Abstract
A critical step in cell division is formation of the mitotic spindle, which is a bipolar array of microtubules that mediates chromosome separation. Here, we report that the SCL-interrupting locus (SIL), a vertebrate-specific cytosolic protein, is necessary for proper mitotic spindle organization in zebrafish and human cells. A homozygous lethal zebrafish mutant, cassiopeia (csp), was identified by a genetic screen for mitotic mutant. csp mutant embryos have an increased mitotic index, have highly disorganized mitotic spindles, and often lack one or both centrosomes. These phenotypes are caused by a loss-of-function mutation in zebrafish sil. To determine if the requirement for SIL in mitotic spindle organization is conserved in mammals, we generated an antibody against human SIL, which revealed that SIL localizes to the poles of the mitotic spindle during metaphase. Furthermore, short hairpin RNA knockdown of SIL in human cells recapitulates the zebrafish csp mitotic spindle defects. These data, taken together, identify SIL as a novel, vertebrate-specific regulator of mitotic spindle assembly.
During mitosis, a dramatic reorganization of the cellular microtubules occurs to create a bipolar mitotic spindle. The microtubules attach to the kinetochores of the duplicated chromosomes and direct the chromosomes as they separate and migrate to opposite poles (17). In animal cells, the spindle poles contain the centrosomes, which play a critical role in nucleating and organizing the microtubules (7). There are hundreds of proteins involved in mitotic spindle formation, including microtubule motor (kinesins and dynein), centrosome, and kinetochore proteins (14, 27). The importance of accurate mitotic spindle formation is emphasized by the existence of the mitotic spindle checkpoint, a signaling network that ensures proper assembly of the spindle prior to anaphase (30).
The zebrafish, Danio rerio, has emerged as a powerful model organism in which to study vertebrate development. It is possible to examine the cell cycle in the developing zebrafish embryos through a variety of assays (38). The zebrafish is also genetically tractable, and large-scale forward genetic screens for embryonic lethal mutations have been conducted (12, 16). Some of these embryonic lethal mutations are predicted to be in cell cycle regulatory genes and are called the early-arrest mutant (25). These mutant, such as speed bump, ogre, zombie, specter, and poltergeist, produce phenotypes that include mitotic bridges, mitotic arrest, absence of cytokinesis, and abnormal nuclei accompanied by cell lysis. As these characteristics typically result from cell division defects, it is likely that the mutations responsible are in genes required for cell cycle progression. More recently, other zebrafish mutants with cell cycle defects have been characterized. The maternal-effect mutation futile cycle, for example, is characterized by failure to form a mitotic spindle in the first cell division (10). A forward genetic screen for mitotic mutant in zebrafish uncovered eight mutant lines, including the crash&burn line, a b-myb mutant, and the cease&desist line, which harbored a mutation in separase (36, 37). An insertional mutagenesis screen uncovered several hundred mutant lines, some of which bore mutations in cell cycle regulatory genes (1, 2, 15). One of the mutant identified by this screen, denticleless (dtl/cdt2), exhibited DNA rereplication and failed to initiate a cell cycle checkpoint following irradiation (34). These zebrafish mutants highlight the strength of the system for studying cell cycle defects in vivo during embryogenesis.
The SCL-interrupting locus (sil) is a vertebrate-specific gene involved in a chromosome 1 deletion that occurs in 25% of T-cell acute lymphoblastic leukemias (T-ALL) (4). The 150-kDa SIL protein lacks homology to any known protein families or motifs (5). SIL mRNA and protein are expressed specifically during mitosis (20), and Sil−/− mice die at embryonic day 10.5 with midline and left-right asymmetry defects (22). SIL is overexpressed in several tumor types, including melanoma and lung carcinomas, and Sil gene expression has been computationally correlated with that of spindle checkpoint genes (13). Additionally, Sil was identified as 1 of 128 genes predictive of the metastatic potential of adenocarcinomas (33). It remains a gene of unknown function, although genetic and biochemical studies have provided insights into potential pathways. Genetic analysis of mice double mutant for Sil and Patched (Ptch), a Sonic Hedgehog (Shh) pathway member, indicated that SIL may be in the Shh pathway and act downstream of Patched (21). Biochemical studies have revealed that SIL is phosphorylated during mitosis and that this phosphorylation is required for interaction with Pin1, which is currently the only identified binding partner of SIL (8). Expression of phosphorylation site mutant SIL or knockdown of SIL by RNA interference (RNAi) causes defects in the spindle checkpoint (8). These data support a potential role for SIL in the cell cycle or the cell cycle checkpoints, though a mechanism remains elusive.
Here, we identify and characterize a new zebrafish mutant, the cassiopeia (csp) line, which has a loss-of-function mutation in zebrafish sil. csp homozygotes have an embryonic lethal defect and exhibit increased numbers of mitotic cells, as detected by phospho-H3 immunostaining. Antibody staining with α-tubulin and γ-tubulin antibodies revealed that mitotic cells of csp mutant embryos have extremely disorganized mitotic spindles and they often lack one or both centrosomes. Immunostaining with an antibody that we generated against human SIL revealed that SIL localizes to the poles of the mitotic spindle in metaphase cells, strongly supporting a role for SIL in microtubule dynamics. Additionally, knockdown of SIL by short hairpin RNA (shRNA) in HeLa cells resulted in dividing cells with disorganized mitotic spindles. These data suggest that SIL plays a conserved critical role in organizing the mitotic spindle in vertebrate cells.
MATERIALS AND METHODS
Zebrafish maintenance and screening.
Zebrafish were maintained and staged as previously described (26, 39). The cspcz65 and cspcz299 mutants were identified by an F2 haploid screen performed in the AB strain background as previously described (37). The Children's Hospital Boston Institutional Animal Care and Use Committee approved all of the zebrafish protocols used in this study.
Genetic mapping and mutation analysis.
csp AB heterozygotes were crossed to polymorphic WIK wild-type fish to generate mapping strains. Embryos from mapping strain heterozygote incrosses were scored at 36 h postfertilization (hpf) by morphology and collected for DNA extraction as previously described (42). Bulk segregant analysis was performed with PCR primers for CA microsatellite markers throughout the genome, and linkage was established on linkage group 22. Linkage analysis of a meiotic panel of 885 diploid csp mutants genetically mapped the mutation to a 4-cM interval flanked by CA microsatellite markers z63239 and z13966. Novel microsatellite and single-strand conformation polymorphism marker regions were identified from ZV4 Sanger assembly sequence and tested to close the interval. Overlapping oligonucleotide probes were designed to the closest markers and used for hybridization to DanioKey BAC library filters. To identify the mutations in sil, overlapping halves of sil were isolated from 30-hpf wild-type, cspcz65, and cspcz299 embryos by reverse transcription-PCR with primers F1 (5′-ATGAACCGTGTACAAGTGGA-3′), R3 (5′-CACTGCTATGACCTGATGTTGG-3′), F3 (5′-TGAGACCTCTAGTACAGGGAAG-3′), and R1 (5′-TTAAAAGAGCTTGGGGAGCTGC-3′) and sequenced directly. Mutations were confirmed by single-embryo genomic PCR amplification of the locus of interest.
