Abstract
Eukaryotic release factor 3 (eRF3) is a GTPase associated with eRF1 in a complex that mediates translation termination in eukaryotes. Studies have related eRF3 with cell cycle regulation, cytoskeleton organization, and tumorigenesis. In mammals, two genes encode two distinct forms of eRF3, eRF3a and eRF3b, which differ in their N-terminal domains. eRF3a is the major factor acting in translation termination, and its expression level controls termination complex formation. Here, we investigate the role of eRF3a in cell cycle progression using short interfering RNAs and flow cytometry. We show that eRF3a depletion induces a G1 arrest and that eRF3a GTP-binding activity, but not the eRF3a N-terminal domain, is required to restore G1-to-S-phase progression. We also show that eRF3a depletion decreases the global translation rate and reduces the polysome charge of mRNA. Finally, we show that two substrates of the mammalian TOR (mTOR) kinase, 4E-BP1 and protein kinase S6K1, are hypophosphorylated in eRF3a-depleted cells. These results strongly suggest that the G1 arrest and the decrease in translation induced by eRF3a depletion are due to the inhibition of mTOR activity and hence that eRF3a belongs to the regulatory pathway of mTOR activity.
Translation termination in eukaryotes requires two release factors, eukaryotic release factor 1 (eRF1) and eRF3, to complete protein synthesis. eRF1 recognizes all three stop codons by direct interaction at the decoding A site and triggers the hydrolysis of the peptidyl-tRNA bond by activation of the peptidyl transferase center (7, 12). eRF3 is a GTPase which associates with eRF1 in its GTP-bound form and stimulates eRF1 release activity in a GTP-dependent manner (13, 26, 47). In connection with its role in translation termination, eRF3 has also been implicated in ribosome recycling and mRNA decay through its interaction with the poly(A)-binding protein PABP (9, 19) and in the nonsense-mediated mRNA decay pathway (31). The SUP35 gene, encoding eRF3, is essential in the yeast Saccharomyces cerevisiae, and eRF3 depletion promotes translational readthrough of stop codons in yeast and mammals (6, 24, 40). The C-terminal regions of eRF3 proteins are highly conserved through evolution and carry the four canonical GTP-binding motifs of the GTPase superfamily. This domain is essential for translation termination and interaction with eRF1. The N-terminal region varies in both length and sequence among species. In yeast, it is neither essential for cell viability nor required for termination but is responsible for prion-like [PSI+] factor formation (33, 34).
However, eRF3 was first identified in a screen for mutants affected in the G1-to-S-phase transition in S. cerevisiae (25). Afterwards, other findings related eRF3 to cell cycle regulation and other cellular processes, such as cytoskeleton organization and tumorigenesis. In S. cerevisiae, mutations in the SUP35 gene increase sensitivity to the microtubule-poisoning drug benomyl and affect chromosome segregation at anaphase (3, 44). In addition, SUP35 repression results in the accumulation of cells with increased sizes and large buds, in the disappearance of actin cytoskeleton structures, and in the impairment of the mitotic spindle structure (46). Furthermore, depletion of eRF3 from Drosophila melanogaster disrupts spindle assembly, chromosome segregation, and cytokinesis during male meiosis (2). Taken together, these data suggest that eRF3 may have nontranslational functions. However, to date, whether these phenotypes, resulting from eRF3 depletion or mutation, are an indirect consequence of translation disruption or reflect the direct involvement of eRF3 in cytoskeletal functioning and cell cycle progression remains unknown.
In mammals, two distinct genes encoding eRF3, eRF3a/GSPT1 and eRF3b/GSPT2, have been identified (20, 21, 23). The products of these genes, eRF3a and eRF3b, share 87% sequence identity and differ only in their N-terminal domains. At the mRNA and protein levels, eRF3a is expressed in all tissues tested, with an abundance correlated with expression of eRF1, whereas eRF3b is poorly expressed in most tissues tested except in brain (6, 20). Both eRF3a and eRF3b can bind eRF1 and have GTPase activities which stimulate eRF1 release activity in vitro (20, 23, 47). In vivo, eRF3a depletion induces an important increase in readthrough whereas eRF3b silencing has no significant effect. In addition, eRF3a depletion reduces the intracellular level of eRF1 protein by affecting its stability. These results suggest that eRF3a is the major factor acting in translation termination in mammals and that its expression level controls the formation of the termination complex by modulating eRF1 protein stability (6). In addition, it has been shown that eRF3a is expressed in a proliferation-dependent manner (20, 21). Whereas eRF3a mRNA is poorly expressed in fibroblastic cells in their quiescent state, its expression level increases largely after stimulation of cell entry in G1 phase by addition of serum or phorbol ester. This increase is followed by a gradual decrease, from the onset of DNA replication in S phase up to 24 h after the beginning of the stimulation. In contrast, eRF3b mRNA does not fluctuate over the same period, showing that, at the mRNA level, eRF3a and eRF3b expression levels differ during cell cycle progression (20). Variations of eRF3a expression were also reported to occur in gastric tumors (29) and during chondrocyte differentiation (43).
Despite the accumulation of numerous observations, the mechanisms connecting eRF3 to cytoskeleton organization, cell cycle progression, and tumorigenesis are far from being understood. In the present work, we investigate the role of eRF3a in cell cycle progression. For this purpose, we silence the expression of the eRF3a gene in human HCT116 cells, using short interfering RNAs (siRNAs). We show that eRF3a depletion induces a G1 arrest which is not due to the elevated level of readthrough in eRF3a-depleted cells. We further show that eRF3a binding to GTP is required to restore G1-to-S-phase progression. Moreover, eRF3a depletion leads to a decrease in the global rate of translation and to the loss of polyribosomes. The influence of translation on cell cycle progression is well documented for the initiation step of translation. Indeed, it has been reported that the activation of cap-dependent translation through eukaryotic initiation factor 4F (eIF4F) complex assembly and eIF4E phosphorylation plays an important role in G1-to-S-phase transition (38). A large number of reports have identified the protein kinase TOR (target of rapamycin) as a major effector of cell growth via the regulation of protein synthesis (reviewed in references 8 and 17). Mammalian TOR (mTOR) controls protein synthesis through the phosphorylation and inactivation of initiation factor 4E-binding protein 1 (4E-BP1) and through the phosphorylation and activation of ribosomal protein S6 kinase 1 (S6K1). Here, we show that these two direct targets of mTOR are hypophosphorylated in eRF3a-depleted cells. These results suggest that eRF3a depletion inhibits mTOR activity. Thus, apart from its function in the translation termination process, eRF3a could belong to the regulatory pathway of mTOR activity.
MATERIALS AND METHODS
Plasmids.
Plasmids expressing siRNAs si-3a1 (targeting eRF3a mRNA) and si-1Y (targeting eRF1 mRNA); plasmids pBK-heRF3a and pBK-heRF3b, expressing human eRF3a and eRF3b, respectively; and plasmid pBK-heRF3aNN, carrying a double mutation in the G1 motif of the eRF3a GTPase domain, have been previously described (6). Plasmids ptRNAam, ptRNAop, and ptRNAoc, expressing amber, opal, and ochre suppressor tRNAs, respectively, have been described previously (27). Plasmid pHC is a derivative of pBK-heRF3a in which the 5′ sequence encoding the N and M domains of eRF3a was deleted. In this construct, the translation start of the eRF3a C-terminal domain is the AUG codon at position 799 of eRF3a mRNA (GenBank accession no. NM_002094). This plasmid was constructed by amplification of eRF3a cDNA, using a pair of oligonucleotides containing NheI and PstI restriction sites for cloning in the NheI and PstI sites of the pBK-CMV expression vector (Stratagene).
Antibodies.
