Abstract
To investigate the contribution of DNA replication initiation and elongation to the intra-S-phase checkpoint, we examined cells treated with the specific topoisomerase I inhibitor camptothecin. Camptothecin is a potent anticancer agent producing well-characterized replication-mediated DNA double-strand breaks through the collision of replication forks with topoisomerase I cleavage complexes. After a short dose of camptothecin in human colon carcinoma HT29 cells, DNA replication was inhibited rapidly and did not recover for several hours following drug removal. That inhibition occurred preferentially in late-S-phase, compared to early-S-phase, cells and was due to both an inhibition of initiation and elongation, as determined by pulse-labeling nucleotide incorporation in replication foci and DNA fibers. DNA replication was actively inhibited by checkpoint activation since 7-hydroxystaurosporine (UCN-01), the specific Chk1 inhibitor CHIR-124, or transfection with small interfering RNA targeting Chk1 restored both initiation and elongation. Abrogation of the checkpoint markedly enhanced camptothecin-induced DNA damage at replication sites where histone γ-H2AX colocalized with replication foci. Together, our study demonstrates that the intra-S-phase checkpoint is exerted by Chk1 not only upon replication initiation but also upon DNA elongation.
In response to DNA damage and/or replication stress, cells induce checkpoint pathways to arrest DNA replication and cell cycle progression (24). Such inhibition is critical for DNA repair and to prevent additional genomic alterations resulting from replication of a damaged DNA template. Cells with deficient replication checkpoints were first identified as undergoing radioresistant DNA synthesis (commonly referred to as RDS) (38). The paradigm for such a defect is the cancer-predisposing hereditary syndrome ataxia telangiectasia (38), which is due to a genetic defect of the ATM gene (53). The checkpoint response is well characterized as a preferential inhibition of initiation for the late-firing origins (27, 32, 46, 50, 54).
The contribution of DNA elongation in the replication checkpoint has been difficult to establish since the agents that induce the checkpoint directly arrest replication fork progression when forks encounter the DNA lesions that elicit the checkpoint. Stabilization of the stalled forks is controlled by checkpoint proteins (13), as the stalled fork itself is recognized as a signal for checkpoint induction (69). To differentiate between passive replication inhibition due to direct collision of DNA replication complexes with the DNA lesions and active replication inhibition due to checkpoint activation, we have utilized single-cell and single-DNA molecule nucleotide pulse-labeling using immunofluorescence microscopy. We also took advantage of the fact that camptothecin (CPT) induces the S-phase checkpoint within minutes of addition and that the majority of topoisomerase I lesions induced by camptothecin reverse within minutes after drug removal (12, 49). These molecular characteristics make CPT a sharp tool for studying the DNA replication checkpoint.
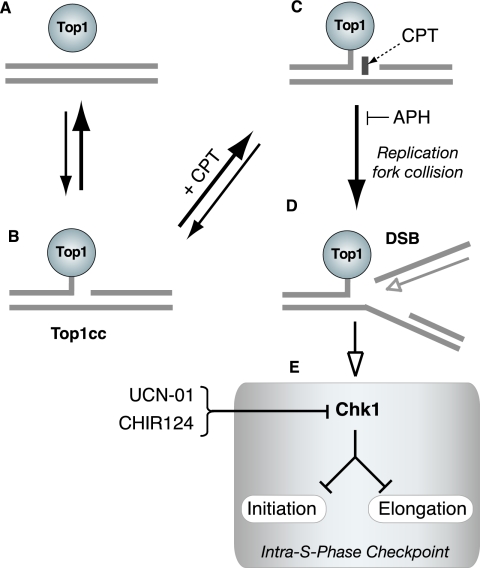
CPT is a selective inhibitor of topoisomerase I (Top1) (4, 21, 36), an enzyme which relaxes DNA supercoiling generated during replication, transcription and chromatin assembly and probably during chromatin remodeling and DNA repair (8, 39, 63). Top1 produces transient single-strand nicks in the DNA by forming catalytic intermediates that are referred to as Top1 cleavage complexes (Top1cc) (Fig. 1). CPT binds at the interface of the DNA-Top1cc as Top1 cleaves the DNA (39) and prevents the religation of the Top1cc, thereby stabilizing the Top1-linked single-stranded DNA nick (7, 21, 39) (Fig. 1C). Top1cc can also be trapped by a wide range of endogenous and exogenous DNA alterations (39, 40). Endogenous lesions that induce Top1cc include nicks, base mismatches introduced during DNA replication and repair or resulting from cytosine deamination, abasic sites (40), and oxidative damage generated by apoptotic stimuli (56, 57). Top1cc can also be induced by a variety of DNA adducts produced by carcinogens such as benzo[a]pyrene diol epoxides, vinyl chloride and ethyl alcohol and by DNA-damaging drugs besides CPTs commonly used for treating human cancers (40).
