Abstract
The MyoD family of basic helix-loop-helix (bHLH) transcription factors has the remarkable ability to induce myogenesis in vitro and in vivo. This myogenic specificity has been mapped to two amino acids in the basic domain, an alanine and threonine, referred to as the myogenic code. These essential determinants of myogenic specificity are conserved in all MyoD family members from worms to humans, yet their function in myogenesis is unclear. Induction of the muscle transcriptional program requires that MyoD be able to locate and stably bind to sequences present in the promoter regions of critical muscle genes. Recent studies have shown that MyoD binds to noncanonical E boxes in the myogenin gene, a critical locus required for myogenesis, through interactions with resident heterodimers of the HOX-TALE transcription factors Pbx1A and Meis1. In the present study, we show that the myogenic code is required for MyoD to bind to noncanonical E boxes in the myogenin promoter and for the formation of a tetrameric complex with Pbx/Meis. We also show that these essential determinants of myogenesis are sufficient to confer noncanonical E box binding to the E12 basic domain. Thus, these data show that noncanonical E box binding correlates with myogenic potential, and we speculate that the myogenic code residues in MyoD function as myogenic determinants via their role in noncanonical E box binding and recognition.
Basic helix-loop-helix (bHLH) transcription factors are critical for regulating the developmental programs of nearly every tissue in metazoans (17, 20). The majority of bHLH proteins function as dimers and bind to the consensus DNA sequence CANNTG, known as an E box (4, 20). In spite of the importance of this large family of transcription factors in development, it remains unclear how individual bHLH proteins are able to discriminate specific target genes in the vast milieu of genomic sequence, which contains millions of potential E box binding sites. Members of the MyoD family of muscle regulatory factors (MRFs) are bHLH proteins required for skeletal muscle development, and MyoD function has been studied extensively as a model for regulating cell fate specification. Genetic studies have indicated that the four members of this family (MyoD, Myf5, myogenin, and MRF4) play distinct but overlapping roles in the embryo and participate in a mutually reinforcing transcriptional network to activate myogenesis appropriately (9, 37, 41). In general, MyoD and Myf5 serve as myogenic specification factors, while myogenin functions downstream and is essential for muscle differentiation (4, 8). MRF4 has recently been identified to play a dual role in specification and differentiation of myoblasts (21).
Each of the members of the MyoD family possesses the remarkable ability to dominantly induce myogenesis in a wide variety of different cell types (4, 29, 43). MRF proteins bind to E boxes as heterodimers with bHLH proteins of the E2A family, such as E12, which on their own are nonmyogenic (6, 7, 11, 23, 30, 33, 43, 44). Activation of the muscle transcriptional program by MyoD exquisitely depends on two amino acids in the basic domain, an alanine and threonine (AT), referred to as the myogenic code (6, 12, 45). These essential determinants of myogenic specificity are conserved in all muscle bHLH proteins from worms to humans and are absent from all nonmuscle bHLH proteins (34). The myogenic code is absolutely required for dominant induction of myogenesis by the MRFs, since replacement of these residues with the asparagines found in the corresponding positions in E12 renders MyoD nonmyogenic (11). Furthermore, when the AT along with a lysine in the junctional region of the first helix of MyoD are substituted for the corresponding amino acids in E12, these myogenic code amino acids are sufficient to induce myogenesis (12, 44). However, the mechanism through which these two amino acids determine myogenic specificity has remained elusive.
In order to initiate the muscle transcriptional program, MyoD must be capable of locating the correct muscle genes in the complicated organization of the genome. myogenin is among the essential immediate early genes activated by MyoD and is known to be a direct transcriptional target of MyoD and other MRF family members (2, 15, 47). Following activation by MyoD, myogenin eventually becomes the main bHLH factor that activates transcription of late muscle genes (10, 32). Recent work has shown that the regulatory elements governing the activation of myogenin are more complex than previously appreciated and that the earliest steps in MyoD activation of myogenin involve the binding of MyoD to imperfect E boxes near the promoter (3). These binding sites are juxtaposed to a binding site for Pbx/Meis HOX-TALE transcription factors that are known to be resident on the promoter in a repressive chromatin state, prior to the expression of MyoD (3, 13). The proposed model suggests that interaction between Pbx/Meis and MyoD/E12 on the myogenin promoter facilitates MyoD binding to the imperfect E boxes and allows for MyoD-dependent recruitment of molecules that remodel chromatin and subsequent stable binding of MyoD/E12 heterodimers to the promoter (3, 16, 26, 35, 36, 38, 39). The ability of MyoD to bind to noncanonical E boxes in condensed chromatin therefore provides an explanation for the ability of MyoD to initiate the transcription of myogenin and other key immediate early genes, and it may also explain the dominant induction of myogenesis by MyoD.
In the present study, we examine the requirement of the myogenic code of MyoD for transcriptional activation, dimerization with E proteins, and DNA binding to both canonical and noncanonical E boxes. We show that the myogenic code is required for MyoD to bind to the noncanonical E boxes in the myogenin promoter and that these two residues are sufficient to confer noncanonical E box binding to the E12 basic domain. Thus, the data presented here demonstrate that noncanonical E box binding correlates with myogenic potential and suggest that the two residues of the myogenic code function as myogenic determinants via their role in noncanonical E box recognition and binding.
MATERIALS AND METHODS
Plasmids.
A portion of the MyoD cDNA containing the entire coding region was excised from pEMSV-MyoD (23) with BspEI and HindIII and cloned into pBluescript II SK(+) to generate pBS-MyoD. Mutations of the alanine and threonine in the MyoD basic domain to asparagines were generated by PCR mutagenesis using the mutagenic primers 5′-GCTGATCGCCGCAAGGCCAACAACA-3′ and 5′-GCGGCGCTCGCGCATGTTGTTG-3′. A PmlI/MluI fragment of the cDNA that contained the mutation was swapped into pBS-MyoD to generate the myogenic code mutant, pBS-MyoD(NN). These cDNAs were subsequently cloned into the pRK5 expression vector, kindly provided by R. Derynck (UCSF), as BamHI/XhoI fragments to generate plasmids pRK5-MyoD and pRK5-MyoD(NN). pRK5-MyoD∼E47, which contains a tethered heterodimer of MyoD and E47 (31), was also provided by R. Derynck (24). The myogenic code mutation was swapped into this tethered heterodimer construct as a PmlI/MluI fragment to generate plasmid pRK5-MyoD(NN)∼E47. The MyoD-E12basic chimeric cDNA contains the E12 basic domain in place of the MyoD basic domain (11). Plasmid pRK5-MyoD-E12basic was generated by cloning the full-length MyoD-E12basic cDNA from plasmid EMSV-MyoD-E12basic (11) into the pRK5 expression vector. Plasmid pRK5-MyoD-E12basic(AT) was generated by introducing the AT in place of the NN in the E12 basic domain in plasmid pRK5-MyoD-E12basic using the mutagenic primers 5′-GGAGCGCCGCGTGGCCGCCACCGCCC-3′ and 5′-GCTCGCGGGCGGTGGCGGCCACGCGG-3′.
All MyoD fusions to the GAL4 DNA binding domain (DBD) were generated in the expression vector pSG424, which contains the GAL4 DBD downstream of the SV40 early promoter (5, 27, 28). pSG424-MyoD-bHLH and pSG424-MyoD(NN)-bHLH were cloned as EcoRI fragments as described previously (5). Fusions of the GAL4 DBD and full-length versions of wild-type MyoD or MyoD(NN) were generated by cloning the MRF cDNAs from pBS-MyoD and pBS-MyoD(NN) into the pSG424 expression vector as blunted BamHI/SalI fragments, to create plasmids pSG424-FL-MyoD and pSG424-FL-MyoD(NN), respectively. The reporter plasmid 4R-TK-luciferase has been described previously (3). pG5E1b-luciferase, a GAL4-dependent reporter construct that contains five copies of the consensus GAL4 binding site and the E1b promoter driving expression of the luciferase gene, was kindly provided by H. Bernstein (UCSF).
Mgn184-luciferase was generated by cloning a PstI/KpnI fragment from the previously described myogenin reporter plasmid pMyo184-lacZ (15) into pGL3-basic (Promega). Mutation of the myogenin E1 E box in Mgn184-luciferase was generated by PCR mutagenesis with the mutagenic primers 5′-AATGGCACCCAGCAGTCGACGTG-3′ and 5′-AGCCCCTCACGTCGACTGCTGGG-3′. Mutation of the myogenin E2 E box was achieved by truncating the distal end of the promoter by 40 base pairs such that the enhancer sequence ended just prior to the second E box.