Zebrafish whole-mount staining.
Whole-mount immunostaining on 24- to 30-hpf zebrafish embryos was performed as previously described (37), with polyclonal Ser-10 phospho-histone H3 (Santa Cruz), polyclonal γ-tubulin (Sigma), and monoclonal α-tubulin (Sigma) antibodies. Phospho-H3 staining was quantified by counting positive cells in defined regions of the tail and with ImageJ software. Bromodeoxyuridine (BrdU) incorporation was performed as previously described (37) and detected with tetramethyl rhodamine-conjugated antibodies (Molecular Probes) on embryos costained with phospho-histone H3 (Santa Cruz) detected with fluorescein-conjugated antibodies (Jackson ImmunoResearch). Apoptosis was detected by acridine orange and terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling with the ApoTag in situ detection kit (Chemicon) as previously described (37). Whole-mount in situ hybridization was performed as previously described (24).
Flow cytometry.
Pools of 20 csp and wild-type 30-hpf embryos were disaggregated, stained with propidium iodide as previously described (38), and analyzed with a Becton Dickinson FACScalibur flow cytometer. Quantitative analysis and cell cycle profiling was performed with FlowJo.
Western blotting.
Protein lysates were extracted from 24- to 30-hpf zebrafish embryos with radioimmunoprecipitation assay buffer supplemented with complete protease inhibitor tablets (Roche). Forty micrograms of total protein was subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose for immunoblotting with polyclonal γ-tubulin (Sigma).
Morpholino oligonucleotides.
Two splice site morpholino oligonucleotides designed against zebrafish sil, moA (5′-CCCACTCAACCATCTGTCAACAT GA-3′) and moB (5′-GTATGGGCTACTC-ACTTGTCCTGGA-3′), were generated by Gene Tools LLC, reconstituted in nuclease-free water, and maintained as a 4 mM stock. Tü or AB strain embryos were injected at the one- to two-cell stage with 1 nl of 0.2 mM moA or moB or 0.1 mM each moA and moB. Injected embryos were maintained at 28.5°C, fixed at 24 hpf, and processed for phospho-H3 or mitotic spindle immunostaining as described above.
Allele-specific genotyping.
The G-to-A mutation at nucleotide 820 in cspcz65 was used to create distinguishing reverse primers (wild-type product, 5′-TGAATGTGTGTAACCCCACTCAAC-3′; mutant product, 5′-TGAATGTGTGTAACCCCACTCAAT-3′) for allele-specific PCRs. One intronic forward primer (5′-ACCATGGATCAGACCAGGAA-3′) was used with each reverse primer to generate a 330-bp product.
Cloning and plasmids.
Full-length zebrafish sil was obtained by reverse transcription-PCR from 30-hpf embryos and cloned into pTOPO-Blunt (Invitrogen). Full-length digoxigenin antisense riboprobe was transcribed from linearized plasmid with a T7 RNA labeling kit (Roche). Full-length human sil was subcloned from Image Clone CS0DG001YK07 (Genoscope) into pCS2 and pCS2-HA (provided by S. Rankin). Five pairs of hairpin primers with target sizes ranging from 20 to 28 bp (SH1, nt 110 to 129 from the translational start site; SH2, nt 338 to 349; SH3, nt 501 to 520; SH4, nt 615 to 642; SH6, nt 2666 to 2686) and either a nonamer loop for SH1, SH2, and SH3 (5′-TTCAAGAGA-3′) or a trimer loop for SH4 and SH6 (5′-ATG-3′) were designed with Bbs1/Xba1 overhangs and cloned into the pMU6pro vector (provided by S. Rankin) as previously described (41).
Cell lines, transfections, and immunostaining.
HEK293T and HeLa (ATTC CCL-2) cell lines were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and 50 μg/ml penicillin-streptomycin. Cells were transfected with the FuGENE 6 reagent (Roche) in Optimem-1 (Invitrogen) supplemented with 2% fetal bovine serum in accordance with the manufacturer's protocol. For immunostaining, cells were cultured on coverslips, fixed for 3 min in −20°C methanol, and permeabilized with 0.4% Triton X-100-phosphate-buffered saline for 10 min. Monoclonal α-tubulin (Sigma) and anti-dynactin (p50; BD Transduction Laboratories) antibodies and polyclonal γ-tubulin antibody (Sigma) were used at a 1:1,000 dilution. Anti-SIL antibody (described below) was used at a 1:100 dilution. The secondary antibodies used, all at a 1:500 dilution, were tetramethyl rhodamine-conjugated anti-mouse (Molecular Probes), tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit (Jackson ImmunoResearch), fluorescein isothiocyanate-conjugated anti-mouse (Jackson ImmunoResearch), and fluorescein isothiocyanate-conjugated anti-rabbit (Jackson ImmunoResearch) antibodies. The shRNA phenotype was quantified by scoring the microtubule morphology for all mitotic cells and assessment for the presence of SIL staining (n = >50 cells per experiment; each shRNA was analyzed in at least three separate experiments). For statistical analysis of these data, a two-tailed t test was performed.
Antiserum production and immunoblotting.
A sil fragment corresponding to the C-terminal 348 amino acids was generated and cloned into pDONR201 with Gateway BP reaction mix (Invitrogen). Gateway LR reactions were used to subclone the sil C terminus into Gateway compatible pGEX2TK (Amersham) and pMAL (New England Biolabs). Purified, glutathione S-transferase-tagged protein was used to immunize rabbits according to standard protocols by Covance, Inc. Nitrocellulose carrying ∼1 mg maltose-binding protein-tagged C-terminal SIL was incubated with antiserum and washed, and SIL-specific antibodies were eluted with 100 mM glycine-HCl (pH 2.5). For immunoblotting, HEK293T cells were lysed with radioimmunoprecipitation assay buffer supplemented with complete protease inhibitors (Roche). Thirty micrograms of total protein was subjected to 6% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Standard techniques were used for Western blotting with 1:2,000 antihemagglutinin (anti-HA) (12CA5; Roche) or 1:1,000 anti-SIL. Horseradish peroxidase-conjugated anti-mouse and anti-rabbit secondary antibodies (Jackson ImmunoResearch) were used and detected with ECL reagent (Amersham).
Microscopy.
Embryos were mounted in glycerol and photographed on a Nikon Eclipse E600 compound microscope with a Nikon Coolpix 4500 camera. Fluorescently stained zebrafish embryos and mammalian cells were mounted in Vectashield (Vector Labs), examined with a Zeiss Axioskop2 Plus compound microscope, and photographed with a Hamamatsu camera with OpenLab software (Improvision). For confocal microscopy, the mounting medium was 0.1% N-propyl gallate in 50% glycerol and 50% 2× phosphate-buffered saline. Confocal images were captured at the Harvard Medical School Department of Cell Biology Nikon Imaging Center on a Nikon Eclipse Spinning Disk Confocal Microscope.
RESULTS
Identification of the zebrafish cassiopeia mutant.
