003. Epidermal growth factor receptor-stimulated activation of phospholipase Cgamma-1 promotes invasion of head and neck squamous cell carcinoma. Cancer Res. 63:5629-5635. [PubMed] [Google Scholar]
- 67.Turner, C. E. 2000. Paxillin interactions. J. Cell Sci. 113:4139-4140. [DOI] [PubMed] [Google Scholar]
- 68.Turner, T., M. V. Epps-Fung, J. Kassis, and A. Wells. 1997. Molecular inhibition of phospholipase Cgamma signaling abrogates DU-145 prostate tumor cell invasion. Clin. Cancer Res. 3:2275-2282. [PubMed] [Google Scholar]
- 69.Tvorogov, D., X. J. Wang, R. Zent, and G. Carpenter. 2005. Integrin-dependent PLC-gamma1 phosphorylation mediates fibronectin-dependent adhesion. J. Cell Sci. 118:601-610. [DOI] [PubMed] [Google Scholar]
- 70.Wells, A. 2000. Tumor invasion: role of growth factor-induced cell motility. Adv. Cancer Res. 78:31-101. [DOI] [PubMed] [Google Scholar]
- 71.Wolf, K., I. Mazo, H. Leung, K. Engelke, U. H. von Andrian, E. I. Deryugina, A. Y. Strongin, E. B. Brocker, and P. Friedl. 2003. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 160:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wonerow, P., A. C. Pearce, D. J. Vaux, and S. P. Watson. 2003. A critical role for phospholipase Cgamma2 in alphaIIbbeta3-mediated platelet spreading. J. Biol. Chem. 278:37520-37529. [DOI] [PubMed] [Google Scholar]
- 73.Yin, G., Q. Zheng, C. Yan, and B. C. Berk. 2005. GIT1 is a scaffold for ERK1/2 activation in focal adhesions. J. Biol. Chem. 280:27705-27712. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, H., D. J. Webb, H. Asmussen, and A. F. Horwitz. 2003. Synapse formation is regulated by the signaling adaptor GIT1. J. Cell Biol. 161:131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, X., A. Chattopadhyay, Q. S. Ji, J. D. Owen, P. J. Ruest, G. Carpenter, and S. K. Hanks. 1999. Focal adhesion kinase promotes phospholipase C-gamma1 activity. Proc. Natl. Acad. Sci. USA 96:9021-9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao, Z. S., E. Manser, T. H. Loo, and L. Lim. 2000. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol. Cell. Biol. 20:6354-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]