EMSAs.
DNA binding assays on canonical E boxes were completed at room temperature using binding buffer conditions described previously (14). Proteins were generated by coupled in vitro transcription-translation reactions (TNT; Promega). MyoD and mutants of MyoD were generated from plasmid pRK5. E12 was generated from plasmid pCITE-E12, which has been described previously (5). Pbx1A and Meis1 were generated from plasmids pGEM-Pbx1A and pBluescript-Meis1, which were kindly provided by S. Tapscott (FHCRC, Seattle, WA) and have been described previously (3). Addition of protein to each electrophoretic mobility shift assay (EMSA) was normalized by quantitation of [35S]methionine-labeled proteins generated in parallel reactions. Quantitation was conducted by phosphorimager analysis using ImageQuant v. 1.2 software (Molecular Dynamics). Radiolabeled, double-stranded oligonucleotide probes were generated using Klenow fragments to fill in overhanging ends with [α-32P]dCTP. The mef2c E2 E box and mutants of that E box have been described previously (14). A total of 4 μl of lysate, consisting of 1 μl of E12 and a combination of MRF and unprogrammed lysate totaling 3 μl, was incubated in 1× binding buffer (40 mM KCl, 15 mM HEPES pH 7.9, 1 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol) with 0.5 μl poly(dI-dC) (1 mg/ml) for 10 min prior to addition of labeled probe (20,000 cpm total). After a 20-min incubation with labeled probe, the entire reaction was loaded on a 6% nondenaturing polyacrylamide-Tris-borate-EDTA (TBE) gel and run at 150 V for 2 h at room temperature. Kinetic EMSAs were performed as above, except that after the addition of probe the reactions were allowed to equilibrate at 25°C for 1 h. To check that the system had reached equilibrium, the experiment was repeated with 2-h and 4-h incubations with nearly identical results. Unlabeled oligonucleotide competitor was added at approximately 100-fold excess at indicated times before loading the reaction on a 6% nondenaturing polyacrylamide-TBE gel. Percent binding over time was calculated by phosphorimager analysis (Molecular Dynamics).
EMSAs of noncanonical E boxes were conducted as described previously (3) with minor modifications. Briefly, each binding reaction contained a total of 12 μl of reticulocyte lysate (TNT; Promega) consisting of various combinations of 2 μl Pbx1A, 2 μl Meis1, 2 μl E12, 6 μl MyoD, or 6 μl of mutant versions of MyoD. In reactions that omitted a member of the binding complex, unprogrammed lysate was used to adjust the reaction to a final volume of 12 μl. This mixture was then incubated at 37°C for 20 min. Labeled oligonucleotide probe was added (100,000 cpm/binding reaction) to the lysate mixture in 1× binding buffer (20 mM HEPES [pH 7.6], 5% glycerol, 1.5 mM MgCl2, 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol) and 0.2 μg poly(dI-dC) in a total volume of 30 μl and incubated on ice for 15 min. A MEF2 binding site (14) was added to every reaction as a nonspecific competitor oligonucleotide. The probe/lysate mixture was run on a 6% nondenaturing polyacrylamide-TBE gel at 4°C for 3 h. The sense strand sequence of the oligonucleotide used to assess binding to the noncanonical E box in the presence and absence of Pbx/Meis was 5′-CGTCTTGATGTGCAGCAACAGCTTAGA-3′, which contains the Pbx/Meis binding consensus and two adjacent noncanonical E boxes, as described previously (3).
Cell culture and transfections.
C3H10T1/2 cells were grown in 10% fetal bovine serum (FBS) in Dulbecco modified Eagle medium (DMEM). A total of 2 μg of plasmid DNA was transfected into 35-mm dishes at 50% confluence using FuGENE6 (Roche) as recommended by the manufacturer. Cells were allowed to recover for 18 h and changed to differentiation media containing 2% horse serum in place of FBS. Transfections of the Mgn184 series of plasmids into C3H10T1/2 cells were conducted in 35-mm dishes using the Superfect transfection reagent (QIAGEN) as recommended by the manufacturer. Following transfection, cells were cultured for 16 h in growth medium (DMEM plus 10% FBS), followed by 48 h in differentiation medium (DMEM, 2% horse serum, and 1× insulin/transferrin/selenium). Cells were harvested by scraping the plates into 100 mM NaPO4 (pH 7.4). Cells were then lysed by three freeze-thaw cycles of 1 min in liquid nitrogen followed by 1 min in a 37°C water bath. Cell debris was removed by centrifugation, and the supernatant was used to assess luciferase activity. Luciferase assays were performed using the Promega luciferase detection assay, according to the manufacturer's recommendations, and relative light units were determined using a Tropix TR717 microplate luminometer. All luciferase activity was normalized to the total protein in the cell lysates as determined by Bradford assay (Bio-Rad), and data are expressed as n-fold activation over the baseline activity of each reporter plasmid.
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP assay kit (Upstate/Millipore, Billerica, MA) according to the manufacturer's instructions with minor modifications. C3H10T1/2 cells grown in 100-mm tissue culture dishes were processed for ChIP experiments 36 h after transfection with expression plasmids encoding Myc-tagged MyoD or MyoD(NN) and the myogenin reporter plasmids Mgn184(WT)-luciferase, Mgn184(mE1)-luciferase, Mgn184(mE1/E2)-luciferase, or Mgn184(mNC)-luciferase. The cells were then treated with 1% formaldehyde in phosphate-buffered saline for 10 min at 37°C to cross-link protein-DNA complexes. The buffer was aspirated, and the cells were washed twice with ice-cold phosphate-buffered saline containing 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml pepstatin A as general protease inhibitors. Cells were then scraped into conical tubes, pelleted, and resuspended in sodium dodecyl sulfate (SDS) lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris, pH 8.1) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, and 1 μg/ml pepstatin A). Following lysis, the cellular debris was sonicated under conditions that consistently resulted in 300- to 1,000-bp DNA fragments. Following sonication, the cellular lysates were digested for 4 h at 37°C with PciI and DraI in restriction endonuclease buffer (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol, pH 7.9) to cleave DNA fragments such that the E1 and E2 E boxes in the myogenin promoter would be present on different restriction fragments from the noncanonical E boxes in the promoter. Immunoprecipitation was performed with 4 μg of mouse monoclonal anti-Myc antibody (clone 9E10) (Upstate/Millipore, Billerica, MA) per sample. Parallel samples were prepared without the addition of antibody as negative controls. Following immunoprecipitation and extensive washing, formaldehyde cross-links were reversed by heating the samples at 65°C in 0.2 M NaCl for 4 h. Immunoprecipitated fragments and raw lysates (input samples) were analyzed by PCR using the following primers: NC forward primer P1, 5′-AATCCACTGGAAACGTCTTGA-3′; and NC reverse primer P2, 5′-CTCCCTGCTGGCATGAAC-3′.
RESULTS
The myogenic code is not required for the inherent transcriptional activity of MyoD.
The two amino acids of the myogenic code function as essential determinants of myogenesis, and mutation of these two residues in the basic domain disrupts the ability of MyoD to dominantly induce the muscle transcriptional program (11, 12). The primary function of the myogenic code has previously been proposed to be in transcriptional activation, based on the observation that myogenic code mutants are defective at inducing transcription of muscle-specific reporter genes (1, 5, 18). Activation of a muscle gene requires that MyoD must first dimerize with an E protein partner and must bind to DNA in order for transcriptional activation to occur. Therefore, we hypothesized that the myogenic code might be important for dimerization and DNA binding specificity and that a DNA binding or dimerization defect would manifest itself as a secondary defect in transcriptional activation of E box-dependent reporters.