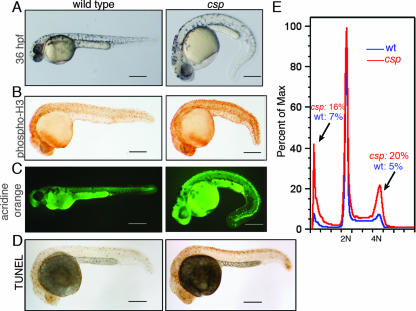
The zebrafish system provides a powerful opportunity to perform large-scale genetic screens for cell division defects in vertebrates. To identify genes regulating the vertebrate cell cycle, we performed a haploid N-ethyl-N-nitrosourea mutagenesis screening of zebrafish for mitotic mutants with phosphorylated histone H3 (phospho-H3) immunostaining. Histone H3 is phosphorylated during the G2 and M phases of the cell cycle and is an informative mitotic marker in zebrafish embryos (18, 37). Twelve mutant lines were recovered from the screen. Two autosomal, recessively inherited, alleles of the cassiopeia (csp) mutant, cspcz65 and cspcz299, were identified. Homozygous csp mutants had a fourfold increase in phospho-H3 staining (Fig. 1B), which was first evident at 16 hpf. Later in development, morphological defects arose, including anterior neural cell death at 24 hpf (Fig. 1A, C, and D), ventral or dorsal tail curvature at 36 hpf (Fig. 1A), and cardiac edema at 36 hpf. Both csp alleles exhibited the same onset and severity of all phenotypes. DNA content analysis by flow cytometry revealed that the mutants possessed increased mitotic 4N and apoptotic sub-G1 populations (Fig. 1E), correlating with the whole-mount staining phenotypes. Unlike previously characterized zebrafish cell cycle mutants that die between 2 and 5 days postfertilization, csp mutants die between 7 and 10 days postfertilization.
FIG. 1.
Characterization of the cassiopeia zebrafish mutant, which has mitotic defects and increased apoptosis. (A) At 36 hpf, csp mutant embryos are ventrally curved, have increased cell death that gives the head an opaque gray appearance, and begin to develop cardiac edema. (B) Phospho-H3 immunostaining on 28-hpf embryos shows an increased number of mitotic cells in csp mutants. (C) Acridine orange staining detects high levels of cell death in the head and dorsal neurons of csp mutant embryos. Note that the bright green of the yolk is autofluorescence. (D) Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) indicates that the dorsal cell death in csp mutants is apoptotic. (E) Representative DNA content analysis of wild-type (wt) and csp mutant embryos showing increased populations of 4N and sub-G1 populations. Scale bars = 250 μm. Max, maximum.
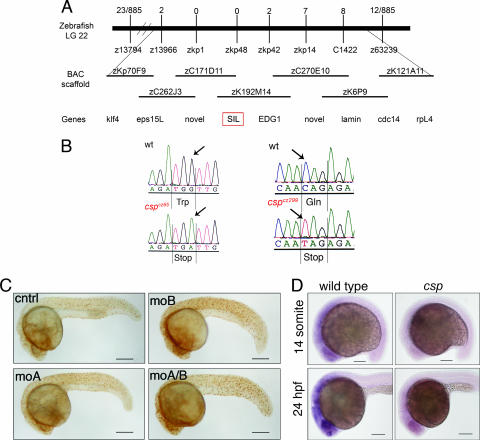
By meiotic linkage analysis, the mutation was genetically mapped to an ∼4-cM interval on linkage group 22 (Fig. 2A). Examination of the fully sequenced bacterial artificial chromosomes (BACs) that spanned this region identified only two genes within the narrowest genetic interval, i.e., the zebrafish homolog of the SCL-interrupting locus (sil) and a completely novel putative transcript. Although it is a gene of unknown function, sil was a logical candidate because it is specifically expressed during mitosis (20). Sequencing of the sil loci of cspcz65 and cspcz299 mutants revealed that each possessed independent nonsense mutations (Fig. 2B). Another csp mutant allele, csphi1262, was identified in a large-scale insertional mutagenesis screen (2, 15) and failed to complement the increased phospho-H3 staining phenotypes of both cspcz65 and cspcz299. The insertion mutation retains about 10% of wild-type levels of sil transcript (15), and the phospho-H3 and morphological phenotypes are less severe than those of cspcz65 and cspcz299.
FIG. 2.
Loss of SIL gene function causes the csp mutant phenotype. (A, top) Schematic representation of the genetic mapping interval containing the csp mutation on zebrafish linkage group 22. Recombinants for each microsatellite or single-strand conformation polymorphism marker across the region are shown. (A, middle) Physical map of the fully sequenced BACs coinciding with the csp interval on linkage group 22. (A, bottom) Genes identified by BLAST searches based on the BAC sequences. Note that microsatellite marker zkp48 falls within the sil locus and has 0 recombinants. (B) Sequencing of the cspcz65 and cspcz299 mutants revealed they possessed independent nonsense mutations. cspcz65 mutants have a G-to-A mutation at nt 820 of the coding sequence, which results in a UGA stop codon corresponding to amino acid 273 replacing the wild-type (wt) UGG (Trp) sequence. A C-to-T mutation at nt 1834 of the cspcz299 locus altered the wild-type CAG (Gln) sequence corresponding to amino acid 612 to a UAG stop codon. (C) Morpholino oligonucleotides targeted against sil cause increased phospho-H3 staining. moA and moB are independent splice site morpholino oligonucleotides that synergize to induce a more robust phenocopy. Scale bars = 250 μm. cntrl, control. (D) Whole-mount in situ hybridization with a sil riboprobe on 14-somite (top) and 24-hpf (bottom) embryos shows that csp mutants (right) lack positive staining, which is the earliest observable phenotype of the csp mutants. Scale bars = 100 μm.
The defects observed in the csp mutants were confirmed to result from sil loss of function by using antisense morpholino oligonucleotide knockdown to phenocopy csp. Antisense morpholino oligonucleotides were designed across the splice acceptor (moA) and donor (moB) flanking exon 6, which contains the cspcz65 mutation. Wild-type embryos injected with each of these morpholino oligonucleotides had increased numbers of phospho-H3-positive cells and were ventrally curved (Fig. 2C). The two splice site morpholino oligonucleotides acted synergistically to induce a more robust phenocopy, although all of the morpholino oligonucleotide knockdowns were less severe than the phenotype evinced by the nonsense mutations. Additionally, we assessed sil expression by in situ hybridization in wild-type and mutant embryos. sil was ubiquitously expressed at low-to-moderate levels in wild-type zebrafish embryos, with the strongest staining present in the eye and anterior neural tissue (Fig. 2D). This expression pattern in the developing zebrafish embryo resembled that of other cell cycle regulators, such as cyclin B1 (37). csp mutants exhibited only background staining, indicating that the mutations in sil likely resulted in nonsense-mediated decay of the mRNA (Fig. 2D). The identification of the nonsense mutations in sil, loss of sil expression in csp mutants, and the fact that knockdown of sil recapitulates the csp phospho-H3 phenotype all verified that the zebrafish csp mutant phenotype resulted from sil inactivation.
cassiopeia mutants exhibit a robust mitotic phenotype.