Antibodies directed against human eRF3a, eRF3b, and eRF1 were previously described (6). Antibodies to human cyclin E and cyclin D were purchased from Upstate (Chemicon Europe). Antibodies against human cyclin A and S6K1 were purchased from Santa Cruz, CA. Antibodies directed against eIF4E, phospho-eIF4E, 4E-BP1, phospho-4E-BP1, phospho-S6, phospho-S6K1, phospho-Akt, and phospho-Erk1/2 were purchased from Cell Signaling Technology. The anti-α-tubulin antibody (DM1A) was from Amersham Biosciences (England). The anti-eRF3a-C antibodies directed against the human eRF3a C-terminal domain were produced by Eurogentec (Belgium) via immunization of rabbits with the synthetic peptides DTNQEERDKGKTVEV and KSGEKSKTRPRFVKQ, derived from the eRF3a protein sequence.
Cell culture and electroporation.
The HCT116 cell line was maintained in McCoy medium (Invitrogen) supplemented with 10% fetal calf serum, 100 μg/ml streptomycin, and 100 units/ml penicillin at 37°C under a 5% CO2 atmosphere. Electroporation was performed as described previously (6) with a gene pulser II electroporation system (Bio-Rad), using 4.8 × 106 cells and 10 to 30 μg of plasmid DNA.
Cell synchronization.
For synchronization in mitosis, exponentially growing HCT116 cells were treated with 50 ng/ml of nocodazole (Sigma) for 14 h. Mitotic cells in prometaphase were collected by shakeoff in phosphate-buffered saline (PBS; 10 mM phosphate buffer, pH 7.4, 140 mM NaCl), washed twice in complete medium, and further cultivated for times ranging from 0 to 16 h. For synchronization in G1/S, HCT116 cells were treated with 2.5 mM of thymidine (Sigma) for 24 h, washed twice in complete medium, and further cultivated for times ranging from 0 to 14 h. In both cases, adherent cells were recovered by trypsinization and treated for either flow cytometry or Western blotting analyses.
Flow cytometry analysis.
Cells were trypsinized, spun down, and resuspended in 100 μl of complete medium. During the vortexing, 2 ml of 70% ethanol in PBS was added and cells were stored at −20°C. Fixed cells were pelleted by centrifugation, resuspended in PBS, and stained at 37°C for 30 min in 40 μg/ml propidium iodide and 100 μg/ml RNase A. Stained cells (104) were analyzed, and the percentages of cells in the G1, S, and G2/M phases of the cell cycle were determined using a Coulter Elite-ESP flow cytometer system (Beckman-Coulter, France). Figures were processed using the WinMDI 2.8 program.
Western blot analysis.
Cell pellets were resuspended in 100 μl of PBS containing a 2× complete EDTA-free cocktail of protease inhibitors (Roche), 1 μg/ml pepstatine, and 10 mM EDTA. Cells were lysed by sonication on ice and centrifuged for 20 min at 16,000 × g, and the supernatant was retained as the cell extract. Protein concentrations of extracts were determined using a Micro BCA protein assay reagent kit (Pierce), bovine serum albumin being used as standard. For each sample, 20 μg of total protein was loaded on 8% polyacrylamide gel and subjected to electrophoresis. Proteins were subsequently electrotransferred onto a Hybond-C extra membrane (Amersham Biosciences), and the membrane was blocked for 1 h in Tris-buffered saline (TBS)-Tween solution (20 mM Tris-HCl, pH 7.6, 140 mM NaCl, 0.2% Tween 20) containing 5% milk. The membrane was incubated overnight with primary antibodies at the appropriate dilution in TBS-Tween, washed five times for 10 min in TBS-Tween, and probed for 45 min with either anti-rabbit or anti-mouse immunoglobulin G peroxidase-linked secondary antibodies at the appropriate dilution in TBS-Tween. The membrane was washed again five times in TBS-Tween and visualized by chemiluminescence and exposure to X-ray film.
Measurement of protein synthesis.
Three days after electroporation, HCT116 cells in 60-mm cell plates were metabolically labeled for 0 to 6 h at 37°C with 0.1 mCi of PRO-MIX l-[35S] in vitro cell labeling mix (Amersham Biosciences) in 3 ml of culture medium. Cells were washed once with PBS, trypsinized, and harvested by centrifugation. Cell pellets were lysed in 50 μl of PBS containing a 2× complete EDTA-free cocktail of protease inhibitors (Roche), 1 μg/ml pepstatine, and 10 mM EDTA. Cells were lysed by sonication on ice and centrifuged for 20 min at 16,000 × g, and the supernatant was retained as the cell extract. Protein concentrations of extracts were determined using a Micro BCA protein assay reagent kit (Pierce). For detection of radiolabeled proteins, 30 μg of total protein was spotted onto Whatman 3MM filter paper, dried, and placed in ice-cold 10% trichloroacetic acid (TCA) for 10 min. Filters were transferred to 5% TCA, boiled for 10 min, washed once with 5% ice-cold TCA for 10 min and once with 95% ethanol, and dried. Radioactivity was determined by scintillation counting.
Polysome analysis.
Isolation of polysomes was performed according to reference 14. Four 150-mm cell plates were used for the gradients. Three days after electroporation, HCT116 cells were incubated for 2 h with 10 ml of fresh medium and cycloheximide was added at 100 μg/ml for 10 min. Cells were then washed with PBS, collected by trypsinization, and pelleted. The dry cell pellet was resuspended in 500 μl of lysis buffer (50 mM Tris-HCl, pH 7.4, 300 mM KCl, 10 mM Mg-acetate, 1 mM dithiothreitol) containing 130 units of RNase inhibitor (Amersham Pharmacia Biotech) and 100 μg/ml of cycloheximide and lysed by adding ∼200 μl of glass beds and vortexing on ice for 30 s. Nuclei and cell debris were removed by centrifugation at 1,000 × g for 10 min, and an aliquot fraction of supernatant corresponding to 30 units of optical density at 260 nm (OD260) was layered onto a 12-ml 15 to 50% (wt/vol) sucrose gradient in 50 mM Tris-acetate (pH 7.5), 50 mM NH4Cl, 12 mM MgCl2, and 1 mM dithiothreitol. The gradient was centrifuged at 39,000 rpm in a SW41 Beckman rotor for 2.75 h at 4°C. The gradient was pumped through a single path UV-1 monitor system (Pharmacia), and the OD254 was recorded.
RESULTS
eRF3a protein level remains constant during cell cycle progression.
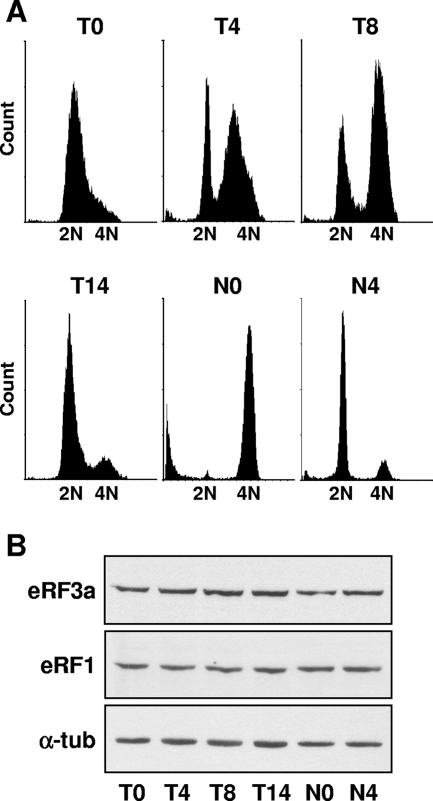
Given the observation that the steady-state level of eRF3a mRNA fluctuated during the cell cycle (20, 21), we used HCT116 cells to determine whether the eRF3a protein level was also subjected to variations during the different phases of the cell cycle. The human colon carcinoma HCT116 cells are currently used for cell cycle analyses because they are well characterized with respect to their intact cell cycle checkpoints and normal p53 responses. HCT116 cells were synchronized either at the G1-to-S-phase transition by thymidine treatment for 24 h and released in the absence of the drug or in the prometaphase stage of mitosis by nocodazole treatment for 14 h. In the latter case, mitotic cells were collected by shakeoff and replated in the absence of the drug. In both cases, synchronized cells were then harvested at various times after release and treated for flow cytometric analysis and Western blot analysis. As illustrated by the flow cytometry profiles (Fig. 1A), a majority of cells were at the G1-to-S-phase transition at time zero after thymidine treatment (Fig. 1A, T0), in S phase 4 h after release (Fig. 1A, T4), in G2/M phase 8 h after release (Fig. 1A, T8), and in G1 phase 14 h after release (Fig. 1A, T14). Nocodazole-treated cells were accumulated in mitosis at time zero (Fig. 1A, N0) and then moved to G1 phase 4 h after release (Fig. 1A, N4). As shown in Fig. 1B, there was no significant variation in the amount of either release factor, eRF3a or eRF1, whatever the phase in which cells were accumulated. This showed that eRF3a and eRF1 protein levels did not fluctuate during the cell cycle. Considering that eRF3a mRNA level increases after stimulation of cell entry in G1 and gradually decreases after G1-to-S-phase transition (20, 21), our result indicated that eRF3 protein level did not follow mRNA fluctuation, suggesting that eRF3, as well as eRF1, is stable throughout the cell cycle, possibly due to its association with eRF1 in a stable translation termination complex. Notice that, as previously shown in various human cell lines (6), eRF3b protein was detected neither in asynchronous nor in synchronized HCT116 cells (data not shown).