FIG. 1.
Proposed mechanism of replication-mediated DSB induction by CPT. (A) Under normal conditions, topoisomerase 1 (Top1) is noncovalently bound to chromatin. (B) Only a small fraction of Top1 creates transient DNA single-strand nicks (Top1cc), allowing unwinding of the DNA to relieve torsional stress. (C) CPT (rectangle) forms a complex with Top1cc, preventing religation of the single-strand break. (D) Replication of the leading strand (top strand) up to the 5′ end of the broken DNA produces a replication-mediated DSB (“replication runoff”). APH prevents the replication DSB by arresting DNA polymerase before it encounters a Top1cc. (E) Replication lesions activate Chk1, which activates the intra-S-phase checkpoint by arresting both DNA replication initiation and DNA elongation. UCN-01 and CHIR124 inhibit Chk1 activity and abrogate the cell cycle checkpoint.
Top1cc are among the best-characterized inducers of replication fork damage. DNA double-strand breaks (DSBs) are created through the collision of DNA replication forks with the trapped Top1cc (Fig. 1D) (20, 22, 45, 58, 59). Replication-mediated DSBs occur on the leading strand of DNA synthesis, and this process is referred to as “replication runoff,” as the polymerase extends the newly synthesized DNA strand up to the last base of the template (58, 59). Accordingly, the DNA polymerase inhibitor aphidicolin (APH) inhibits the formation of replication-mediated DSB and CPT cytotoxicity, without affecting the CPT-induced Top1cc (Fig. 1C and D), highlighting the need for ongoing DNA replication in the production of DNA damage (20, 22, 45, 58).
Top1cc inhibit DNA synthesis by at least two mechanisms. First, the trapped Top1cc can arrest DNA replication forks directly as they create replication-mediated DSBs (Fig. 1D) (25, 55, 60). Second, the replication-mediated DSBs can be sensed as DNA damage and induce checkpoints that halt DNA synthesis to allow DNA repair and prevent further damage (48, 49, 59, 66). DNA replication can be inhibited at doses as low as 0.03 μM CPT that produce a low frequency of Top1cc and minimal cytotoxicity (20, 25, 67). The replication checkpoint elicited by Top1 inhibitors restrains DNA replication initiation primarily through activation of the ATR (ATM and Rad3-related) and Chk1 protein kinases (Fig. 1E) (10, 25, 42, 48, 64, 67, 68). This checkpoint remains effective hours after the removal of CPT (and reversal of the Top1cc) (25, 49, 67) and has recently been proposed to operate both at the level of initiation and replication fork elongation in response to ATR, Hus1, and Chk1 activation (66).
Chk1 kinase activity can be inhibited by the protein kinase inhibitor 7-hydroxystaurosporine (UCN-01) (Fig. 1E) (6, 19), which was previously identified as a potent abrogator of the CPT-induced cell cycle arrest in S phase and as being able to restore DNA synthesis (26, 48, 49, 62). UCN-01 also produces a marked increase in the cytotoxicity of CPT (48), likely due to the increased levels of unrepaired DSBs (16). Recently, a more specific inhibitor of Chk1 (CHIR-124) has been identified (35). The quinolone-based small molecule CHIR-124 abrogates the S and G2/M checkpoints and also synergistically increases the cytotoxicity of CPTs (35, 61).
DSBs induce the phosphorylation of histone H2AX on serine 139 (43, 44, 47). That phosphorylated form, which is referred to as γ-H2AX, can be detected with specific antibodies by immunofluorescence or Western blotting (43). CPT rapidly induces γ-H2AX foci in replicating cells, demonstrating the existence of DSBs associated with replication (16, 17, 59). The CPT-induced γ-H2AX foci have been proposed to result from replication fork collisions with Top1cc (17) and are therefore expected to coincide with DNA replication foci.
Human cells replicate their genome within nuclear sites (replication factories) that can be identified as replication foci by nucleotide incorporation into distinct structural units in the nucleus (34). Replication foci appear in specific patterns throughout the S phase (13, 14). The pattern of early-S-phase cells consists of a large number of small foci distributed evenly throughout the nucleus (14, 29). Cells in mid-S phase are characterized by the presence of replication foci around the periphery of the nucleus and nucleolar regions, while cells in late S phase have a relatively small number of large foci, corresponding to the replication of heterochromatic regions (14, 23, 37). These differential patterns allow the determination of the replication status of individual cells at various stages of S phase.