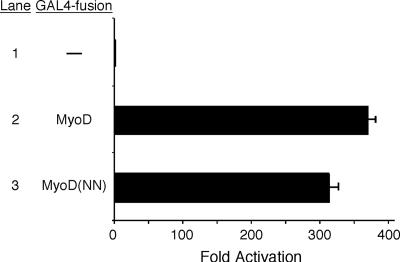
To determine the function of the myogenic code in transcriptional activation, we first examined the transcriptional activity of MyoD and a myogenic code mutant of MyoD, referred to as MyoD(NN), when DNA binding by the bHLH domain was not required (Fig. 1). To generate MyoD proteins with identical DNA binding characteristics, we designed fusion proteins that would bind DNA via the Saccharomyces cerevisiae GAL4 DBD, rather than the MyoD basic domain. Cotransfection of these fusion proteins with an upstream activator sequence-dependent reporter showed that the activation domain of MyoD(NN) induced transcription to nearly equivalent levels as that induced by wild-type MyoD when DNA binding by the basic domain was not a requirement for activation (Fig. 1). While this does not preclude the possibility that the myogenic code may be required for activation potential when bound to DNA through the basic domain, it does suggest that the transcriptional activation domain is not fundamentally defective.
FIG. 1.
Mutation of the myogenic code does not disrupt the inherent transcriptional activation potential of MyoD. Expression plasmids encoding either GAL4(DBD)-MyoD or GAL4(DBD)-MyoD(NN) were cotransfected into C3H10T1/2 cells with pG5E1b, a reporter plasmid that has five GAL4 binding sites and a minimal promoter upstream of luciferase. Approximately 300-fold activation was observed with either MyoD or MyoD(NN), indicating that the two proteins had no difference in their ability to activate transcription when DNA binding was mediated via the GAL4 DBD. Data are expressed as the mean n-fold activation compared to the activation of pG5E1b-luciferase by the GAL4 DBD alone (Lane 1) in three independent experiments. Error bars represent the standard errors of the means.
Determinants of myogenic specificity within MyoD are required for efficient DNA binding and dimerization.
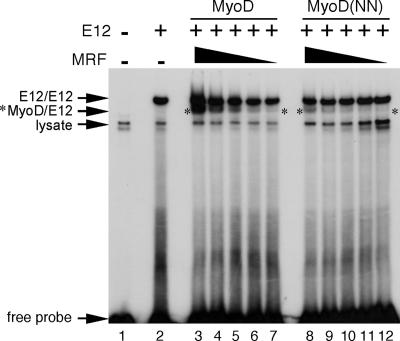
Having established that MyoD(NN) does not have an inherent deficit in transcriptional activation potential, we next tested the notion that the myogenic code is required for efficient DNA binding by comparing the ability of MyoD and MyoD(NN) to bind to a canonical E box (Fig. 2). To compare binding affinities over a range of concentrations, we performed EMSAs over a range of MyoD and MyoD(NN) concentrations in binding reactions that contained an excess of E12 protein (Fig. 2). We also quantified the amount of MyoD and MyoD(NN) that was synthesized in vitro to ensure that equal or nearly equal amounts of the two proteins were present in each binding reaction (see Fig. S1A and B in the supplemental material). It is important to note that E12 homodimers bind less efficiently to the canonical mef2c E box than do MyoD/E12 heterodimers, but E12 is present in drastic excess so it appears to bind efficiently (Fig. 2, lane 2). E12 was provided in this manner to ensure that its availability was not a rate-limiting step to DNA binding. MyoD and MyoD(NN) each bound to the E box probe, but there was clearly less heterodimeric MRF/E12 complex formed when MyoD(NN) was included in the binding reaction (Fig. 2, lanes 8 to 12) versus when MyoD was added (Fig. 2, lanes 3 to 7). Similar results, showing reduced E box binding by MyoD(NN) compared to wild-type MyoD, were observed when E47 was used as the E protein dimerization partner (data not shown).
FIG. 2.
The myogenic code of MyoD is critical for optimal DNA binding to a canonical E box. E12, MyoD, and MyoD(NN) were used in EMSAs with double-stranded oligonucleotides representing the canonical E box from the mef2c skeletal muscle promoter. E12 was included in all of the binding reactions in dramatic excess. Under these conditions, E12 bound to the E box as a homodimer in the absence of MRF protein (lane 2). Either recombinant MyoD (lanes 3 to 7) or MyoD(NN) protein (lanes 8 to 12) was added to the excess E12 as a titration. The amount of recombinant MyoD and MyoD(NN) was matched in lanes 3 and 8, lanes 4 and 9, lanes 5 and 10, lanes 6 and 11, and lanes 7 and 12. MyoD(NN) bound to the canonical E box less efficiently than wild-type MyoD at each concentration of protein. The MRF/E12 heterodimeric DNA-bound species is labeled with an asterisk. Note that the MyoD/E12 band overlaps with the E12 homodimer band, especially at higher concentrations of MyoD (lanes 3 and 4), which makes the E12 homodimer mobility shift appear to change in response to increases in MyoD concentration. Lysate-derived nonspecific mobility shifts are indicated.
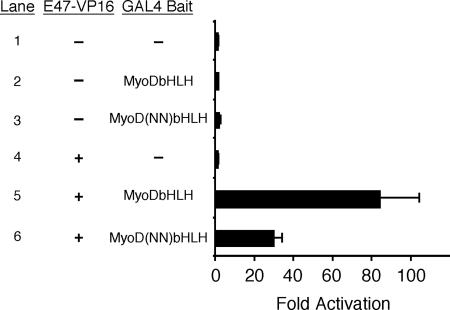
DNA binding by MyoD/E protein heterodimers requires two distinct steps. The proteins must dimerize, and then the heterodimer must bind to the E box sequence. Therefore, we tested whether the MyoD(NN) DNA binding defect observed in Fig. 2 was due all or in part to a requirement of the myogenic code for dimerization. To address this possibility, we utilized a mammalian two-hybrid system, which allowed for transcription of a GAL4-dependent reporter, pG5E1b-luciferase, in a dimerization-dependent manner. We fused the bHLH region of either wild-type MyoD or MyoD(NN) to the GAL4 DBD. These fusion proteins lack the MyoD activation domains and therefore have limited intrinsic capacity to activate the GAL4-dependent reporter on their own. However, if the bHLH fused to the GAL4 DBD heterodimerizes with another bHLH protein that contains an activation domain, the complex will activate transcription and provide an indirect measurement of dimerization. The GAL4-dependent reporter exhibited no activity when cotransfected with the GAL4 DBD alone or with GAL4-MyoD bHLH or GAL4-MyoD(NN) DBD (Fig. 3, lanes 1 to 3). Likewise, the reporter was not activated by E47-VP16 in the presence of the GAL4 DBD alone (Fig. 3, lane 4). By contrast, cotransfection of GAL4-MyoD bHLH and E47-VP16 resulted in strong activation of the reporter due to MyoD-E47 dimerization (Fig. 3, lane 5). Mutation of the myogenic code resulted in approximately threefold less transcriptional activation due to reduced dimerization with E47 (Fig. 3, lane 6), suggesting that a defect in dimerization contributes, at least in part, to the diminished DNA binding of MyoD(NN).
FIG. 3.
The myogenic code is required for optimal dimerization of MyoD with E protein. Plasmids expressing a fusion of the bHLH domain of MyoD and the GAL4 DBD (GAL4-MyoDbHLH) were cotransfected into C3H10T1/2 cells with the reporter plasmid pG5E1b-luciferase, which is a reporter plasmid dependent on five GAL4 binding sites. GAL4, GAL4-MyoDbHLH, and Gal4-MyoD(NN)bHLH all failed to activate the reporter alone, since each of these proteins lacks a transcriptional activation domain (lanes 1 to 3). By contrast, cotransfection of GAL4-MyoDbHLH or GAL4-MyoD(NN)bHLH with pRK5-E47-VP16, a plasmid that expresses the bHLH domain of E47 fused to the viral transcriptional activator protein VP16 (E47-VP16), resulted in potent transactivation of the reporter via dimerization between the MyoD and E47 bHLH domains (lanes 5 and 6). E47-VP16 failed to activate the reporter on its own, since this protein does not bind to the upstream activator sequence (lane 4). Data are expressed as the mean n-fold activation of the reporter when cotransfected with GAL4 DBD alone (lane 1) from three independent experiments. Error bars represent the standard errors of the means.