FIG. 1.
Expression of eRF3a during the cell cycle. HCT116 cells were synchronized in G1/S with thymidine treatment for 24 h, released, and harvested at time zero (T0), 4 h (T4), 8 h (T8), and 14 h (T14) after release. HCT116 cells were synchronized in mitosis with nocodazole treatment for 14 h, shaken off, released, and harvested at time zero (N0) and 4 h (N4) after release. (A) Fluorescence-activated cell sorting (FACS) analysis. Event count (y axis) versus DNA content (x axis) values are shown. (B) Western blotting using anti-eRF3a, anti-eRF1, or anti-α-tubulin (α-tub) antibodies as indicated.
eRF3a silencing induces a G1 arrest in HCT116 cells.
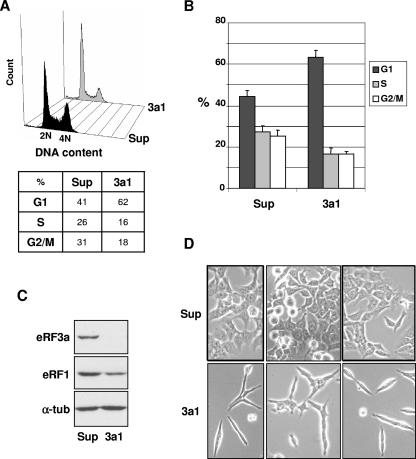
The human eRF3a/GSPT1 gene was first isolated by its ability to complement a temperature-sensitive gst1 mutation of S. cerevisiae (21). Gst1 is a cdc-like mutant arrested at the G1-to-S-phase transition at the nonpermissive temperature. Then, it appeared that the yeast GST1 gene was identical to the SUP35 gene, whose product played an essential role in translation termination (47). Since then, the human GSPT1 gene was widely considered a marker of the cell cycle (18, 43). However, the relationship between the function of the eRF3a/GSPT1 gene product in translation termination and its putative role in cell cycle control remained to be determined. To investigate the role of human eRF3a in cell cycle regulation, we analyzed the effect of an siRNA targeting eRF3a mRNA on cell cycle progression. Parallel cultures of HCT116 cells were electroporated with either the empty vector pSuper as a control or the plasmid expressing si-3a1, and the cell cycle was analyzed 3 days after electroporation by flow cytometry. As shown in Fig. 2A and B, a clear increase (∼20%) in the number of cells in G1 phase and a concomitant decrease in the number of cells in S and G2/M phases were reproducibly observed for cells expressing si-3a1, compared to what was observed for pSuper-electroporated control cells. This suggested that eRF3a depletion altered cell cycle progression, likely at the G1 phase. Western blot analysis confirmed the depletion of eRF3a in cells expressing 3a1 siRNA targeting eRF3a mRNA (Fig. 2C). Notice that eRF3a depletion also induced a reduction in the intracellular level of eRF1 protein, as previously shown in 293 cells (6). Using phase-contrast microscopy, we also observed profound modifications of cell morphology when eRF3a was silenced (Fig. 2D). 3a1-electroporated HCT116 cells changed from polygonal to spindle-shaped cells with extending pseudopodia. Some cells exhibited ruffles localized to the distal ends of the pseudopodia. These cell shape modifications likely reflected cell suffering.
FIG. 2.
eRF3a depletion alters cell cycle progression and cell morphology. HCT116 cells were electroporated with either empty pSuper vector (Sup) as a control or plasmid expressing si-3a1 targeting eRF3a mRNA (3a1) and analyzed 3 days after electroporation. (A) FACS analysis. Event count (y axis) versus DNA content (x axis) values are shown. The percentages of cells in the G1, S, and G2/M phases of the cell cycle are indicated below the graph. (B) The percentages of cells in G1, S, and G2/M were determined for nine independent electroporation experiments and are expressed as means and standard deviations. The significance of the 3a1 sample values with respect to the Sup basal value was determined by a two-tailed t test for G1, S, and G2/M cells. The P values were less than 0.0001 for the three cell types. (C) Western blot analysis using anti-eRF3a, anti-eRF1, or anti-α-tubulin (α-tub) antibodies as indicated on the left. (D) Phase-contrast microscopy images.
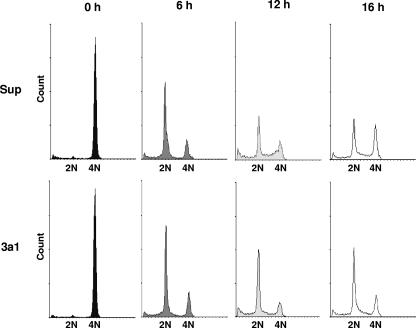
The cell cycle defect caused by eRF3a depletion was further characterized using HCT116 cells synchronized in mitosis. For this purpose, HCT116 cells were electroporated either with the plasmid expressing si-3a1 or with the pSuper vector and synchronized 30 h later by nocodazole treatment for 14 h. Mitotic cells were collected by shakeoff, released in the absence of the drug, and treated for flow cytometry analysis at various times after release. At time zero, pSuper- as well as 3a1-electroporated cells were almost exclusively at the M phase, as shown by the single peak of cells with 4-N DNA content (Fig. 3). Six hours after release, the majority of pSuper- and 3a1-electroporated cells were found in G1 phase. However, whereas some pSuper-electroporated cells entered S phase 12 h after release and G2/M phase 16 h after release, most 3a1-electroporated cells remained in G1 phase 12 and 16 h after release. These results clearly showed that the cell cycle of eRF3a-depleted cells was arrested or at least significantly slowed down during G1 phase.
FIG. 3.
eRF3a-depleted cells are arrested in the G1 phase of the cell cycle. HCT116 cells electroporated with either pSuper vector (Sup, upper panels), or plasmid expressing si-3a1 (3a1, lower panels) were synchronized in mitosis by nocodazole treatment for 14 h. Mitotic cells were shaken off, released for the time indicated above each panel, and analyzed by flow cytometry. Event count (y axis) versus DNA content (x axis) values are shown.