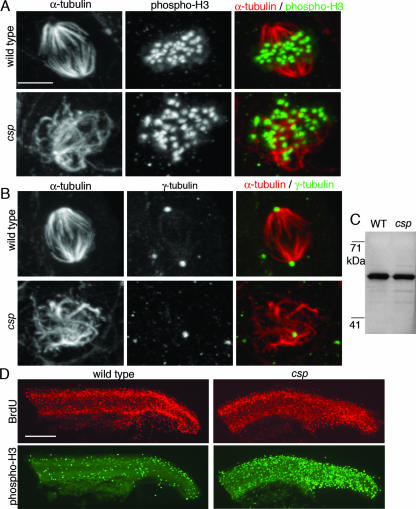
To characterize the mitotic defect in csp mutants, we analyzed the mitotic spindle by whole-mount immunostaining with α-tubulin in wild-type and mutant embryos. At 24 hpf, wild-type embryos have 10 to 20 metaphase spindles detectable in the tail (Fig. 3A and B). Nearly all metaphase cells are bipolar, although there is a low frequency (<1%) of multipolar cells in wild-type embryos (n = >300 mitotic cells). csp mutants at the same time point had approximately 200 mitotic cells with very disorganized mitotic spindles (Fig. 3A and B). Some of these abnormal spindles (15%, 49/330) were monopolar, such that an array of microtubules emanated from a single point. Most of the spindles (81%, 268/330) completely lacked polarity, and the chromosomes were not aligned along a metaphase plate. A small number of mitotic spindles (4%, 13/330) in csp mutants were bipolar, although they appeared to have broadened poles. Interphase cells of csp mutants possessed a normal microtubule cytoskeletal structure, suggesting that loss of sil specifically causes defects during mitosis.
FIG. 3.
The mitotic spindles of csp mutants are numerous and highly disorganized. (A) Microtubule staining detected by immunostaining against α-tubulin (red) and phospho-H3 (green) shows that all of the phospho-H3-positive cells in the csp mutants have an abnormal spindle (bottom) compare to wild-type bipolar spindles (top). The phospho-H3 staining demonstrates that the DNA is not properly aligned along a metaphase plate in the mitotic cells of the csp mutant. Scale bar = 5 μm. (B) Centrosome immunostaining with γ-tubulin (green) reveals that the disorganized mitotic cells of csp mutants identified with α-tubulin (in red) often have only one centrosome (bottom), which often stains with weaker intensity than wild-type centrosomes (top). (C) Immunoblotting for γ-tubulin shows that csp mutants have wild-type (WT) levels of γ-tubulin protein. (D) csp mutants are positive for BrdU incorporation. BrdU-labeled (red) embryos were costained for phospho-H3 (green) to distinguish the wild type from csp mutants. Scale bar = 200 μm.
To further examine the organization of csp mutant mitotic spindles, γ-tubulin immunostaining was used to detect the centrosomes. Mutant mitotic cells lacked spindle poles and often lacked one or both centrosomes by γ-tubulin staining. The foci of γ-tubulin were fainter than wild-type centrosomes and were usually within the microtubules rather than adjacent to them (Fig. 3B). This phenotype was not caused by loss of γ-tubulin itself, as immunoblotting showed that csp mutants expressed wild-type levels of γ-tubulin (Fig. 3C). Of eight zebrafish cell cycle mutants analyzed for α- and γ-tubulin staining, these mitotic spindle disorganization and centrosome loss phenotypes are unique to the csp mutant embryos. This combination of phenotypes provides insight into possible mechanisms of sil function during mitosis. It is possible that sil is required for proper centrosome duplication, and a primary centrosome defect results in abnormally formed spindles. Alternatively, sil may have a primary role in microtubule stability or organization that, when disrupted, subsequently results in centrosome dissociation from the mitotic spindle. These zebrafish mitotic spindle immunostaining experiments implied a novel role for SIL in mitotic spindle dynamics.
The csp mutant embryos had a striking increase in mitotic cells with extremely disorganized mitotic spindles, and it seemed likely that the cells of the mutant embryos were arrested in mitosis. To test this, we examined BrdU incorporation, a marker of S phase, in wild-type and mutant embryos. It has previously been shown that the crash&burn line, a zebrafish loss-of-function mutant with a mutation in b-myb, exhibits mitotic arrest and lacks BrdU incorporation (37). Surprisingly, csp mutant embryos were positive for BrdU incorporation (Fig. 3D), implying that the cells can continue to proliferate despite loss of SIL. The increased number of mitotic cells in csp mutants may therefore represent a delay, rather than an arrest, in mitotic progression resulting from the abnormal spindle morphology. csp mutants had wild-type numbers of anaphase and telophase mitotic figures, as detected by α-tubulin staining, further indicating that some cells in the csp mutants are capable of completing mitosis. The presence of BrdU incorporation in csp mutants likely explains why they have a less severe morphological phenotype and live longer than other zebrafish cell cycle mutants. Normal BrdU incorporation as also seen in Sil−/− mice (22), highlighting a similarity between the zebrafish and mouse defects resulting from SIL loss of function.
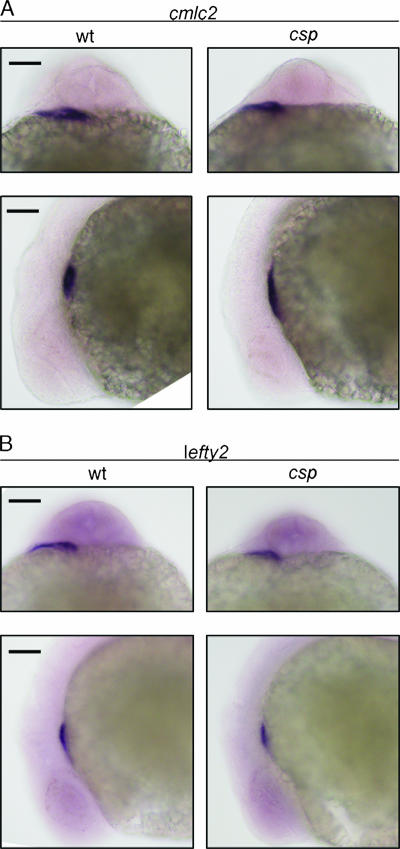
Sil−/− mice are characterized by defects in left-right asymmetry. Several genes, including nodal, pitx2, and lefty, are normally expressed in the left lateral plate mesoderm but demonstrate bilateral expression in Sil−/− mice upon in situ hybridization (22). In addition, Sil−/− mice have randomized looping of the embryonic heart tube (22). To determine if csp homozygous mutant zebrafish exhibit similar defects in left-right asymmetry, we performed in situ hybridization with genes that are expressed in the heart, which is on the left side of a 24-hpf zebrafish embryo. csp homozygous mutant and wild-type embryos both showed the same left-side expression of cardiac myosin light chain 2 (cmlc2) and lefty2 (Fig. 4A and B). The cardiac edema that develops in the csp mutants prevented the analysis of cardiac looping in the zebrafish embryos. It is surprising that csp mutant zebrafish do not exhibit the same left-right defects as found in the Sil−/− mouse model because left-right development is typically well conserved among vertebrates.