In the present study we used a short exposure to CPT to inhibit DNA replication. By monitoring individual cells before and after CPT treatment, we sought to determine whether a difference existed between early- and late-S-phase cells in their ability to arrest DNA replication. Labeling of replication foci and DNA fibers with halogenated nucleotides and specific antibodies was also used to examine checkpoint control exerted both at the DNA replication initiation and elongation levels. The Chk1 inhibitors UCN-01 and CHIR-124 both induced new replication foci and restored replication in preexisting foci, as well as DNA initiation and elongation in DNA fibers. Similar results were obtained in cells transfected with small interfering RNA (siRNA) targeting Chk1. γ-H2AX intensity was also increased dramatically by UCN-01, suggesting that Chk1 prevents replication-mediated DNA damage by inhibiting both DNA initiation and elongation (Fig. 1E).
MATERIALS AND METHODS
Cell lines, chemicals, and drugs.
HT29 colon carcinoma cells were grown in Dulbecco modified Eagle medium complemented with 10% fetal bovine serum (Gibco-BRL, Grand Island, NY) at 37°C and 5% CO2. HT29 cells, camptothecin, and UCN-01 were obtained from the Developmental Therapeutics Program (DTP, DCTD, National Cancer Institute, Bethesda, MD). CHIR-124 was obtained from Chiron Corp.
[3H]TdR incorporation assay.
HT29 cells were prelabeled for 48 h with 0.01 μCi of [14C]TdR (Perkin-Elmer, Wellesley, MA)/ml and pulse-labeled for 10 min with 1 μCi of [3H]TdR (MP Biochemicals-ICN, Irvine, CA)/ml to measure DNA synthesis. Incorporation was stopped by washing the cells twice with cold Hanks buffered saline solution (CellGro; Mediatech, Inc., Herndon, VA). After the cells were scraped into 4 ml of Hanks balanced salt solution, aliquots were precipitated with 100% trichloroacetic acid in triplicate. Samples were kept on ice and mixed vigorously with a vortex mixer every 10 min for 2 h. After centrifugation at 9,400 × g for 10 min at 4°C, the supernatants were removed, and 0.5 ml of 0.4 M NaOH was added overnight at 37°C to dissolve the precipitates. Samples were counted by using dual-label liquid scintillation, and the amount of [3H]TdR was normalized to the amount of [14C]TdR in each sample. DNA synthesis was measured as a ratio of [3H] to [14C] in the treated samples divided by the ratio of [3H] to [14C] in the untreated control samples.
BrdU incorporation assay.
HT29 cells were incubated with 50 μM bromodeoxyuridine (BrdU; CalBiochem) for 30 min (either before or after the addition of CPT). Cells were fixed in cold 70% ethanol at the indicated times and stored at 4°C. DNA was denatured by using 2 N HCl and 0.5% Triton X-100 and then neutralized with 0.1 M sodium borate (pH 8.5). After two washes with 0.5% Tween 20 and 0.5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS), anti-BrdU fluorescein isothiocyanate (Becton Dickinson, Franklin Lakes, NJ) was added for 1 h. After two washes, samples were incubated with RNase-propidium iodide and analyzed on a FACScan flow cytometer (Becton Dickinson).
The percentage of cells in early S phase versus late S phase was determined by using CellQuest software (BD Biosciences, San Jose, CA). The number of BrdU-positive cells was divided evenly into early- and late-S-phase populations in the untreated control samples. These parameters were also used to determine the number of BrdU-positive cells after CPT treatment. The number of BrdU-positive cells in early S phase after drug treatment was expressed as a percentage of untreated early-S-phase cells; the same was done for late-S-phase cells. The results represent the average ± the standard error of the mean (SEM) of three independent experiments.
Protein extracts and immunoblotting.
Cells were grown to 70 to 80% at the time of drug treatment. Cells were harvested and washed twice with PBS and then incubated on ice for 30 min in lysis buffer (1% sodium dodecyl sulfate, 1 mM sodium orthovanadate, 10 mM Tris [pH 7.4], and phosphatase inhibitor cocktail [Sigma Chemical Co., St. Louis, MO]) and protease inhibitor (Roche Applied Science, Indianapolis, IN). Cell extracts were sonicated, incubated on ice for 10 min, and then boiled for 10 min. The protein concentration was determined by using a DC Bio-Rad protein assay.