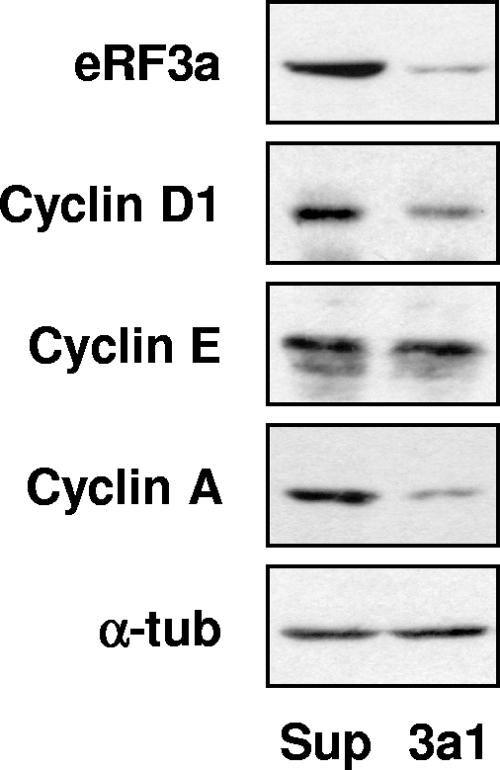
G1 phase is the longest phase of the cell cycle and can be divided into two functionally different periods, early and late G1, which are separated by the restriction point. The progression of cells in G1 is largely controlled by the expression of different cyclins: cyclin D1 level is the highest in early G1, whereas cyclin E level is at its maximum in late G1. Both are degraded at the beginning of S phase, cyclin D1 just before cyclin E (reviewed in reference 30). Cyclin A is synthesized at the G1-to-S-phase transition and degraded in prometaphase of mitosis. To determine the period of G1 phase in which 3a1-electroporated cells were arrested, the protein levels of these different cyclins were analyzed by Western blotting. Compared to what was found for pSuper-electroporated control cells, the decrease in eRF3a protein level in 3a1-electroporated cells was accompanied by a decrease in cyclin A level, confirming that eRF3a-depleted cells were arrested before S phase (Fig. 4). We also observed a decrease in cyclin D1 level, whereas cyclin E level remained unchanged. Together with the cell cycle profiles of synchronized cells, these observations clearly demonstrated that eRF3a-depleted cells were blocked in late G1. These data suggested that eRF3a played a role in cell cycle regulation and particularly in G1 phase progression.
FIG. 4.
eRF3a-depleted cells are arrested in late G1 phase. HCT116 cells were electroporated with either pSuper vector (Sup) or plasmid expressing si-3a1 (3a1) and analyzed 3 days after electroporation by Western blotting using anti-eRF3a, anti-cyclin D1, anti-cyclin E, anti-cyclin A, and anti-α-tubulin (α-tub) antibodies as indicated on the left.
eRF1 depletion and other factors inducing readthrough do not affect cell cycle progression and cell morphology.
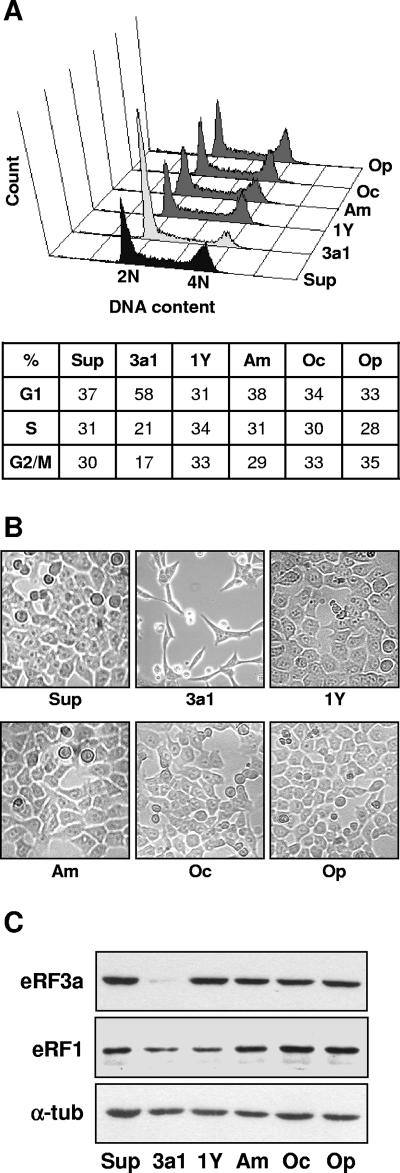
We have previously shown that eRF3a depletion induces an increase in stop codon readthrough (6). Depending on its efficiency, translational readthrough results in the abnormal C-terminal extension of variable amounts of proteins. The presence of such an extension could impair the functioning of some of these proteins, including proteins essential for cell cycle progression. In addition to eRF3a depletion, eRF1 silencing and suppressor tRNA overexpression also promote translational readthrough of stop codons (5, 36), the latter being the most efficient (11). To determine whether the G1 arrest induced by eRF3a depletion was the result of an increase in stop codon readthrough, we compared the effects of eRF3a silencing on cell cycle progression and cell morphology with those of eRF1 depletion and suppressor tRNA overexpression. For this purpose, empty pSuper vector, plasmids directing the synthesis of siRNAs specifically targeting either eRF3a (si-3a1) or eRF1 (si-1Y) mRNAs, and plasmids overexpressing amber, ochre, or opal suppressor tRNAs were electroporated in parallel cultures of HCT116 cells. Three days after electroporation, we analyzed cell cycle distribution by flow cytometry and cell morphology by contrast phase microscopy; pSuper-electroporated cells were used as a reference for the normal cell cycle profile and cell morphology. Depletion of eRF3a and eRF1 was confirmed by Western blotting. As shown in Fig. 5A, cell accumulation in G1 phase was observed only for si-3a1-expressing cells. Cells expressing si-1Y or suppressor tRNAs did not show any significative modification of the cell cycle (Fig. 5A). Moreover, in contrast to eRF3a depletion, suppressor tRNA overexpression or eRF1 depletion had no effect on cell morphology (Fig. 5B). As previously reported for eRF1 silencing in 293 cells (5, 6), the expression of si-1Y in HCT116 cells induced a moderate decrease in eRF1 level compared with that in eRF3a level induced by si-3a1 (Fig. 5C). This moderate effect of si-1Y on eRF1 level was explained by the high stability of eRF1 protein (6). Given that readthrough levels obtained by suppressor tRNA overexpression were ∼50-fold higher than the readthrough level obtained with eRF3a depletion (data not shown), the absence of effect of suppressor tRNAs on cell cycle profile and cell morphology strongly suggested that the modifications observed in eRF3a-depleted cells were not the consequence of the deleterious effect of stop codon readthrough on proteins essential for cell cycle progression.
FIG. 5.
eRF1 depletion and suppressor tRNA expression do not induce G1 arrest. HCT116 cells were electroporated with either pSuper vector (Sup); plasmids expressing si-3a1 targeting eRF3a mRNA (3a1); plasmids expressing si-1Y targeting eRF1 mRNA (1Y); or the plasmid ptRNAam, ptRNAoc, or ptRNAop, expressing amber (Am), ochre (Oc), or opal (Op) suppressor tRNA, respectively, and analyzed 3 days after electroporation. (A) FACS analysis. Event count (y axis) versus DNA content (x axis) values are shown. The percentages of cells in the G1, S, and G2/M phases of the cell cycle are indicated below the graph. (B) Phase-contrast microscopy images. (C) Western blot analyses of eRF3a, eRF1, and α-tubulin (α-tub).
The eRF3a C-terminal domain and its ability to bind GTP are required to restore cell cycle progression in eRF3a-depleted cells.