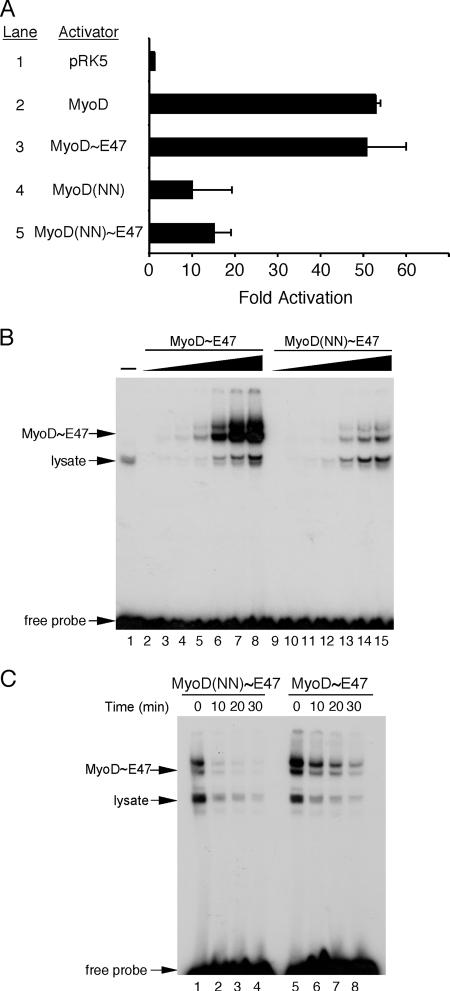
The data presented in Fig. 3 suggest that the myogenic code is required for efficient dimerization between MyoD and its E protein partner. However, the relatively small change in dimerization efficiency seemed unlikely to account for the complete loss of myogenic capacity of MyoD(NN) (5, 11, 12), suggesting that the myogenic code may be required for DNA binding independent of dimerization. To test this possibility and to overcome the dimerization defect in MyoD(NN), we used tethered heterodimers of either MyoD or MyoD(NN) and E47. These tethered dimers have the two bHLH partners covalently joined via a flexible linker, which drastically increases the local concentration of E protein. The increase in local concentration of E protein and MyoD relative to each other is thought to make dimerization no longer rate limiting, and the wild-type version of this molecule, MyoD∼E47, is fully functional since it has been shown previously to be capable of converting fibroblasts into skeletal muscle (31). To discriminate between the roles of the myogenic code in DNA binding and dimerization, we cotransfected a reporter that contains four multimerized copies of an E box driving luciferase expression (4R-TK-luciferase) with expression vectors for the tethered dimers, MyoD∼E47 or MyoD(NN)∼E47 (Fig. 4A). MyoD∼E47 activated the E box-dependent reporter as efficiently as untethered MyoD (Fig. 4A, compare lanes 2 and 3). MyoD(NN) also activated the reporter but less efficiently than wild-type MyoD (Fig. 4A, compare lanes 2 and 4), which is consistent with previous studies (1, 5, 44). Importantly, we found that tethering MyoD(NN) to E47 resulted in a slight increase in transcriptional activation compared to untethered MyoD(NN), but this did not restore wild-type levels of activation (Fig. 4A, lane 5).
FIG. 4.
A tethered heterodimer of MyoD(NN) and E47 exhibits defective DNA binding. (A) Plasmids expressing MyoD, MyoD(NN), or tethered heterodimers of these proteins with E47, MyoD∼E47, and MyoD(NN)∼E47 were cotransfected into C3H10T1/2 cells with the MyoD-responsive reporter plasmid 4R-TK-luciferase. MyoD and MyoD∼E47 each activated this reporter plasmid approximately 50-fold (lanes 2 and 3). Both MyoD(NN) alone and the tethered heterodimer MyoD(NN)∼E47 exhibited reduced activation of the reporter to only 10- and 15-fold, respectively (lanes 4 and 5). Data are expressed as the mean n-fold activation over the activation of 4R-TK-luciferase by pRK5 alone (lane 1) from three independent transfections. Error bars represent the standard errors of the means. (B) The myogenic code is required for DNA binding independent of its function in dimerization. Recombinant MyoD∼E47 or MyoD(NN)∼E47 was used in EMSA with a radiolabeled, double-stranded oligonucleotide representing the mef2c E2 E box. Lanes 2 to 8 show a titration of MyoD∼E47 protein binding to the E box probe. There was significantly less DNA-bound species when identical amounts of MyoD(NN)∼E47 were used (lanes 9 to 15). Lane 1 contains reticulocyte lysate without recombinant MyoD∼E47 protein, represented by a minus sign. Nonspecific lysate-derived bands are indicated. (C) Kinetic analysis of recombinant MyoD∼E47 fusion protein bound to the canonical E box from mef2c. Recombinant MyoD∼E47 or MyoD(NN)∼E47 was used in EMSA with a radiolabeled, double-stranded oligonucleotide representing the mef2c E2 E box. After MyoD∼E47 protein and radiolabeled oligonucleotide reached equilibrium at room temperature, unlabeled competitor oligonucleotides were added to the samples prior to loading the complex on a gel. Time in minutes indicates the length of competition at room temperature. MyoD(NN)∼E47 (lanes 1 to 4) displayed a faster off rate from DNA than did MyoD∼E47 (lanes 5 to 8), as indicated by the rapid loss of binding to radiolabeled probe exhibited by the mutant (compare the change from lane 1 to lane 2 to the change from lane 5 to lane 6).
To determine if the defective activation of the E box-dependent reporter by tethered MyoD(NN)∼E47 was due to a deficiency in DNA binding, we examined the ability of the tethered heterodimers to bind to DNA over a range of concentrations (Fig. 4B). MyoD(NN)∼E47 bound to the canonical E box in a dose-dependent manner (Fig. 4B, lanes 9 to 15), but the affinity was substantially lower than that of wild-type MyoD tethered to E47 (Fig. 4B, lanes 2 to 8). To determine if the decrease in DNA binding observed in Fig. 2A and 4B was due to an increased rate of dissociation of the MyoD(NN)/E protein heterodimer and DNA, we examined the kinetics of MyoD(NN)∼E47 dissociation from the E box in comparison to the dissociation of wild-type MyoD∼E47 from the E box (Fig. 4C). Tethered heterodimers were incubated with radiolabeled oligonucleotides representing a canonical E box, followed by competition with an excess of unlabeled oligonucleotide for various amounts of time before loading the complex on a gel. MyoD∼E47 bound robustly to the E box (Fig. 4C, lane 5), and this binding diminished as time with excess competitor increased (Fig. 4C, lanes 6 to 8). As expected, MyoD(NN)∼E47 bound with reduced efficiency in the initial binding reaction (time 0; Fig. 4C, lane 1). Interestingly, however, the reduction in the amount of bound heterodimer was faster for MyoD(NN)∼E47 (Fig. 4C, lanes 2 to 4) than for wild-type MyoD∼E47 (Fig. 4C, lanes 6 to 8). Indeed, the initial off rate, as measured by the slope of the decrease in signal over the first ten minutes, was twofold faster for MyoD(NN) than for MyoD (data not shown). For the EMSA shown in Fig. 4B and C, we quantified the amount of MyoD∼E47 and MyoD(NN)∼E47 included in each reaction by phosphorimager analysis of SDS-polyacrylamide gel electrophoresis, and in each case, nearly identical amounts of the two proteins were added (see Fig. S1C in the supplemental material). Taken together, the data presented in Fig. 4 demonstrate that the myogenic code is required for efficient E box binding and stabilization.
The myogenic code is required for E box-independent activation of the myogenin promoter by MyoD.
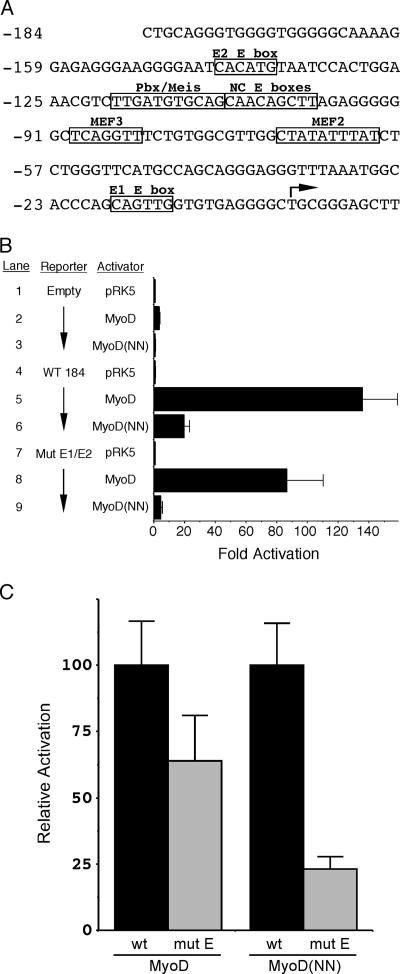
The data presented above demonstrate that the myogenic code of MyoD is a critical determinant of DNA binding and that mutation of these two residues results in decreased affinity for DNA. This decreased affinity is due in part to reduced dimerization affinity, a reduced affinity for E box binding, and an increased off rate from DNA. However, an intact myogenic code was not absolutely required for canonical E box binding (Fig. 2), and therefore, the requirement of these two amino acid determinants for dominant myogenic induction by MyoD is left unexplained. Recently, the discovery of critical noncanonical E boxes in the myogenin promoter (Fig. 5A) suggested that the DNA binding ability of MyoD is tightly regulated to optimize binding to this critical locus in the muscle transcriptional program (3). Therefore, to address the role of the myogenic code in activation of myogenin via the noncanonical E boxes in the promoter, we tested the ability of MyoD and MyoD(NN) to activate the myogenin promoter via E box-dependent and E box-independent mechanisms (Fig. 5). Both wild-type MyoD and MyoD(NN) activated the myogenin promoter significantly (Fig. 5B, lanes 5 and 6). These data and data presented elsewhere (6, 12, 46) clearly demonstrate that MyoD(NN) exhibits significant activation of the myogenin promoter, although it is substantially less robust than wild-type MyoD.