Cell extracts were electrophoresed in 4 to 20% Tris-glycine precast gels (Invitrogen, Carlsbad, CA) and transferred onto Immobilon-P membranes (Millipore, Bedford, MA) by using a semidry apparatus. Immunoreactive bands were visualized by using enhanced chemiluminescence (ECL Super Signal; Pierce).
Anti-Chk1 and antiactin monoclonal antibodies were obtained from Santa Cruz Biotech (Santa Cruz, CA), and polyclonal anti-Chk1-S317 was obtained from Bethyl Laboratories (Montgomery, TX). Anti-Chk2 and anti-Chk2T68 were obtained from Cell Signaling (Danvers, MA).
Chk1 knockdown with siRNA.
siRNA targeting Chk1 was obtained from Dharmacon (Lafayette, CO) (20 μM stock; SMARTpool mixture of four siRNA duplexes, catalog no. M-003255-02-0010). Control siRNA was obtained from QIAGEN, Inc. (Valencia, CA) [20 μM stock; r(UUCUCCGAACGUGUCACGU)dTdT and r(ACGUGACACGUUCGGAGAA)dTdT strands]. Two hundred nanomolar of siRNA per transfection or Lipofectamine 2000 (Invitrogen) (5 μl per six-well plate for fiber and lysate analysis and 2 μl per four-well chamber for foci analysis) was incubated separately in prewarmed (37°C) OptiMem medium (Gibco/Invitrogen) for 15 min. Each siRNA mixture was added to the appropriate amount of Lipofectamine/OptiMem and incubated for an additional 15 min. Then, 500 μl of each siRNA-Lipofectamine mixture was added to each plate or chamber. After 24 h, the medium was replaced with fresh Dulbecco modified Eagle medium and incubated for a further 48 h, for a total 72 h of transfection, at which time the experiments were performed.
Immunofluorescence microscopy staining of replication foci using CldU and IdU.
DNA replication sites were visualized by incorporation of chlorodeoxyuridine (CldU) and iododeoxyuridine (IdU) into DNA. HT29 cells were grown in four-well chamber slides (Nalge-Nunc International, Rochester, NY) and labeled with 100 μM CldU (ICN, Irvine, CA) or IdU (Sigma Chemical Co., St. Louis, MO) for 45 min at different time intervals. Cells were washed with PBS, fixed with cold 70% ethanol, and stored at 4°C.
For antibody staining, the ethanol was removed, and 100% methanol was added for 5 min. Cells were washed twice with PBS and incubated with 1.5 M HCl for 30 min to denature the DNA. Cells were washed with PBS, permeabilized with 0.5% Tween 20 in PBS for 5 min, and then incubated in 5% normal goat serum (NGS; Sigma Chemical Co., St. Louis, MO), 0.5% Tween 20, and 0.1% BSA (Jackson Immunoresearch, West Grove, PA) in PBS for 20 min to reduce nonspecific binding. Primary antibodies CldU (rat anti-BrdU; Accurate Chemical and Science Co., Westbury, NY) and IdU (mouse anti-BrdU; BD Biosciences) were diluted in NGS buffer, added to the slides, and incubated in a humid environment for 2 h. Slides were washed with PBS-Tween 20 and then in a high-salt buffer (200 mM NaCl, 0.2% Tween 20, and 0.2% NP-40 in PBS) for 15 min. The samples were incubated in NGS buffer a second time for 20 min, followed by incubation with secondary antibodies (CldU, donkey anti-rat Alexa Fluor 488 [Molecular Probes/Invitrogen]; IdU, goat anti-mouse Cy3 [Jackson Immunoresearch]) for 1 h. Finally, slides were washed with PBS-Tween 20, mounted with Vectashield antifade mounting media (Vector Laboratories, Inc., Burlingame, CA), and stored at 4°C. Images were visualized by using a Nikon Eclipse TE-300 confocal microscope.
DNA fiber assay for DNA replication studies.