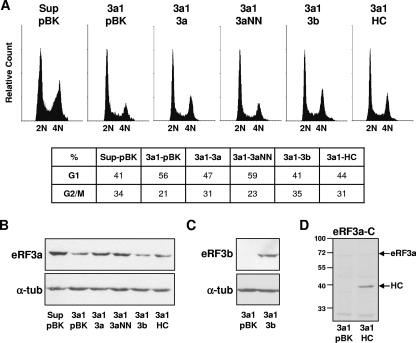
It has been shown that the eRF3 C-terminal domain carrying the GTPase activity is sufficient for translation termination (37, 47). By its N-terminal domain carrying the PABP-binding site, eRF3 is involved in mRNA decay and ribosome recycling (9, 19, 22, 45). In mammals, eRF3b, which differs from eRF3a in its N-terminal domain, can fulfill all the functions of eRF3a in translation termination (6, 23). To determine which of the eRF3 functions are required to complement the G1 arrest induced by eRF3a depletion, we performed coelectroporation of HCT116 cells with both the plasmid expressing si-3a1 and a set of pBK-CMV derivatives expressing either eRF3a (plasmid pBK-heRF3a), eRF3b (plasmid pBK-heRF3b), an N-terminally truncated form of eRF3a (plasmid pHC), or a mutated form of eRF3a carrying a double mutation in the G1 motif of the GTP-binding domain (plasmid pBK-heRF3aNN). Thirty hours after electroporation, cells were synchronized in mitosis by nocodazole treatment and released as described above. Cells were fixed at different times after release and analyzed by flow cytometry (shown only for 16 h after release) (Fig. 6A). Cells coelectroporated with plasmids pSuper and pBK-CMV served as a reference for the normal cell cycle (Fig. 6A, Sup-pBK). Cells coelectroporated with plasmids expressing si-3a1 and pBK-CMV were used as a reference for the G1 arrest observed in eRF3a-depleted cells (Fig. 6A, 3a1-pBK). For each electroporation experiment, we verified the amounts of endogenous eRF3a and overexpressed release factors by Western blot analysis (Fig. 6B, C, and D). Compared to eRF3a-depleted cells which were coelectroporated with the plasmid expressing si-3a1 and the empty expression vector pBK-CMV (Fig. 6A, 3a1-pBK), cells overexpressing exogenous eRF3a (Fig. 6A, 3a1-3a) exhibited a reduced number of cells in G1 (from 56% to 47%) and concomitantly a 10% increase in the number of cells in G2/M. This complementation effect on G1 arrest was even more pronounced when eRF3b was overexpressed in eRF3a-depleted cells (Fig. 6A, 3a1-3b). These results indicated that eRF3a as well as eRF3b overexpression alleviated the G1 arrest induced by eRF3a depletion. Thus, eRF3b, which can replace eRF3a in its function in translation termination in human cells (6), can also fulfill the function of eRF3a in cell cycle progression.
FIG. 6.
eRF3a GTP-bound form is required to restore cell cycle progression in eRF3a-depleted cells. HCT116 cells were coelectroporated with either pSuper vector (Sup) or plasmid expressing si-3a1 (3a1) and with either the empty expression vector pBK-CMV (pBK) or plasmids overexpressing either human eRF3a (3a), human eRF3a mutated in its GTPase domain (3aNN), human eRF3b (3b), or a truncated form of human eRF3a containing only the C-terminal domain (HC). Thirty hours after electroporation, cells were synchronized in mitosis by nocodazole treatment, shaken off, and released for 16 h. (A) Flow cytometry analysis of cells electroporated with the plasmids indicated in abbreviation above each panel. Relative event count (y axis) versus DNA content (x axis) values are shown. Cells with DNA content values lower than 2N were excluded from the count. Percentages of cells in G1 (2N DNA content) and G2/M (4N DNA content) are indicated in the lower panel. (B) Western blot analysis of eRF3a and α-tubulin (α-tub) in each cell sample as indicated below. (C) Western blot analysis of eRF3a-depleted cells overexpressing eRF3b (3a1-3b) or not (3a1-pBK) using anti-eRF3b and anti-α-tubulin (α-tub) antibodies as indicated on the left. (D) Western blot analysis of eRF3a-depleted cells overexpressing N-terminally truncated eRF3a (3a1-HC) or not (3a1-pBK) using anti-eRF3a-C antibodies as indicated above. Entire eRF3a and N-terminally truncated eRF3a (HC) proteins are indicated by arrows on the right.
Overexpression of an N-terminally truncated form of eRF3a, having only the C-terminal domain carrying the GTPase activity, also restored G1 and G2/M cell counts identical to those obtained with eRF3a overexpression (Fig. 6A, 3a1-HC). This indicated that the function of eRF3a in cell cycle progression did not require the N-terminal region of the protein which harbors the PABP-binding domain and hence that the association of eRF3a with PABP did not participate in the effect of eRF3a on cell cycle progression. This finding corroborated the results of genetic studies with the yeast S. cerevisiae, where the mammalian eRF3a C-terminal domain was sufficient to compensate for the growth arrest of the gst1 mutant (21, 45). Furthermore, eRF3a-depleted cells overexpressing an eRF3a form carrying point mutations in the GTP-binding site (Fig. 6A, 3a1-3aNN) exhibited a cell cycle profile very similar to that of eRF3a-depleted cells overexpressing the empty vector pBK-CMV (Fig. 6A, 3a1-pBK). This result indicated that the mutated form of eRF3a did not alleviate the G1 arrest induced by eRF3a depletion and suggested that the effect of eRF3a on cell cycle progression required the presence of its GTP-bound form. It has been recently shown in in vitro experiments that eRF1 has a strong stimulatory effect on the binding of eRF3 to GTP, indicating that, in cells, eRF3 will predominantly be bound to GTP in an eRF1-eRF3-GTP ternary complex (16, 37). Thus, the involvement of eRF3a in the control of the cell cycle could rely on the formation of the eRF1-eRF3-GTP ternary complex. Alternatively, it cannot be excluded that, in addition to eRF1, another, not-yet-identified factor could be able to promote eRF3 binding to GTP.
eRF3a depletion induces a decrease in global translation.
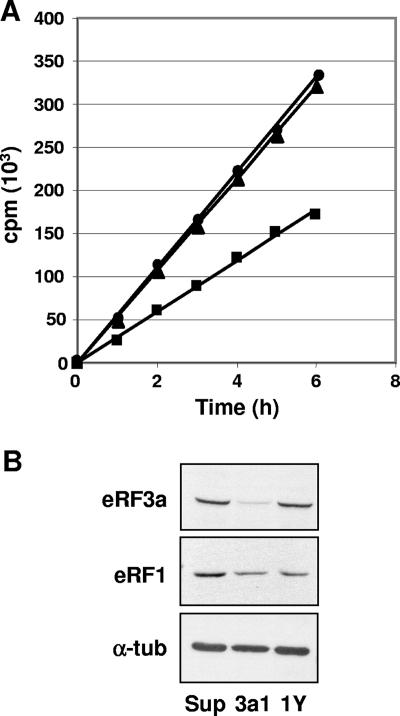
The results presented above and the fact that inhibition of translation initiation causes inhibition of protein synthesis and G1 arrest (15) prompted us to examine the status of translation in eRF3a-depleted HCT116 cells. We first studied the time course of incorporation of radiolabeled amino acids to compare the translation rate of cells expressing either si-3a1 targeting eRF3a mRNA or si-1Y targeting eRF1 mRNA with that of pSuper-expressing cells. For this purpose, 3 days after electroporation, cellular proteins were labeled with a mixture of [35S]methionine and [35S]cysteine for various times and TCA precipitated. As shown in Fig. 7A, eRF3a depletion induced a twofold decrease in the rate of amino acid incorporation into proteins whereas eRF1 depletion had no effect. As shown by Western blot analysis of the 6-h-radiolabeled cells (Fig. 7B), the reduction of eRF1 in si-3a1-expressing cells was almost identical to the decrease of eRF1 induced by si-1Y, suggesting that the effect of si-3a1 expression on translation rate was not due only to eRF1 depletion.