FIG. 5.
The myogenic code is required for canonical E box-independent activation of the myogenin promoter. (A) Sequence of the minus 184 myogenin promoter (Mgn-184), which extends from nucleotides −184 to +11, relative to the transcriptional start site at +1 (arrow). Canonical E boxes 1 and 2 (denoted as E1 and E2, respectively) flank a bipartite sequence composed of a binding site for Pbx/Meis and two overlapping noncanonical (NC) E boxes. MEF2 and MEF3 binding sites are also indicated. (B) Plasmids expressing either MyoD or MyoD(NN) were cotransfected with a reporter plasmid that contained the wild-type myogenin promoter (WT 184) or a mutant form lacking both canonical E boxes (Mut E1/E2) driving expression of luciferase. Neither protein, MyoD or MyoD(NN), significantly activated transcription of the promoterless parent plasmid (lanes 2 and 3). MyoD and MyoD(NN) both significantly activated the wild-type myogenin promoter, approximately 130-fold and 20-fold, respectively (lanes 5 and 6). Mutation of both the E1 and E2 canonical E boxes resulted in a marginal loss of transactivation by MyoD to 90-fold (lane 8). By contrast, mutation of the E1 and E2 E boxes resulted in loss of activation by MyoD(NN) to background or near-background levels (lane 9). Data are expressed as the mean n-fold activation of each reporter by the parent expression vector pRK5 in three independent transfections and analyses. Error bars represent the standard errors of the means. (C) Normalized activation of the mutant E1/E2 Mgn-184 myogenin reporter plasmid. The activation of wild-type (wt) Mgn-184 by both MyoD and MyoD(NN) was set to 100%, and the activation of the mutant reporter was expressed as a percentage of the activation of the wild-type reporter. MyoD activated the reporter lacking canonical E boxes to 64% of the level that it activated the wild-type Mgn-184 reporter. By contrast, the myogenic code mutant MyoD(NN) was only 23% as effective at activation of the mutant reporter compared to the wild-type myogenin reporter. Error bars represent the standard errors of the means from three independent transfections performed in triplicate.
Interestingly, wild-type MyoD also efficiently activated a mutant form of the myogenin promoter in which both canonical E boxes (E1 and E2) were mutated (Fig. 5B, lane 8). These observations are consistent with the notion that activation of myogenin by MyoD occurs via the noncanonical E boxes in the promoter (3, 13). In contrast to wild-type MyoD, the MyoD(NN) myogenic code mutant failed to activate the myogenin promoter in which the two canonical E boxes were mutated (Fig. 5B, lane 9). If the ability to bind to and utilize noncanonical E boxes was similar between MyoD and MyoD(NN), then both proteins should have efficiently activated the mE1/E2 reporter since this construct lacks high-affinity binding sites for MyoD. We normalized the activation of the mutant E1/E2 myogenin reporter by MyoD and MyoD(NN) to the activation of the wild-type reporter (Fig. 5C). Compared to the wild-type reporter, MyoD was 64% as effective at activation of the mutant reporter. By contrast, MyoD(NN) was only able to activate the mutant myogenin promoter to 23% of the level that it activated the wild-type promoter (Fig. 5C). Thus, even when the overall deficit in myogenin activation by MyoD(NN) is taken into account, this myogenic code mutant of MyoD is significantly less efficient than wild-type MyoD at activation through the noncanonical E boxes in the promoter. These data strongly support the notion that the myogenic code of MyoD is required for activation of myogenin via the noncanonical E boxes in the promoter.
The MyoD myogenic code is essential for noncanonical E box binding.
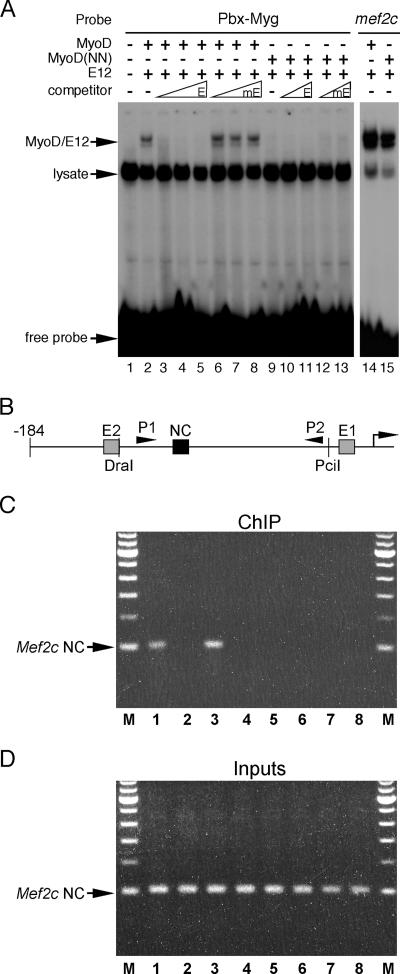
The complete inability of MyoD(NN) to activate the myogenin promoter via the noncanonical E boxes suggested that the myogenic code may be essential for noncanonical E box binding by MyoD. Therefore, we tested the requirement of the myogenic code for MyoD binding to these noncanonical E boxes (Fig. 6). Under conditions where we observed binding to the myogenin noncanonical E boxes by wild-type MyoD (Fig. 6A, lane 2), we never observed any binding by MyoD(NN) (Fig. 6A, lane 9). The binding by MyoD to the noncanonical E boxes was specific, as an unlabeled wild-type E box was able to compete fully for binding to the noncanonical E boxes (Fig. 6A, lanes 3 to 5) while an unlabeled mutant E box oligonucleotide was unable to compete for MyoD binding (Fig. 6A, lanes 6 to 8). The lysates used in this experiment contained equivalent levels of MyoD and MyoD(NN) proteins (see Fig. S1D in the supplemental material), and both MyoD and MyoD(NN) exhibited strong binding to a canonical E box from mef2c as heterodimers with either E12 (Fig. 6A, lanes 14 and 15, respectively) or E47 (data not shown). Thus, these data indicate that the residues of the myogenic code, which are essential determinants of myogenic specificity within MyoD, are required for noncanonical E box binding.
FIG. 6.