FIG. 4.
csp mutant zebrafish do not exhibit left-right symmetry defects. In situ hybridization for cardiac myosin light chain 2 (cmlc2) (A) and lefty2 (B) showed robust staining in the heart field of 24-hpf wild-type (wt) and csp mutant embryos. Top, dorsal views of embryos with the anterior surface to the top. Bottom, left-side lateral views of the same embryos. Scale bars = 100 μm.
We have shown that the csp phenotype is caused by a mutation in sil. Phenotypic analysis of the csp mutant demonstrated that SIL functions in zebrafish to organize the mitotic spindle. SIL is evolutionarily conserved yet vertebrate specific and is likely to have a role in mitosis in other vertebrates.
Human SIL localizes to mitotic spindle poles.
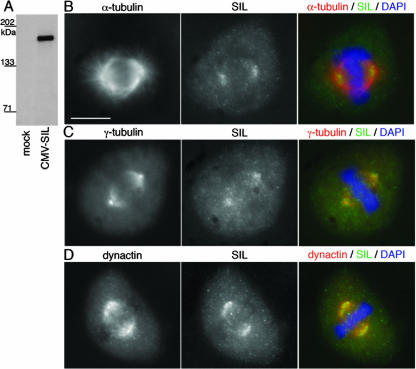
As the human and zebrafish SIL sequences are only 34% identical (see Fig. S1 in the supplemental material), we sought to determine if the role of SIL in mitotic spindle organization is conserved in mammals. To this end, and because the previously described SIL antibody (AP243) (20) was unavailable, we generated an anti-SIL polyclonal antibody by using the C terminus of human SIL as an immunogen. After affinity purification, the anti-SIL antibody specifically detected a single band of approximately 150 kDa in SIL-transfected HEK293T cells (Fig. 5A). Consistent with previous observations (20), HeLa and HEK293T cells transfected with SIL showed strong cytoplasmic expression of the protein (Fig. 6A). This anti-SIL antibody did not detect endogenous SIL by immunoblotting but did reveal increased expression of endogenous protein in mitotic cells by immunostaining.
FIG. 5.
SIL localizes to the mitotic spindle in HeLa cells. (A) Affinity-purified anti-SIL detects a single band of roughly 150 kDa in transfected HEK293T cells. (B) Immunostaining for α-tubulin (red) and endogenous SIL (green) in mitotic HeLa cells shows accumulation of SIL at the mitotic spindle poles during metaphase. The hazy cytoplasmic staining of anti-SIL is not seen in interphase cells and is likely detecting cytosolic SIL protein expression during mitosis. (C) Costaining for centrosomes with γ-tubulin (red) indicates that SIL localizes to the pericentriolar region but does not completely colocalize with the centrosome. (D) Immunostaining for dynactin (red) and SIL (green) reveals that the spindle pole localization of SIL has some overlap with that of dynactin. Scale bar = 10 μm. DAPI, 4′,6′-diamidino-2-phenylindole.
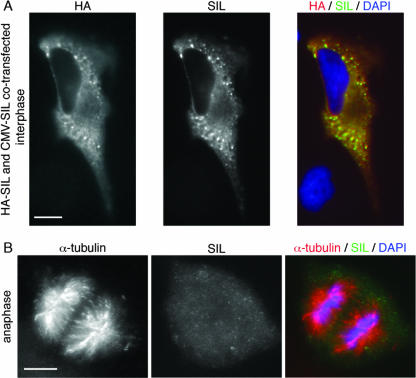
FIG. 6.
SIL is expressed in the cytoplasm when overexpressed, and SIL does not localize to the spindle poles during anaphase. (A) HeLa cells were cotransfected with pCMV-HA-SIL and pCMV-SIL and immunostained with anti-HA (red) and anti-SIL (green) antibodies. SIL was present in the cytoplasm and stains positively with both antibodies. As in the cell shown, high levels of overexpression resulted in the formation of positively stained foci, which may be accumulation of proteins at the ribosomes or protein aggregates. CMV, cytomegalovirus. (B) A cell in anaphase stained with the anti-SIL antibody reveals that SIL does not localize to the spindle poles when cells are in anaphase. Scale bars = 10 μm.
To assess the subcellular localization of SIL throughout the cell cycle, we examined immunostained HeLa cells in a time course following thymidine block and release. In metaphase cells, endogenous SIL protein was detected at the poles of the mitotic spindle where the microtubules coalesce adjacent to the centrosome (Fig. 5B). The polar accumulation of SIL surrounded the pericentriolar region but did not completely colocalize with γ-tubulin (Fig. 5C). Dynactin, a dynein adaptor complex that localizes to mitotic spindle poles (31, 32), showed some colocalization with SIL (Fig. 5D). SIL was associated with the spindle only in metaphase cells; cells undergoing anaphase lacked SIL localization to the spindle poles (Fig. 6B). Interphase cells had virtually no SIL expression, consistent with the known cell cycle regulation of SIL expression (20), and SIL did not accumulate around interphase centrosomes. The spindle poles are a dynamic structure to which many microtubule regulatory components localize during mitosis (14, 27). The presence of SIL at this site strongly suggests that SIL plays a role in regulating mitotic spindle formation or maintenance in vertebrate cells.
SIL is required for mitotic spindle organization in human cells.
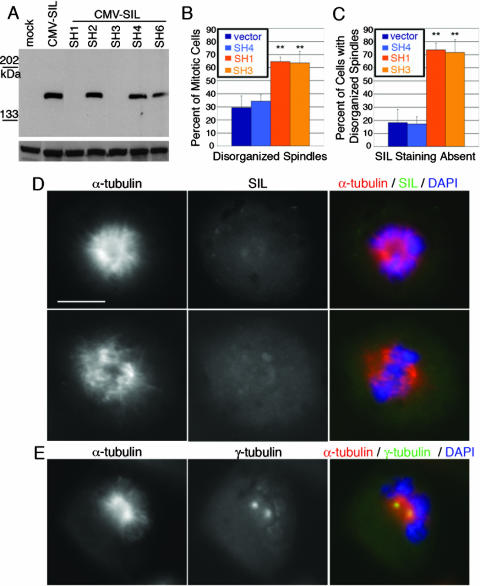
On the basis of the csp mutant phenotype and the localization of SIL to the mitotic spindle poles, we next evaluated SIL loss of function in human cells. shRNAs were targeted against SIL in HEK293T and HeLa cells to determine if loss of SIL in human cell culture causes mitotic defects. Immunoblotting showed that two shRNAs (SH1 and SH3) efficiently knocked down SIL overexpression (Fig. 7A). Transfected HeLa cells were examined for spindle morphology defects. There was a significant basal level of spindle abnormalities in HeLa cells; on average, ∼30% of mitoses appeared abnormal in both vector-transfected and untransfected cells (Fig. 7B). SH1- and SH3-transfected cells exhibited a significantly increased number of abnormal spindles (Fig. 7B); about 60% of the mitotic cells possessed abnormal mitotic spindles. Costaining for SIL showed that 44% of mitoses in SH1-transfected cells and 49% of mitoses in SH3-transfected cells lost SIL staining. These percentages reflect a combination of transfection and knockdown efficiencies of the shRNAs. Less than 20% of the control cells with disorganized spindles lacked SIL staining, whereas approximately 70% of SIL knockdown cells with disorganized spindles lacked SIL expression (Fig. 7C). These data indicate that the increase in cells exhibiting disorganized spindles results from loss of SIL. As with the csp mutant embryos, the abnormal spindles had a spectrum of morphologies, including monopolar and completely disorganized spindles, in which the chromosomes failed to form a metaphase plate (Fig. 7D). Unlike the csp mutants, in which only 4% of the mitotic cells were bipolar, most (∼65%) of the mitoses in the SIL knockdown cells formed a metaphase plate but the spindles failed to coalesce, or focus, at the spindle pole (Fig. 7D). The shRNA knockdown results demonstrate that SIL plays an evolutionarily conserved role in regulating mitotic spindle dynamics in vertebrates.