FIG. 7.
eRF3a depletion causes a decrease in the rate of amino acid incorporation into proteins. HCT116 cells electroporated with either pSuper vector, plasmid expressing si-3a1 targeting eRF3a mRNA, or plasmid expressing si-1Y targeting eRF1 mRNA were labeled 3 days after electroporation for 0 to 6 h with a mixture of [35S]methionine and [35S]cysteine. Total proteins (30 μg) were spotted on Whatman 3MM filter paper and TCA precipitated. (A) Radioactivity (counts per minute [cpm]) was determined by scintillation counting. Results are shown for pSuper vector (filled circles), plasmid expressing si-3a1 (filled squares), and plasmid expressing si-1Y (filled triangles). (B) Western blot analysis of the radiolabeled cells expressing pSuper vector (Sup), si-3a1 (3a1), or si-1Y (1Y) using anti-eRF3a, anti-eRF1, or anti-α-tubulin (α-tub) antibodies as indicated on the left.
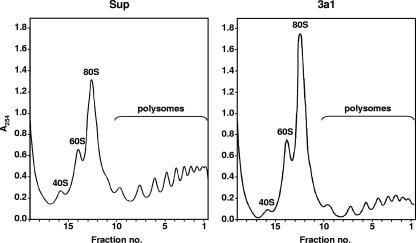
We then compared the polysomal distributions of si-3a1- and pSuper-electroporated cells. Three days after electroporation, cell extracts corresponding to 30 OD260 units were fractionated by sucrose gradient centrifugation as described in Materials and Methods. Compared to that for pSuper control cells, the absorbance profile of the gradient for eRF3a-depleted cells revealed a reduction of the number of polysome peaks (from 10 to 8), a global reduction of the intensities of the polysome peaks (Fig. 8, fractions 1 to 10), and an increase in the level of free 80S ribosomes (Fig. 8, fractions 10 to 13). The disaggregation of polysomes into free ribosomes is characteristic of a defect in the initiation stage of translation (1, 32). Thus, surprisingly, the modification of the polysome profile in eRF3a-depleted cells indicated that eRF3a depletion caused inhibition of translation initiation. These results prompted us to investigate whether this effect on translation initiation was an indirect consequence of the decrease in translation termination efficiency or a direct effect of eRF3a depletion on the control of translation initiation.
FIG. 8.
eRF3a depletion causes a loss of polysomes. Cytoplasmic extracts of HCT116 cells electroporated with either pSuper vector (Sup) or plasmid expressing si-3a1 (3a1) were fractionated by 15 to 50% sucrose gradient centrifugation as described in Materials and Methods. The absorbance profile of the gradient is shown (sedimentation was from left to right), and the collected fractions are indicated on the x axis.
eRF3a depletion inhibits the mTOR pathway.
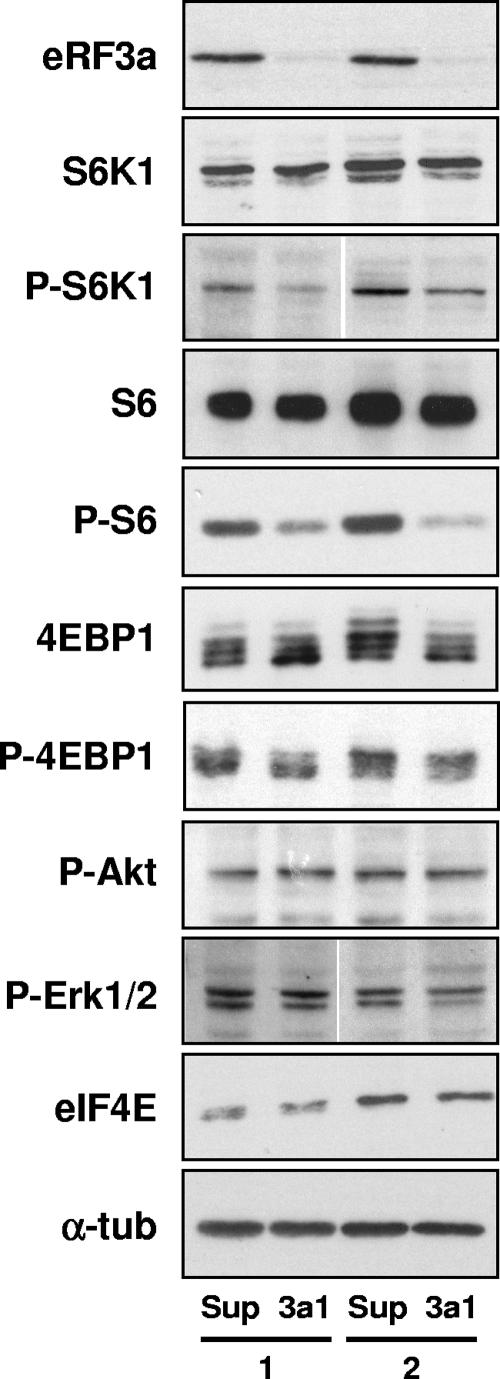
Phosphorylation events controlled via the mTOR signaling pathway clearly play a central role in regulating translation rates and translation initiation in a number of cell systems (17). In addition, the inhibition of mTOR activity is responsible for the arrest of cell cycle progression in G1 phase (1). The ribosomal protein S6K1 and the eukaryotic initiation factor 4E-BP1 are both well-characterized targets of mTOR. mTOR phosphorylation of S6K1 increases its kinase activity and results in an elevated phosphorylation of ribosomal protein S6 (rpS6). Phosphorylation of 4E-BP1 by mTOR results in the dissociation of 4E-BP1 from eIF4E, leading to the activation of cap-dependent translation initiation (reviewed in reference 17). Thus, to check the status of the mTOR pathway when eRF3a is depleted, we have examined the phosphorylation statuses of 4E-BP1, S6K1, and ribosomal protein S6 in si-3a1- and pSuper-expressing cells. The results of Western blot experiments performed on two different electroporation experiments clearly showed a hypophosphorylation of S6K1 (Thr389) and rpS6 (Ser240 and Ser244) in eRF3a-depleted cells compared to what was found for the pSuper-expressing control cells, whereas there was no change in the global levels of these proteins and of eIF4E (Fig. 9). Although the dephosphorylation of 4E-BP1 on Thr70 was not complete, as shown with the anti-phospho-4E-BP1 antibodies (Fig. 9, P-4E-BP1), the hypophosphorylation of 4E-BP1 was clearly visible with the anti-4E-BP1 antibodies, which showed a decrease in the upper, phosphorylated form of 4E-BP1 with a concomitant increase in the lower, nonphosphorylated form of 4E-BP1 (Fig. 9, 4E-BP1).
FIG. 9.
eRF3a depletion induces the hypophosphorylation of mTOR targets. Cytoplasmic extracts of HCT116 cells from two independent electroporation experiments with either pSuper vector (Sup) or plasmid expressing si-3a1 (3a1) were analyzed by Western blotting using antibodies directed against eRF3a, S6K1, phospho-S6K1 Thr389 (P-S6K1), ribosomal protein S6 and phospho-S6 Ser240/244 (P-S6), 4E-BP1 and phospho-4E-BP1 Thr70 (P-4E-BP1), phospho-Akt Ser473 (P-Akt), phospho-Erk1/2 Ser202 and Tyr204 (P-Erk1/2), eIF4E, and α-tubulin (α-tub), which served as a loading control.
We also investigated the phosphorylation statuses of two other kinases that are involved in phosphorylation cascades which modulate translation initiation, i.e., the protein kinase Akt, one of the upstream effectors of mTOR, and the mitogen-activated protein kinases Erk1 and Erk2, implicated in a growth regulatory pathway parallel to the mTOR pathway. As shown in Fig. 9, eRF3a depletion did not modify the phosphorylation of Akt on Ser473 and of Erk1/2 on Ser202 and Tyr204. Altogether, these results suggest that eRF3a depletion inhibited translation initiation and cell cycle progression via mTOR signaling pathway inhibition, downstream of Akt activation and independently of the Erk1/2 signaling pathway.