The myogenic code of MyoD is required for noncanonical E box binding. (A) Recombinant MyoD, MyoD(NN), and E12 proteins were transcribed and translated in vitro and used in EMSAs with a radiolabeled, double-stranded oligonucleotide representing the Pbx/Meis and noncanonical E box binding sites from the myogenin promoter (Pbx-Myg). Wild-type MyoD bound to the two juxtaposed noncanonical E boxes in the myogenin promoter (lane 2). This binding was specific, as it was efficiently competed by unlabeled competitor oligonucleotide containing the canonical E box from the mef2c gene (E, lanes 3 to 5) but not with a mutant version of the mef2c E box (mE, lanes 6 to 8). MyoD(NN) failed to exhibit any binding to the Pbx-Myg noncanonical E boxes under the same condition (lane 9). This complete lack of binding was specific to noncanonical E boxes, as the same MyoD(NN) lysate used for the noncanonical EMSA was incubated with a radiolabeled oligonucleotide representing the canonical mef2c E E2 box (lane 15) and exhibited binding similar to that of wild-type MyoD (lane 14). (B) Schematic representation of the myogenin promoter construct analyzed by ChIP. The canonical, E1 and E2, and noncanonical (NC) E boxes are denoted. The location of the DraI and PciI restriction sites are also indicated. The primers, P1 and P2, used in the ChIP assays are indicated by arrowheads. The transcriptional start site is depicted as a bent arrow at the right of the schematic. (C) C3H10T1/2 cells were cotransfected with expression plasmids for Myc-tagged versions of either MyoD or MyoD(NN) and various Mgn-184 reporter constructs and were subjected to ChIP and PCR with primers P1 and P2 depicted in panel B. Prior to ChIP, samples were digested with DraI and PciI. (D) Transfections were performed identically to those shown in panel C, except that PCR was performed prior to ChIP to analyze the amount of input DNA in each sample. Cotransfections noted in each lane for panels C and D are as follows: lane 1, MyoD plus Mgn-184(wt); lane 2, MyoD(NN) plus Mgn-184(wt); lane 3, MyoD plus Mgn-184(mutE1/E2); lane 4, MyoD(NN) plus Mgn-184(mutE1/E2); lane 5, MyoD plus Mgn-184(mutNC); lane 6, MyoD(NN) plus Mgn-184(mutNC); lane 7, MyoD plus Mgn-184(wt) with nonspecific antibody in the ChIP reaction; and lane 8, MyoD(NN) plus Mgn-184(wt) with nonspecific antibody in the ChIP reaction. M, marker.
To determine if the myogenic code is also required for noncanonical E box binding in vivo, we determined the ability of wild-type MyoD and MyoD(NN) to bind to the noncanonical E boxes in the myogenin promoter in transfected cells (Fig. 6B). C3H10T1/2 cells were cotransfected with Myc-tagged expression plasmids for MyoD or MyoD(NN) and various myogenin reporter constructs (Fig. 6B). Following transfection, cells were prepared for ChIP analysis (described in detail in Materials and Methods), and DNA was digested with DraI and PciI to separate the noncanonical E boxes from the canonical E1 and E2 E boxes in the myogenin promoter (Fig. 6B). Immunoprecipitated products were then amplified with the primers P1 and P2 depicted in Fig. 6B and described in detail in Materials and Methods. MyoD efficiently bound the noncanonical E boxes in the myogenin promoter, whereas MyoD(NN) exhibited no detectable binding (Fig. 6C, lanes 1 and 2, respectively). The precipitation of the DraI-PciI fragment of the myogenin promoter by MyoD specifically depended on the presence of intact noncanonical E boxes, since mutation of that element ablated the detection of the product by ChIP with wild-type MyoD (Fig. 6C, lane 5). As expected, mutation of both canonical E boxes in the myogenin promoter had no affect on the binding of wild-type MyoD to the noncanonical E boxes or detection of the DraI-PcI fragment of the promoter (Fig. 6C, lane 3). Importantly, the DraI-PciI fragment of the myogenin promoter was not detected in ChIP reactions including MyoD(NN) under any condition (Fig. 6C, lanes 2, 4, and 6). ChIP reactions in which nonspecific antibody was included failed to detect the DraI-PciI band from the myogenin promoter in the presence of both MyoD and MyoD(NN) (Fig. 6C, lanes 7 and 8). All reactions had comparable amounts of input DNA (Fig. 6D). Taken together, these data further demonstrate that the myogenic code is essential for MyoD interaction with the noncanonical E boxes in the myogenin promoter in vivo and that this interaction occurs with the noncanonical E boxes in a sequence-specific manner.
The myogenic code is sufficient to confer noncanonical E box binding to the E12 basic domain.
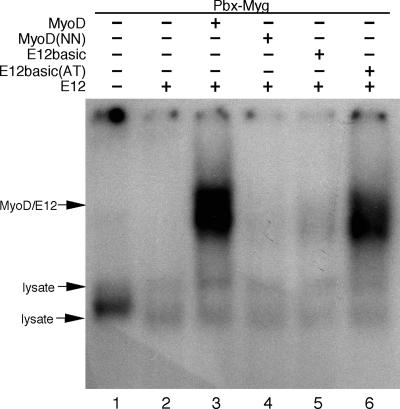
The results presented in Fig. 6 showed the requirement of the myogenic code in MyoD for noncanonical E box binding. We next wanted to determine if these essential determinants of myogenic specificity from MyoD were sufficient to confer noncanonical E box binding specificity. Previous studies have shown that substitution of the E12 basic domain into MyoD (MyoD-E12basic) disrupts the dominant inducing myogenic capability of MyoD and that substitution of the AT back into the E12 basic domain restores myogenic ability (11, 12). Therefore, we compared MyoD-E12basic and MyoD-E12basic(AT) to MyoD and MyoD(NN) for the ability to bind to the myogenin noncanonical E boxes (Fig. 7). Approximately equal amounts of each of the four MyoD proteins were used in each relevant reaction (see Fig. S1E in the supplemental material). As expected, wild-type MyoD/E12 heterodimers bound to the noncanonical E box (Fig. 7, lane 3). Also as expected, mutation of the AT to NN in MyoD(NN) disrupted binding to the noncanonical E box (Fig. 7, lane 4). Similarly, MyoD-E12basic [E12basic] was unable to bind to the myogenin noncanonical E boxes (Fig. 7, lane 5). Importantly, substitution of the AT residues back into MyoD-E12basic [E12basic(AT)] restored the ability to bind to the myogenin noncanonical E boxes (Fig. 7, lane 6). Taken together with the results presented in Fig. 6, these data demonstrate that the myogenic code amino acids, which are essential determinants of myogenesis, are also essential for noncanonical E box binding. Furthermore, these data suggest an explanation for how the myogenic code may provide myogenic specificity to MyoD by allowing binding to noncanonical E boxes in myogenin and probably other key immediate early targets in the muscle transcriptional program.
FIG. 7.
The myogenic code is sufficient to confer noncanonical E box binding to a nonmyogenic bHLH protein containing the E12 basic domain. Equal amounts of recombinant MyoD, MyoD(NN), MyoD-E12basic, or MyoD-E12basic(AT) were included in EMSA containing recombinant E12 and a single radiolabeled, double-stranded oligonucleotide representing the Pbx/Meis and noncanonical E box binding sites from the myogenin promoter (Pbx-Myg). Wild-type MyoD/E12 bound efficiently to the Pbx-Myg noncanonical E boxes (lane 3). MyoD(NN) and MyoD-E12basic were unable to bind to the noncanonical E boxes in Pbx-Myg (lanes 4 and 5, respectively). MyoD-E12basic(AT), which contains the E12 basic domain with AT in place of NN in the context of MyoD, bound to the noncanonical E boxes in Pbx-Myg along with E12 (lane 6). E12 alone did not bind to the Pbx-Myg probe (Lane 2). Lane 1 contains reticulocyte lysate without recombinant protein, represented by a minus sign. Included proteins in each reaction are denoted by a plus sign. Nonspecific, lysate-derived bands are denoted.
The MyoD myogenic code is required for formation of a higher order complex on DNA with Pbx/Meis.
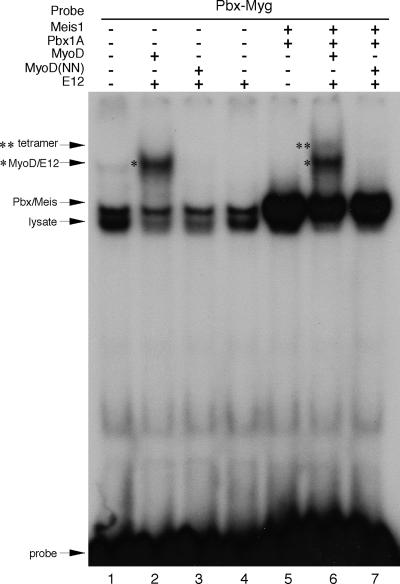
While myogenic code mutants are incapable of directly binding to the noncanonical E boxes in the myogenin promoter, it is likely that MyoD binding to these noncanonical E boxes is aided in vivo by a resident Pbx/Meis heterodimer that is bound at a site adjacent to the noncanonical E boxes (Fig. 5A) prior to myogenic induction (13). Therefore, we compared the abilities of equivalent amounts of MyoD and MyoD(NN) (see Fig. S1D in the supplemental material) to bind to the noncanonical E boxes when the adjacent Pbx/Meis binding site was occupied (Fig. 8). A radiolabeled probe representing both the Pbx/Meis and noncanonical E box binding sites was readily bound by Pbx1A/Meis1 heterodimers (Fig. 8, lane 5) and by MyoD/E12 heterodimers (Fig. 8, lane 2). As expected, MyoD(NN)/E12 heterodimers displayed no binding to the noncanonical E box in the absence of Pbx/Meis (Fig. 8, lane 3). When all four proteins (Pbx1A, Meis1, MyoD, and E12) were added to the binding reaction, a higher order complex was formed that migrated slightly slower than the MyoD/E12 heterodimer (Fig. 8, lane 6). However, neither binding by MyoD(NN)/E12 nor the higher order complex containing Pbx1A/Meis1 was ever observed when MyoD(NN) was included in the binding reaction (Fig. 8, lane 7). Thus, we conclude that the myogenic code is completely required for binding to noncanonical E boxes in the myogenin gene, an essential target in the muscle transcriptional program, even in the presence of Pbx/Meis.