FIG. 7.
shRNA knockdown of SIL in HeLa cells causes disorganized spindles. (A) Two hairpins, SH1 and SH3, knock down cytomegalovirus (CMV) SIL overexpression in HEK293T cells. The loading control a the bottom is a 70-kDa band detected with the 12CA5 anti-HA antibody. (B) Quantification of mitotic cells stained for α-tubulin reveals that SH1 and SH3 have a statistically significant increase in cells with disorganized mitotic spindles. (C) The majority of SIL knockdown cells with disorganized spindles lack SIL staining. SH4, a nonfunctional shRNA, was used as a secondary negative control in panels B and C. **, P < 0.001. (D) Examples of SH1-transfected cells with abnormal spindles and loss of SIL staining. (Top) A cell in which the chromatin fails to form a metaphase plate, the spindle lacks proper organization, and spindle poles are not defined. (Bottom) A cell in which a metaphase plate is formed and the bipolar mitotic spindle is not focused at the spindle pole. Scale bar = 10 μm. DAPI, 4′,6′-diamidino-2-phenylindole. (E) An SH1-transfected cell stained for α-tubulin and γ-tubulin shows an example of a knockdown cell in which the centrosomes are associated with the spindle but fail to localize to spindle poles.
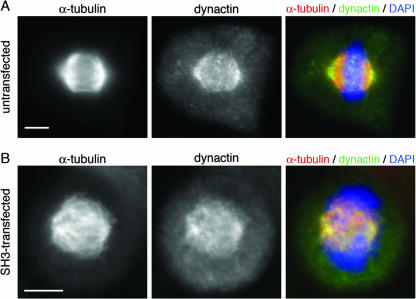
The shRNA knockdown of SIL in mammalian cells phenocopied the microtubule organization defects characterized in the zebrafish csp mutant; however, SIL knockdown cells did not reveal centrosome loss, as seen in the zebrafish mutants. Examination of γ-tubulin staining in SIL knockdown cells showed that the centrosomes did not localize to the spindle poles in cells with severely disorganized mitotic spindles. Rather, the centrosomes remained closely associated with each other and the microtubule clusters at the center of the cell (Fig. 7E). In knockdown cells in which a bipolar spindle was formed but was unfocused at the poles, the centrosomes appeared to be properly localized to the spindle poles (data not shown). Dynactin properly localized to the mitotic spindle in SIL knockdown cells but failed to accumulate at spindle poles in severely disrupted mitotic cells and remained dispersed across the microtubules (Fig. 8). Additionally, the SIL knockdown cells did not exhibit the same increase in mitotic index as in the zebrafish csp mutants. It is likely that the degree of phenotypic severity between the zebrafish csp mutants and the SIL knockdown cells reflects the difference between complete and partial loss of SIL. Furthermore, the fact that the mitotic index was unaffected in the SIL knockdown cells is consistent with an earlier study in which RNAi of SIL caused cells to progress abnormally through the nocodazole-induced mitotic spindle checkpoint but did not affect the cell cycle of unchallenged cells (8). Because mitotic spindle assembly and spindle checkpoint signaling are inherently linked, it is possible that the previously reported checkpoint response defect following SIL RNAi is connected to the spindle organization phenotype described here.
FIG. 8.
Dynactin localizes to the spindle poles and remains associated with the mitotic spindle in SIL knockdown cells. (A) Costaining with α-tubulin (red) and dynactin (green) shows that dynactin localizes to the spindle pole during metaphase and has a broader region of expression than SIL. Note that dynactin is highly expressed on the astral side of the spindle. SIL expression is not seen on the astral side of the spindle. (B) A representative SH3-transfected cell with disorganized spindles shows that SIL knockdown does not result in dynactin dissociation from the spindle. Greater than 100 knockdown cells were examined, and none of them lost dynactin expression on the mitotic spindle. Scale bars = 10 μm. DAPI, 4′,6′-diamidino-2-phenylindole.
DISCUSSION
The zebrafish cassiopeia mutant was isolated from a genetic screen for mitotic mutants in zebrafish. Two alleles exhibiting equal phenotypic severity were recovered along with 10 other mutant lines. Positional cloning of the csp mutation determined that a mutation in SIL was responsible for the csp phenotype. Each allele possessed a nonsense mutation in the coding region of SIL, suggesting that SIL loss of function resulted in the csp phenotype. This was confirmed with morpholino antisense oligonucleotides targeted against SIL to phenocopy csp. In addition, in situ hybridization with SIL demonstrated that csp homozygous mutant embryos failed to express SIL, indicating that the nonsense mutations led to nonsense-mediated decay of the mRNA. These data, taken together, demonstrate that the csp mutant results from loss of function of zebrafish SIL.
Like all of the mitotic mutants recovered from the screen, the csp mutation is recessive and embryonic lethal. csp homozygous mutant embryos exhibit an increased mitotic index, as detected by whole-mount immunostaining for phosphorylated histone H3 (phospho-H3). In addition, the embryos have increased numbers of apoptotic cells and remain positive for BrdU incorporation. These phenotypes indicate that loss of SIL results in defects that slow progression through mitosis. To further characterize the mitoses of csp mutant embryos, we used mitotic spindle immunostaining with antibodies against α-tubulin and γ-tubulin. These experiments revealed a striking phenotype in the csp mutants. Their mitotic cells consistently had a very disorganized mitotic spindle. In addition, one or both centrosomes were often missing from the mitotic cells. These findings establish a function for SIL in mitotic spindle dynamics.
Several lines of evidence have suggested that SIL could have a role in mitosis, although the precise mechanism of action is unknown. SIL mRNA and protein expression is regulated with the phases of the cell cycle, and peak expression of SIL coincides with mitosis (20). Additionally, SIL is hyperphosphorylated during mitosis and the phosphorylated form of SIL interacts with the peptidyl-prolyl isomerase Pin1 (8). Pin1 is a pleiotropic regulator of mitotic phosphoproteins, and the cis/trans isomerization of the proline residues on its target proteins is proposed to determine substrate specificity (40). These data imply a role for SIL in mitotic progression. The mitotic defects of the zebrafish csp mutants indicate that SIL is required for proper organization of the mitotic spindle. Immunostaining with a SIL antibody in HeLa cells revealed that SIL localizes to the poles of the mitotic spindle apparatus during metaphase. This is a region to which many regulators of the mitotic spindle localize during mitosis (29). The localization of SIL to the mitotic spindle during metaphase, combined with the mitotic spindle defects of the csp mutants, implied a novel role for SIL in mitotic spindle organization. These finding were further substantiated by targeted knockdown of SIL in HeLa cells by shRNA technology. Loss of SIL from dividing HeLa cells resulted in disorganized mitotic spindles. The similarity between the SIL loss-of-function phenotypes in the zebrafish csp embryos and the knockdown cells indicated that SIL is an evolutionarily conserved protein essential for proper mitotic spindle assembly.