DISCUSSION
Several findings have related eRF3 with cytoskeleton organization, cell cycle regulation (2, 46), and tumorigenesis (4, 29). However, it was not determined whether the phenotypes promoted by eRF3 depletion or mutation were the result of a translation termination defect or reflected a direct involvement of eRF3 in the cell cycle. The aim of this study was to gain insights into the function of eRF3a in cell cycle progression. We show here, using siRNAs targeting eRF3a mRNA, that eRF3a depletion in asynchronous or in synchronized HCT116 cells induces an arrest of the cell cycle in late G1 phase (Fig. 2, 3, and 4). These data are in good agreement with that obtained for S. cerevisiae, in which thermosensitive mutants of eRF3 (gst1 mutants) are arrested after the START point, at the G1-to-S-phase transition (21, 25).
We subsequently show that the G1 arrest induced by eRF3a depletion is accompanied by a decrease in the rate of translation (Fig. 7) and by a disaggregation of polysomes (Fig. 8), the latter effect being indicative of a defect in the initiation step of translation. In mammalian cells, inhibition of mTOR activity by rapamycin also leads to a block in translation which causes cell arrest in G1. The mTOR protein kinase acts through two physically and functionally distinct complexes, mTORC1 and mTORC2. mTORC1, which is sensitive to rapamycin, controls translation and cell growth in response to nutrients via phosphorylation of S6K and 4E-BP. In contrast, mTORC2, which is rapamycin insensitive, has been shown to control actin cytoskeleton dynamics (8). The serine/threonine protein kinase Akt, which is a positive regulator of mTOR activity, is activated by direct phosphorylation on two sites, Thr308 and Ser473. The 3-phosphoinositide-dependent protein kinase PDK1 phosphorylates Akt on Thr308 (42), and in a positive feedback loop, the complex mTORC2 is required for Akt Ser473 phosphorylation (41). In eRF3a-depleted cells, the hypophosphorylation of key phosphorylation sites in 4E-BP1 and S6K1, which are both direct targets of mTORC1, and the absence of modification of Akt phosphorylation on Ser473 (Fig. 9) suggested that eRF3a depletion inhibited translation initiation and cell cycle progression via mTORC1 inhibition but did not interfere with Akt activation involving mTORC2. Both Erk1 and Erk2 mitogen-activated protein kinases function in a protein kinase cascade that plays a critical role in the regulation of cell growth and differentiation (10) and have been shown to regulate mTORC1 in response to growth stimulators (28, 39). The absence of effect on Erk1/2 phosphorylation indicated that eRF3a depletion did not affect upstream effectors of the Erk1/2 signaling pathway.
Altogether, our results support the idea that eRF3a, and possibly eRF3b, which complements the effect of eRF3a depletion on the cell cycle (Fig. 6), is involved in the control of mTOR activity. One of the possibilities is that eRF3a is directly acting in the control of mTOR activity. As a consequence, by regulating translation initiation through mTOR activity, eRF3a would exert a retrocontrol on the whole translation process, adjusting the rate of translation initiation to the efficiency of translation termination. Alternatively, eRF3a depletion could indirectly affect mTOR activity through a general stress response to slowed protein synthesis. However, the role of eRF3a in the mTOR pathway seems to be dependent on GTP binding, which is strongly stimulated by the association with eRF1. A role for eRF1 in cell cycle regulation has been suggested for S. cerevisiae (46) and for Arabidopsis thaliana (35). However, none of the effects induced by eRF3a depletion in HCT116 cells were observed for eRF1 depletion. A possible explanation is that, due to the high stability of eRF1 in human cells (6), the level of eRF1 depletion which was obtained by si-1Y RNA expression was not sufficient to induce a decrease in the rate of translation and cell cycle arrest. Nevertheless, because eRF3a depletion induced eRF1 depletion by decreasing eRF1 stability (6), the effect on cell cycle and on translation could be due to the concomitant reduction of both factors. It is also possible that another factor, not yet identified, promotes GTP binding to eRF3a. The precise role and the partners of eRF3a in mTOR signaling pathway remain to be determined.
The involvement of eRF3 in the TOR signaling pathway could explain the pleiotropic effect described for yeast and Drosophila eRF3 mutants (2, 3, 46) and particularly the arrest of S. cerevisiae gst1 mutants at G1-to-S-phase transition (21, 25). Furthermore, the signaling pathways that regulate mTOR activity are frequently activated in human cancers and it has been postulated that the activation of mTOR contributes to the genesis of cancer (reviewed in reference 17). Thus, the overexpression of the eRF3a/GSPT1 gene that is observed in some histological types of gastric tumors (29) could be responsible for the activation of mTOR and hence for malignant transformation.
Acknowledgments
This work was supported by the Association Française contre les Myopathies and by the Association pour la Recherche sur le Cancer (grant no. 3784). Céline Chauvin held fellowships from the French Ministère de la Recherche et de l'Enseignement Supérieur, from the Fondation pour la Recherche Médicale, and from the Société Française du Cancer.
We thank Michel Kress, Sophie Bellanger, and Olivier Ganier for their help in videomicroscopy and cell cycle analyses. We thank Isabelle Dusanter-Fourt and Mario Pende for the generous gift of the antibodies directed against proteins involved in the mTOR signaling pathway.
Footnotes
Published ahead of print on 11 June 2007.
REFERENCES
- 1.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu, J., B. C. Williams, Z. Li, E. V. Williams, and M. L. Goldberg. 1998. Depletion of a Drosophila homolog of yeast Sup35p disrupts spindle assembly, chromosome segregation, and cytokinesis during male meiosis. Cell Motil. Cytoskeleton 39:286-302. [DOI] [PubMed] [Google Scholar]
- 3.Borchsenius, A. S., A. A. Tchourikova, and S. G. Inge-Vechtomov. 2000. Recessive mutations in SUP35 and SUP45 genes coding for translation release factors affect chromosome stability in Saccharomyces cerevisiae. Curr. Genet. 37:285-291. [DOI] [PubMed] [Google Scholar]
- 4.Brito, M., J. Malta-Vacas, B. Carmona, C. Aires, P. Costa, A. P. Martins, S. Ramos, A. R. Conde, and C. Monteiro. 2005. Polyglycine expansions in eRF3/GSPT1 are associated with gastric cancer susceptibility. Carcinogenesis 26:2046-2049. [DOI] [PubMed] [Google Scholar]
- 5.Carnes, J., M. Jacobson, L. Leinwand, and M. Yarus. 2003. Stop codon suppression via inhibition of eRF1 expression. RNA 9:648-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chauvin, C., S. Salhi, C. Le Goff, W. Viranaicken, D. Diop, and O. Jean-Jean. 2005. Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol. Cell. Biol. 25:5801-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavatte, L., L. Frolova, L. Kisselev, and A. Favre. 2001. The polypeptide chain release factor eRF1 specifically contacts the s(4)UGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem. 268:2896-2904. [DOI] [PubMed] [Google Scholar]
- 8.Corradetti, M. N., and K. L. Guan. 2006. Upstream of the mammalian target of rapamycin: do all roads pass through mTOR? Oncogene 25:6347-6360. [DOI] [PubMed] [Google Scholar]
- 9.Cosson, B., N. Berkova, A. Couturier, S. Chabelskaya, M. Philippe, and G. Zhouravleva. 2002. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol. Cell 94:205-216. [DOI] [PubMed] [Google Scholar]
- 10.Cowley, S., H. Paterson, P. Kemp, and C. J. Marshall. 1994. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell 77:841-852. [DOI] [PubMed] [Google Scholar]