FIG. 8.
The myogenic code is required for MyoD/E12 and Pbx/Meis heterodimers to form a tetrameric complex on the myogenin promoter. Recombinant MyoD, MyoD(NN), E12, Pbx1A, and Meis1 proteins were used in EMSAs with a single radiolabeled, double-stranded oligonucleotide representing the Pbx/Meis and noncanonical E box binding sites from the myogenin promoter (Pbx-Myg). Lane 1 contains reticulocyte lysate without recombinant protein, represented by a minus sign. MyoD/E12 bound to the noncanonical E boxes in the Pbx-Myg oligonucleotide (lane 2, denoted by an asterisk). This complex was not formed when MyoD was replaced with MyoD(NN) (lane 3) or with E12 alone (lane 4). Pbx1A/Meis1 heterodimers bound efficiently to Pbx-Myg (lane 5) and formed a higher order species representing a tetrameric complex with MyoD and E12 (lane 6). The tetrameric complex is denoted by a double asterisk. By contrast, an equivalent amount of MyoD(NN) did not form a tetrameric complex with Pbx/Meis (lane 7).
DISCUSSION
The two amino acids of the myogenic code, an alanine and threonine in the basic domain, are essential determinants of myogenic specificity and provide the critical distinction between the MRF family of bHLH transcription factors and all other bHLH family members. Mutation of these amino acids results in a bHLH protein that is competent to activate transcription but that is no longer capable of converting nonmuscle cells into muscle. It has remained unclear what properties these amino acids afford to the MRF family for conferring myogenic-inducing ability. In the current study, we presented work that establishes the myogenic code as a critical determinant that allows MyoD to bind DNA with high affinity. We showed that these amino acid determinants of myogenic specificity within MyoD are required for efficient dimerization with E proteins and are required for high-affinity DNA binding. The requirement of the myogenic code for DNA binding is especially pronounced at low-affinity sites within the myogenin promoter, where the myogenic code is completely required for MyoD to bind directly to noncanonical E boxes and to form a tetrameric DNA-bound complex with Pbx/Meis.
Previous studies have come to different conclusions about the function of the myogenic code in transcriptional activation and the induction of myogenesis. Some studies have suggested that the primary role of the myogenic code is for proper display of the transcriptional activation domains of the MRFs (5, 18). These models suggest that the activation domain of MyoD is masked or the signal to activate is not transmitted properly to the activation domain in the absence of an intact myogenic code. By contrast, the initial work on the myogenic code suggested that it functioned to recruit a “recognition factor” that was essential for myogenic activation (40, 44). The existence of this factor was based on the observation that myogenic code mutants for several of the MRFs activated transcription in certain cell types better than in others (40, 44). Our data presented here suggest that the primary requirement of the myogenic code is for DNA binding and dimerization, but the data do not preclude the possibility that an intact myogenic code may result in a conformation of MyoD that enables interaction with a factor that aids in activation, or perhaps more likely, a factor that helps to stabilize DNA binding. Indeed, our data support a model in which Pbx/Meis serves as a myogenic pioneer factor that functions to stabilize the initial binding of a small amount of MyoD to myogenin and possibly other essential targets, which would then allow more robust binding subsequently to canonical E boxes, as has been suggested previously (3).
MyoD mutants that lack a region that is enriched for cysteines and histidines, or a C-terminal region known as helix III, were originally shown to be defective in chromatin remodeling at the myogenin promoter (16). Subsequently, it was shown that the chromatin remodeling domains of MyoD are also required for efficient binding to noncanonical E boxes, suggesting that the original stabilization of binding at the promoter through interaction with Pbx/Meis is a critical step in the formation of a stably remodeled locus and transcriptional activation (3). An important distinction between the chromatin remodeling domains of MyoD and the myogenic code is that the residues of the myogenic code are essential for myogenesis, whereas the chromatin remodeling domains are not (16). Chromatin remodeling domain mutants retain the ability to induce myogenesis, albeit less potently than wild-type MyoD (16). We predict that this retained ability to induce myogenesis is the result of the residual capacity to bind noncanonical E boxes, which is in contrast to the complete inability of MyoD(NN) to bind to noncanonical E boxes and to induce myogenesis.
The existence of distinct mutations in diverse regions of MyoD that affect interaction with Pbx/Meis further supports the notion that there is a particular conformation that is critical to inducing the muscle transcriptional program. Indeed, recent work has shown that the carboxy- and amino-terminal portions of MyoD function together to activate transcription, suggesting that the positioning of these domains in relation to one another is a critical aspect of gene activation (19). In addition, mutation of the critical alanine of the myogenic code in MyoD is known to cause the protein to have an altered conformation when bound to DNA, as shown by increased sensitivity to protease cleavage (18). It is possible that a DNA binding defect could contribute, at least in part, to the increased protease sensitivity of the myogenic code mutants since stable binding to DNA by wild-type MyoD might provide some protection against protease cleavage.
A conformation role for the myogenic code is appealing, since the MyoD crystal structure shows that the two residues of the myogenic code do not make direct contact with DNA (25). The crystal structure also suggests that replacing the AT with bulkier amino acids, such as the asparagines found in E12, would displace an arginine slightly C-terminal to the myogenic code (25). However, a second site suppressor screen to identify substitutions at the position of this arginine that could restore myogenic activity did not yield any proteins with dominant inducing activity (18). This, and altered protease sensitivity in myogenic code mutants bound to DNA, suggests that the AT may be required to set the basic domain in a particular conformation specific to the MRF family of bHLH transcription factors (18). The crystal structure of MyoD further indicates that the basic domain forms an α-helix when bound to DNA, and the stability of this helix is a determinant in the binding affinity for a particular site (25, 42). The myogenic code affects not only the DNA binding affinity of MyoD but also the binding specificity, as substitution of AT into the E12 basic domain confers the DNA binding specificity of MyoD (22). A role in establishing proper conformation of the basic domain would explain how seemingly subtle mutations in the MyoD basic domain have such drastic effects on numerous aspects of MyoD function, including myogenic specificity.
The ability of MyoD to induce myogenesis in cell culture and to initiate muscle specification in vivo requires that MyoD bind to select, immediate early targets. The ability to bind stably at critical muscle loci, such as myogenin, requires the ability to interact with Pbx/Meis and to bind to noncanonical E boxes (3, 13). We hypothesize that the myogenic code is the critical determinant for optimal dimerization and DNA binding, which are required to stabilize MyoD on noncanonical binding sites in critical muscle loci to allow for subsequent chromatin remodeling, transcriptional activation, and myogenic induction. Examples such as this, of highly fine-tuned binding specificity, are likely to be a common mechanism by which individual members of large families of transcription factors are able to discriminate appropriate binding sites in the context of the genome and activate distinct transcriptional programs.
Supplementary Material
Acknowledgments
We are grateful to Anthony Gerber for helpful discussions and critical comments on the manuscript. We thank Stephen Tapscott, Harold Bernstein, and Rik Derynck for providing plasmids used in these studies.
A.B.H. was supported in part by a predoctoral fellowship from the Howard Hughes Medical Institute. This work was supported by grants HL64658 and AR52130 from the NIH to B.L.B.