The most common appearance of the mitotic spindle apparatus in SIL knockdown cells was a bipolar spindle with unfocused poles. The defects in mitotic spindle focusing observed in the SIL knockdown cells resemble those seen when dynein, dynactin, or NuMA is inhibited. Inhibition of any of these proteins results in loss of spindle pole focus such that the microtubules fail to converge at the spindle pole and splay outward (9, 29). The loss of spindle pole focus subsequently causes displacement of the centrosomes from the spindle pole (9, 29). The similarity between the SIL knockdown phenotype and the loss-of-function phenotypes of dynein, dynactin, and NuMA indicates that SIL may act similarly to these known regulators of microtubule stability and organization. The mitotic spindle defects of the csp mutant also resemble that of the Drosophila abnormal spindle (asp) mutant (11, 35). A clear mammalian homolog of the Asp protein remains elusive. Although an invertebrate orthologue of SIL may exist despite sequence divergence, the example of Drosophila Asp illustrates that species-specific factors can participate in mitotic spindle organization.
Defects in the mitotic spindle can cause missegregation of chromosomes and lead to genomic instability, which is a hallmark of cancer cells (23). SIL was initially identified by (and named for) its involvement in an interstitial 1p33 deletion that occurs in 25% of T-ALL patients (4). The deletion juxtaposes the ubiquitous SIL promoter with the protooncogene SCL, resulting in aberrant expression of SCL in T cells (28). The hemizygous loss of SIL has not been eliminated as a factor contributing to the development of T-ALL, and loss of heterozygosity of 1p33 has been reported in T-ALL patients (19). Given our findings that SIL plays a role in mitotic spindle organization, it is tempting to speculate that loss of SIL may play a role in T-ALL. It is further possible that sil is involved in other cancers as it is one of the genes in a metastatic signature (33) and a tumor array revealed that sil is misexpressed in many tumor types (13). It will be important to further elucidate the effects of sil misexpression and their putative role in cancer. The csp mutant may be a good model in which to study this, as other zebrafish mutants have been shown to exhibit a predisposition to cancer (3, 6, 37). The mitotic defects exhibited by the zebrafish csp mutant, combined with our shRNA analysis in mammalian cells, define a novel function for SIL in mitotic spindle organization that is conserved among vertebrates.
Supplementary Material
Acknowledgments
We thank Praise Opara for technical assistance with zebrafish maintenance and cell culture, K. Rose Finley and James Ziai for technical support with the haploid screening, Matthew Keefe for embryo microinjections, Bruce Barut for assistance with the positional cloning project, Susannah Rankin and Marc Kirschner for reagents and advice on the mammalian cell experiments, Marnie Halpern and Joseph Yost for plasmids, and Craig Ceol for reagents and advice on the antibody production. We also thank Jim Amatruda, Caroline Burns, Alan Davidson, Michael Dovey, David Langenau, Ryan Murphey, Jennifer Shepard, and Howard Stern for helpful conversations throughout the course of this work. We are grateful to Craig Ceol, Jenna Galloway, Marc Kirschner, Trista North, and David Pellman for critical reading of the manuscript.
This work was supported by National Institutes of Health grant 5R01 CA103846-02 (to L.I.Z) and an Albert J. Ryan Fellowship (to K.L.P). L.I.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 18 June 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Amsterdam, A., S. Burgess, G. Golling, W. Chen, Z. Sun, K. Townsend, S. Farrington, M. Haldi, and N. Hopkins. 1999. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 13:2713-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amsterdam, A., R. M. Nissen, Z. Sun, E. C. Swindell, S. Farrington, and N. Hopkins. 2004. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA 101:12792-12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amsterdam, A., K. C. Sadler, K. Lai, S. Farrington, R. T. Bronson, J. A. Lees, and N. Hopkins. 2004. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2:E139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aplan, P. D., D. P. Lombardi, A. M. Ginsberg, J. Cossman, V. L. Bertness, and I. R. Kirsch. 1990. Disruption of the human SCL locus by “illegitimate” V-(D)-J. recombinase activity. Science 250:1426-1429. [DOI] [PubMed] [Google Scholar]
- 5.Aplan, P. D., D. P. Lombardi, and I. R. Kirsch. 1991. Structural characterization of SIL, a gene frequently disrupted in T-cell acute lymphoblastic leukemia. Mol. Cell. Biol. 11:5462-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berghmans, S., R. D. Murphey, E. Wienholds, D. Neuberg, J. L. Kutok, C. D. Fletcher, J. P. Morris, T. X. Liu, S. Schulte-Merker, J. P. Kanki, R. Plasterk, L. I. Zon, and A. T. Look. 2005. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. USA 102:407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blagden, S. P., and D. M. Glover. 2003. Polar expeditions—provisioning the centrosome for mitosis. Nat. Cell Biol. 5:505-511. [DOI] [PubMed] [Google Scholar]
- 8.Campaner, S., P. Kaldis, S. Izraeli, and I. R. Kirsch. 2005. Sil phosphorylation in a Pin1 binding domain affects the duration of the spindle checkpoint. Mol. Cell. Biol. 25:6660-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Compton, D. A. 2000. Spindle assembly in animal cells. Annu. Rev. Biochem. 69:95-114. [DOI] [PubMed] [Google Scholar]
- 10.Dekens, M. P., F. J. Pelegri, H. M. Maischein, and C. Nusslein-Volhard. 2003. The maternal-effect gene futile cycle is essential for pronuclear congression and mitotic spindle assembly in the zebrafish zygote. Development 130:3907-3916. [DOI] [PubMed] [Google Scholar]
- 11.do Carmo Avides, M., and D. M. Glover. 1999. Abnormal spindle protein, Asp, and the integrity of mitotic centrosomal microtubule organizing centers. Science 283:1733-1735. [DOI] [PubMed] [Google Scholar]
- 12.Driever, W., L. Solnica-Krezel, A. F. Schier, S. C. Neuhauss, J. Malicki, D. L. Stemple, D. Y. Stainier, F. Zwartkruis, S. Abdelilah, Z. Rangini, J. Belak, and C. Boggs. 1996. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123:37-46. [DOI] [PubMed] [Google Scholar]