- 11.Diop, D., C. Chauvin, and O. Jean-Jean. 2007. Aminoglycosides and other factors promoting stop codon readthrough in human cells. C. R. Biol. 330:71-79. [DOI] [PubMed] [Google Scholar]
- 12.Frolova, L., X. Le Goff, H. H. Rasmussen, S. Cheperegin, G. Drugeon, M. Kress, I. Arman, A. L. Haenni, J. E. Celis, M. Philippe, J. Justesen, and L. Kisselev. 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372:701-703. [DOI] [PubMed] [Google Scholar]
- 13.Frolova, L., X. Le Goff, G. Zhouravleva, E. Davydova, M. Philippe, and L. Kisselev. 1996. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA 2:334-341. [PMC free article] [PubMed] [Google Scholar]
- 14.Gielkens, A. L., T. J. Berns, and H. Bloemendal. 1971. An efficient procedure for the isolation of polyribosomes from tissue culture. Eur. J. Biochem. 22:478-484. [DOI] [PubMed] [Google Scholar]
- 15.Hanic-Joyce, P. J., G. C. Johnston, and R. A. Singer. 1987. Regulated arrest of cell proliferation mediated by yeast prt1 mutations. Exp. Cell Res. 172:134-145. [DOI] [PubMed] [Google Scholar]
- 16.Hauryliuk, V., A. Zavialov, L. Kisselev, and M. Ehrenberg. 2006. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie 88:747-757. [DOI] [PubMed] [Google Scholar]
- 17.Hay, N., and N. Sonenberg. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926-1945. [DOI] [PubMed] [Google Scholar]
- 18.Horrevoets, A. J., R. D. Fontijn, A. J. van Zonneveld, C. J. de Vries, J. W. ten Cate, and H. Pannekoek. 1999. Vascular endothelial genes that are responsive to tumor necrosis factor-alpha in vitro are expressed in atherosclerotic lesions, including inhibitor of apoptosis protein-1, stannin, and two novel genes. Blood 93:3418-3431. [PubMed] [Google Scholar]
- 19.Hoshino, S., M. Imai, T. Kobayashi, N. Uchida, and T. Katada. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 274:16677-16680. [DOI] [PubMed] [Google Scholar]
- 20.Hoshino, S., M. Imai, M. Mizutani, Y. Kikuchi, F. Hanaoka, M. Ui, and T. Katada. 1998. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J. Biol. Chem. 273:22254-22259. [DOI] [PubMed] [Google Scholar]
- 21.Hoshino, S., H. Miyazawa, T. Enomoto, F. Hanaoka, Y. Kikuchi, A. Kikuchi, and M. Ui. 1989. A human homologue of the yeast GST1 gene codes for a GTP-binding protein and is expressed in a proliferation-dependent manner in mammalian cells. EMBO J. 8:3807-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosoda, N., T. Kobayashi, N. Uchida, Y. Funakoshi, Y. Kikuchi, S. Hoshino, and T. Katada. 2003. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278:38287-38291. [DOI] [PubMed] [Google Scholar]
- 23.Jakobsen, C. G., T. M. Segaard, O. Jean-Jean, L. Frolova, and J. Justesen. 2001. Identification of a novel termination release factor eRF3b expressing the eRF3 activity in vitro and in vivo. Mol. Biol. (Moscow) 35:672-681. [PubMed] [Google Scholar]
- 24.Janzen, D. M., and A. P. Geballe. 2004. The effect of eukaryotic release factor depletion on translation termination in human cell lines. Nucleic Acids Res. 32:4491-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kikuchi, Y., H. Shimatake, and A. Kikuchi. 1988. A yeast gene required for the G1-to-S transition encodes a protein containing an A-kinase target site and GTPase domain. EMBO J. 7:1175-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi, T., Y. Funakoshi, S. Hoshino, and T. Katada. 2004. The GTP-binding release factor eRF3 as a key mediator coupling translation termination to mRNA decay. J. Biol. Chem. 279:45693-45700. [DOI] [PubMed] [Google Scholar]
- 27.Le Goff, X., M. Philippe, and O. Jean-Jean. 1997. Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol. Cell. Biol. 17:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, L., Z. Chen, H. Erdjument-Bromage, P. Tempst, and P. P. Pandolfi. 2005. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 121:179-193. [DOI] [PubMed] [Google Scholar]
- 29.Malta-Vacas, J., C. Aires, P. Costa, A. R. Conde, S. Ramos, A. P. Martins, C. Monteiro, and M. Brito. 2005. Differential expression of the eukaryotic release factor 3 (eRF3/GSPT1) according to gastric cancer histological types. J. Clin. Pathol. 58:621-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malumbres, M., and M. Barbacid. 2005. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 30:630-641. [DOI] [PubMed] [Google Scholar]
- 31.Maquat, L. E. 2005. Nonsense-mediated mRNA decay in mammals. J. Cell Sci. 118:1773-1776. [DOI] [PubMed] [Google Scholar]
- 32.Morley, S. J., and S. Naegele. 2002. Phosphorylation of eukaryotic initiation factor (eIF) 4E is not required for de novo protein synthesis following recovery from hypertonic stress in human kidney cells. J. Biol. Chem. 277:32855-32859. [DOI] [PubMed] [Google Scholar]
- 33.Patino, M. M., J. J. Liu, J. R. Glover, and S. Lindquist. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273:622-626. [DOI] [PubMed] [Google Scholar]
- 34.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15:3127-3134. [PMC free article] [PubMed] [Google Scholar]
- 35.Petsch, K. A., J. Mylne, and J. R. Botella. 2005. Cosuppression of eukaryotic release factor 1-1 in Arabidopsis affects cell elongation and radial cell division. Plant Physiol. 139:115-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips-Jones, M. K., F. J. Watson, and R. Martin. 1993. The 3′ codon context effect on UAG suppressor tRNA is different in Escherichia coli and human cells. J. Mol. Biol. 233:1-6. [DOI] [PubMed] [Google Scholar]
- 37.Pisareva, V. P., A. V. Pisarev, C. U. Hellen, M. V. Rodnina, and T. V. Pestova. 2006. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J. Biol. Chem. 281:40224-40235. [DOI] [PubMed] [Google Scholar]
- 38.Pyronnet, S., and N. Sonenberg. 2001. Cell-cycle-dependent translational control. Curr. Opin. Genet. Dev. 11:13-18. [DOI] [PubMed] [Google Scholar]
- 39.Roux, P. P., B. A. Ballif, R. Anjum, S. P. Gygi, and J. Blenis. 2004. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA 101:13489-13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salas-Marco, J., and D. M. Bedwell. 2004. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 24:7769-7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarbassov, D. D., D. A. Guertin, S. M. Ali, and D. M. Sabatini. 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307:1098-1101. [DOI] [PubMed] [Google Scholar]
- 42.Stephens, L., K. Anderson, D. Stokoe, H. Erdjument-Bromage, G. F. Painter, A. B. Holmes, P. R. Gaffney, C. B. Reese, F. McCormick, P. Tempst, J. Coadwell, and P. T. Hawkins. 1998. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science 279:710-714. [DOI] [PubMed] [Google Scholar]
- 43.Tallheden, T., C. Karlsson, A. Brunner, J. Van Der Lee, R. Hagg, R. Tommasini, and A. Lindahl. 2004. Gene expression during redifferentiation of human articular chondrocytes. Osteoarthritis Cartilage 12:525-535. [DOI] [PubMed] [Google Scholar]
- 44.Tikhomirova, V. L., and S. G. Inge-Vechtomov. 1996. Sensitivity of sup35 and sup45 suppressor mutants in Saccharomyces cerevisiae to the anti-microtubule drug benomyl. Curr. Genet. 30:44-49. [DOI] [PubMed] [Google Scholar]
- 45.Uchida, N., S. Hoshino, H. Imataka, N. Sonenberg, and T. Katada. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/poly(A)-dependent translation. J. Biol. Chem. 277:50286-50292. [DOI] [PubMed] [Google Scholar]
- 46.Valouev, I. A., V. V. Kushnirov, and M. D. Ter-Avanesyan. 2002. Yeast polypeptide chain release factors eRF1 and eRF3 are involved in cytoskeleton organization and cell cycle regulation. Cell Motil. Cytoskeleton 52:161-173. [DOI] [PubMed] [Google Scholar]
- 47.Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov, L. Kisselev, and M. Philippe. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14:4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]