Footnotes
Published ahead of print on 11 June 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bengal, E., O. Flores, P. N. Rangarajan, A. Chen, H. Weintraub, and I. M. Verma. 1994. Positive control mutations in the MyoD basic region fail to show cooperative DNA binding and transcriptional activation in vitro. Proc. Natl. Acad. Sci. USA 91:6221-6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergstrom, D. A., B. H. Penn, A. Strand, R. L. Perry, M. A. Rudnicki, and S. J. Tapscott. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell 9:587-600. [DOI] [PubMed] [Google Scholar]
- 3.Berkes, C. A., D. A. Bergstrom, B. H. Penn, K. J. Seaver, P. S. Knoepfler, and S. J. Tapscott. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14:465-477. [DOI] [PubMed] [Google Scholar]
- 4.Berkes, C. A., and S. J. Tapscott. 2005. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 16:585-595. [DOI] [PubMed] [Google Scholar]
- 5.Black, B. L., J. D. Molkentin, and E. N. Olson. 1998. Multiple roles for the MyoD basic region in transmission of transcriptional activation signals and interaction with MEF2. Mol. Cell. Biol. 18:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan, T. J., T. Chakraborty, and E. N. Olson. 1991. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc. Natl. Acad. Sci. USA 88:5675-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan, T. J., and E. N. Olson. 1990. Myogenin resides in the nucleus and acquires high affinity for a conserved enhancer element on heterodimerization. Genes Dev. 4:582-595. [DOI] [PubMed] [Google Scholar]
- 8.Buckingham, M. 2001. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 11:440-448. [DOI] [PubMed] [Google Scholar]
- 9.Buckingham, M., L. Bajard, T. Chang, P. Daubas, J. Hadchouel, S. Meilhac, D. Montarras, D. Rocancourt, and F. Relaix. 2003. The formation of skeletal muscle: from somite to limb. J. Anat. 202:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao, Y., R. M. Kumar, B. H. Penn, C. A. Berkes, C. Kooperberg, L. A. Boyer, R. A. Young, and S. J. Tapscott. 2006. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 25:502-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, R. L., P. F. Cheng, A. B. Lassar, and H. Weintraub. 1990. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell 60:733-746. [DOI] [PubMed] [Google Scholar]
- 12.Davis, R. L., and H. Weintraub. 1992. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science 256:1027-1030. [DOI] [PubMed] [Google Scholar]
- 13.de la Serna, I. L., Y. Ohkawa, C. A. Berkes, D. A. Bergstrom, C. S. Dacwag, S. J. Tapscott, and A. N. Imbalzano. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25:3997-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dodou, E., S. M. Xu, and B. L. Black. 2003. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech. Dev. 120:1021-1032. [DOI] [PubMed] [Google Scholar]
- 15.Edmondson, D. G., T. C. Cheng, P. Cserjesi, T. Chakraborty, and E. N. Olson. 1992. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Mol. Cell. Biol. 12:3665-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerber, A. N., T. R. Klesert, D. A. Bergstrom, and S. J. Tapscott. 1997. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 11:436-450. [DOI] [PubMed] [Google Scholar]
- 17.Hjalt, T. 2004. Basic helix-loop-helix proteins expressed during early embryonic organogenesis. Int. Rev. Cytol 236:251-280. [DOI] [PubMed] [Google Scholar]
- 18.Huang, J., H. Weintraub, and L. Kedes. 1998. Intramolecular regulation of MyoD activation domain conformation and function. Mol. Cell. Biol. 18:5478-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishibashi, J., R. L. Perry, A. Asakura, and M. A. Rudnicki. 2005. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J. Cell Biol. 171:471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, S. 2004. An overview of the basic helix-loop-helix proteins. Genome Biol. 5:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kassar-Duchossoy, L., B. Gayraud-Morel, D. Gomes, D. Rocancourt, M. Buckingham, V. Shinin, and S. Tajbakhsh. 2004. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature 431:466-471. [DOI] [PubMed] [Google Scholar]
- 22.Kophengnavong, T., J. E. Michnowicz, and T. K. Blackwell. 2000. Establishment of distinct MyoD, E2A, and Twist DNA binding specificities by different basic region-DNA conformations. Mol. Cell. Biol. 20:261-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lassar, A. B., R. L. Davis, W. E. Wright, T. Kadesch, C. Murre, A. Voronova, D. Baltimore, and H. Weintraub. 1991. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell 66:305-315. [DOI] [PubMed] [Google Scholar]
- 24.Liu, D., B. L. Black, and R. Derynck. 2001. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 15:2950-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma, P. C., M. A. Rould, H. Weintraub, and C. O. Pabo. 1994. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77:451-459. [DOI] [PubMed] [Google Scholar]
- 26.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 14:763-772. [DOI] [PubMed] [Google Scholar]
- 27.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 28.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1996. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 16:2627-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molkentin, J. D., and E. N. Olson. 1996. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. USA 93:9366-9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murre, C., P. S. McCaw, and D. Baltimore. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777-783. [DOI] [PubMed] [Google Scholar]
- 31.Neuhold, L. A., and B. Wold. 1993. HLH forced dimers: tethering MyoD to E47 generates a dominant positive myogenic factor insulated from negative regulation by Id. Cell 74:1033-1042. [DOI] [PubMed] [Google Scholar]
- 32.Ohkawa, Y., C. G. Marfella, and A. N. Imbalzano. 2006. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 25:490-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson, E. N. 1990. MyoD family: a paradigm for development? Genes Dev. 4:1454-1461. [DOI] [PubMed] [Google Scholar]
- 34.Olson, E. N., and W. H. Klein. 1994. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 8:1-8. [DOI] [PubMed] [Google Scholar]
- 35.Puri, P. L., S. Iezzi, P. Stiegler, T. T. Chen, R. L. Schiltz, G. E. Muscat, A. Giordano, L. Kedes, J. Y. Wang, and V. Sartorelli. 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell 8:885-897. [DOI] [PubMed] [Google Scholar]
- 36.Puri, P. L., V. Sartorelli, X. J. Yang, Y. Hamamori, V. V. Ogryzko, B. H. Howard, L. Kedes, J. Y. Wang, A. Graessmann, Y. Nakatani, and M. Levrero. 1997. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1:35-45. [DOI] [PubMed] [Google Scholar]
- 37.Sabourin, L. A., and M. A. Rudnicki. 2000. The molecular regulation of myogenesis. Clin. Genet. 57:16-25. [DOI] [PubMed] [Google Scholar]
- 38.Sartorelli, V., J. Huang, Y. Hamamori, and L. Kedes. 1997. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 17:1010-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sartorelli, V., P. L. Puri, Y. Hamamori, V. Ogryzko, G. Chung, Y. Nakatani, J. Y. Wang, and L. Kedes. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4:725-734. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz, J. J., T. Chakraborty, J. Martin, J. M. Zhou, and E. N. Olson. 1992. The basic region of myogenin cooperates with two transcription activation domains to induce muscle-specific transcription. Mol. Cell. Biol. 12:266-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tajbakhsh, S. 2003. Stem cells to tissue: molecular, cellular and anatomical heterogeneity in skeletal muscle. Curr. Opin. Genet. Dev. 13:413-422. [DOI] [PubMed] [Google Scholar]
- 42.Turner, E. C., C. H. Cureton, C. J. Weston, O. S. Smart, and R. K. Allemann. 2004. Controlling the DNA binding specificity of bHLH proteins through intramolecular interactions. Chem. Biol. 11:69-77. [DOI] [PubMed] [Google Scholar]
- 43.Weintraub, H., R. Davis, S. Tapscott, M. Thayer, M. Krause, R. Benezra, T. K. Blackwell, D. Turner, R. Rupp, S. Hollenberg, et al. 1991. The myoD gene family: nodal point during specification of the muscle cell lineage. Science 251:761-766. [DOI] [PubMed] [Google Scholar]
- 44.Weintraub, H., V. J. Dwarki, I. Verma, R. Davis, S. Hollenberg, L. Snider, A. Lassar, and S. J. Tapscott. 1991. Muscle-specific transcriptional activation by MyoD. Genes Dev. 5:1377-1386. [DOI] [PubMed] [Google Scholar]
- 45.Weintraub, H., T. Genetta, and T. Kadesch. 1994. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev. 8:2203-2211. [DOI] [PubMed] [Google Scholar]
- 46.Winter, B., T. Braun, and H. H. Arnold. 1992. Co-operativity of functional domains in the muscle-specific transcription factor Myf-5. EMBO J. 11:1843-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yee, S. P., and P. W. Rigby. 1993. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 7:1277-1289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.