- 13.Erez, A., M. Perelman, S. M. Hewitt, G. Cojacaru, I. Goldberg, I. Shahar, P. Yaron, I. Muler, S. Campaner, N. Amariglio, G. Rechavi, I. R. Kirsch, M. Krupsky, N. Kaminski, and S. Izraeli. 2004. Sil overexpression in lung cancer characterizes tumors with increased mitotic activity. Oncogene 23:5371-5377. [DOI] [PubMed] [Google Scholar]
- 14.Gadde, S., and R. Heald. 2004. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 14:R797-R805. [DOI] [PubMed] [Google Scholar]
- 15.Golling, G., A. Amsterdam, Z. Sun, M. Antonelli, E. Maldonado, W. Chen, S. Burgess, M. Haldi, K. Artzt, S. Farrington, S. Y. Lin, R. M. Nissen, and N. Hopkins. 2002. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 31:135-140. [DOI] [PubMed] [Google Scholar]
- 16.Haffter, P., M. Granato, M. Brand, M. C. Mullins, M. Hammerschmidt, D. A. Kane, J. Odenthal, F. J. van Eeden, Y. J. Jiang, C. P. Heisenberg, R. N. Kelsh, M. Furutani-Seiki, E. Vogelsang, D. Beuchle, U. Schach, C. Fabian, and C. Nusslein-Volhard. 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123:1-36. [DOI] [PubMed] [Google Scholar]
- 17.Heald, R. 2000. Motor function in the mitotic spindle. Cell 102:399-402. [DOI] [PubMed] [Google Scholar]
- 18.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348-360. [DOI] [PubMed] [Google Scholar]
- 19.Iolascon, A., M. F. Faienza, B. Coppola, A. Moretti, G. Basso, R. Amaru, G. Vigano, and A. Biondi. 1997. Frequent clonal loss of heterozygosity (LOH) in the chromosomal region 1p32 occurs in childhood T cell acute lymphoblastic leukemia (T-ALL) carrying rearrangements of the TAL1 gene. Leukemia 11:359-363. [DOI] [PubMed] [Google Scholar]
- 20.Izraeli, S., T. Colaizzo-Anas, V. L. Bertness, K. Mani, P. D. Aplan, and I. R. Kirsch. 1997. Expression of the SIL gene is correlated with growth induction and cellular proliferation. Cell Growth Differ. 8:1171-1179. [PubMed] [Google Scholar]
- 21.Izraeli, S., L. A. Lowe, V. L. Bertness, S. Campaner, H. Hahn, I. R. Kirsch, and M. R. Kuehn. 2001. Genetic evidence that Sil is required for the Sonic Hedgehog response pathway. Genesis 31:72-77. [DOI] [PubMed] [Google Scholar]
- 22.Izraeli, S., L. A. Lowe, V. L. Bertness, D. J. Good, D. W. Dorward, I. R. Kirsch, and M. R. Kuehn. 1999. The SIL gene is required for mouse embryonic axial development and left-right specification. Nature 399:691-694. [DOI] [PubMed] [Google Scholar]
- 23.Jallepalli, P. V., and C. Lengauer. 2001. Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer 1:109-117. [DOI] [PubMed] [Google Scholar]
- 24.Jowett, T. 1999. Analysis of protein and gene expression. Methods Cell Biol. 59:63-85. [DOI] [PubMed] [Google Scholar]
- 25.Kane, D. A., H. M. Maischein, M. Brand, F. J. van Eeden, M. Furutani-Seiki, M. Granato, P. Haffter, M. Hammerschmidt, C. P. Heisenberg, Y. J. Jiang, R. N. Kelsh, M. C. Mullins, J. Odenthal, R. M. Warga, and C. Nusslein-Volhard. 1996. The zebrafish early arrest mutants. Development 123:57-66. [DOI] [PubMed] [Google Scholar]
- 26.Kimmel, C. B., W. W. Ballard, S. R. Kimmel, B. Ullmann, and T. F. Schilling. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253-310. [DOI] [PubMed] [Google Scholar]
- 27.Kline-Smith, S. L., and C. E. Walczak. 2004. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell 15:317-327. [DOI] [PubMed] [Google Scholar]
- 28.Lecuyer, E., and T. Hoang. 2004. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp. Hematol. 32:11-24. [DOI] [PubMed] [Google Scholar]
- 29.Merdes, A., and D. W. Cleveland. 1997. Pathways of spindle pole formation: different mechanisms; conserved components. J. Cell Biol. 138:953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musacchio, A., and K. G. Hardwick. 2002. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 3:731-741. [DOI] [PubMed] [Google Scholar]
- 31.Quintyne, N. J., S. R. Gill, D. M. Eckley, C. L. Crego, D. A. Compton, and T. A. Schroer. 1999. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147:321-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quintyne, N. J., and T. A. Schroer. 2002. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J. Cell Biol. 159:245-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramaswamy, S., K. N. Ross, E. S. Lander, and T. R. Golub. 2003. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33:49-54. [DOI] [PubMed] [Google Scholar]
- 34.Sansam, C. L., J. L. Shepard, K. Lai, A. Ianari, P. S. Danielian, A. Amsterdam, N. Hopkins, and J. A. Lees. 2006. DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 20:3117-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saunders, R. D., M. C. Avides, T. Howard, C. Gonzalez, and D. M. Glover. 1997. The Drosophila gene abnormal spindle encodes a novel microtubule-associated protein that associates with the polar regions of the mitotic spindle. J. Cell Biol. 137:881-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shepard, J. L., J. F. Amatruda, D. Finkelstein, J. Ziai, K. R. Finley, H. M. Stern, K. Chiang, C. Hersey, B. Barut, J. L. Freeman, C. Lee, J. N. Glickman, J. L. Kutok, J. C. Aster, and L. I. Zon. 2007. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev. 21:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shepard, J. L., J. F. Amatruda, H. M. Stern, A. Subramanian, D. Finkelstein, J. Ziai, K. R. Finley, K. L. Pfaff, C. Hersey, Y. Zhou, B. Barut, M. Freedman, C. Lee, J. Spitsbergen, D. Neuberg, G. Weber, T. R. Golub, J. N. Glickman, J. L. Kutok, J. C. Aster, and L. I. Zon. 2005. A zebrafish bmyb mutation causes genome instability and increased cancer susceptibility. Proc. Natl. Acad. Sci. USA 102:13194-13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shepard, J. L., H. M. Stern, K. L. Pfaff, and J. F. Amatruda. 2004. Analysis of the cell cycle in zebrafish embryos. Methods Cell Biol. 76:109-125. [DOI] [PubMed] [Google Scholar]
- 39.Westerfield, M. 1993. The zebrafish book: a guide for the laboratory use of zebrafish. University of Oregon Press, Eugene.
- 40.Yaffe, M. B., M. Schutkowski, M. Shen, X. Z. Zhou, P. T. Stukenberg, J. U. Rahfeld, J. Xu, J. Kuang, M. W. Kirschner, G. Fischer, L. C. Cantley, and K. P. Lu. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278:1957-1960. [DOI] [PubMed] [Google Scholar]
- 41.Yu, J. Y., S. L. DeRuiter, and D. L. Turner. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. USA 99:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, J., W. S. Talbot, and A. F. Schier. 1998. Positional cloning identifies zebrafish one-eyed pinhead as a permissive EGF-related ligand required during gastrulation. Cell 92:241-251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.