Approximately 5 × 105 cells were plated in each well of a six-well plate. Cells were pulse-labeled with 100 μM IdU for 45 min, washed with prewarmed (37°C) PBS, and pulsed with 100 μM CldU for 45 min. The medium was prewarmed for both pulses. To investigate the effect of CPT on initiation, 2.5 μM CPT was added to the medium during the last 30 min of the IdU pulse. To study fork progression, 2.5 μM CPT was added during the CldU pulse. The checkpoint kinase inhibitors UCN-01 or CHIRON 124 were added during both pulses at concentrations of 300 and 100 nM, respectively. At the end of the CldU pulse, cells were harvested and resuspended in 50 μl of PBS. Cell suspensions (2.5 μl) were mixed with 7.5 μl of lysis buffer (0.5% sodium dodecyl sulfate, 200 mM Tris-HCl [pH 7.4], 50 mM EDTA). Each mixture was dropped on the top of an uncoated regular glass slide. Slides were inclined at 45° to spread the suspension on the glass. Once dried, DNA spreads were fixed by incubation for 5 min in a 3:1 solution of methanol-acetic acid. The slides were dried and placed in prechilled 70% ethanol at 4°C for at least 1 h or overnight. Slides were then incubated in methanol and washed in PBS. DNA was denatured with 2.5 N HCl for 30 min at 37°C. The slides were rinsed several times in PBS and incubated with the following antibodies: mouse anti-BrdU fluorescein isothiocyanate (Becton Dickinson) and rat anti-CldU (Accurate Chemical and Science Co., Westbury, NY) diluted in 1% BSA. After incubation in a humid chamber for 1 h at 37°C, slides were washed three times, each time for 3 min in PBS containing 0.1% Triton X-100. The slides were incubated with secondary fluorescent antibodies (Alexa anti-mouse 488 [Molecular Probes/Invitrogen] and Alexa anti-rat 594 [Molecular Probes/Invitrogen] diluted in 1% BSA) for 1 h at 37°C. Slides were washed three times for 3 min in PBS-0.1% Triton X-100 and mounted by using Vectashield. Pictures were acquired with the Pathway microscope and Attovision software (Becton Dickinson). Signals were measured by using ImageJ software (NCI/NIH), with some modifications made specifically to measure DNA fibers.
Protein/nucleotide staining.
After incubation with 100 μM IdU for 45 min, with or without CPT for 30 min, HT29 cells were fixed at the indicated times after removal of IdU with 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 10 min. The cells were washed and incubated with methanol for 15 min at −20°C. Fixed cells were stored in 70% ethanol at 4°C for up to a week.
At the time of antibody staining, ethanol was removed, and cells were washed twice with PBS and incubated for 1 h with 8% BSA in PBS to block nonspecific binding. After a 5-min PBS wash, the cells were incubated for 2 h with rabbit anti-γ-H2AX antibody (kindly provided by William Bonner, National Cancer Institute) diluted in 1% BSA in PBS. Slides were washed twice with PBS and then incubated with anti-rabbit antibody conjugated with Alexa Fluor 488 (Molecular Probes, Eugene, OR) for 1 h. After a PBS wash, the cells were again fixed with 4% paraformaldehyde for 5 min, followed by a 10-min incubation with 1.5 M HCl at 37°C to denature the DNA. Cells were washed again, incubated with 0.5% Tween 20 in PBS for 5 min, and incubated with NGS for 20 min. IdU primary antibody (mouse anti-BrdU [BD Biosciences]) was diluted in blocking buffer and incubated for 2 h in a humid environment. Cells were washed and incubated with anti-mouse conjugated with Cy3 (Jackson Immunoresearch) for 1 h, washed, and mounted by using Vectashield mounting medium. Images were visualized by using a Nikon Eclipse TE-300 confocal microscope.
Cells analyzed for γ-H2AX without nucleotide were fixed with 4% paraformaldehyde and stored in 70% ethanol at 4°C. Antibody staining was done according to the protocol outlined above until the secondary antibody; after which cells were washed and incubated with 0.5 mg of RNase/ml and 50 μg of propidium iodide/ml for 30 min. Preparations were mounted and imaged as described above. The γ-H2AX fluorescence intensity was measured as the average pixel intensity (Adobe Photoshop 7.0) of 25 cells from each sample.
RESULTS
Short treatment with CPT induces late-S-phase delay with persistent inhibition of DNA synthesis.
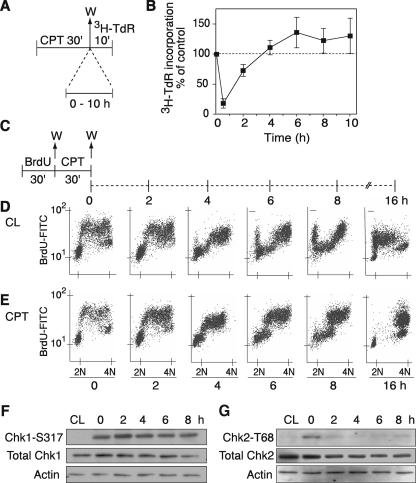
Analysis of [3H]TdR incorporation in human colorectal carcinoma HT29 cells revealed a marked inhibition of DNA synthesis (80%) within 30 min of CPT treatment (Fig. 2B). Overall, [3H]TdR incorporation appeared to recover within a few hours after the removal of CPT (Fig. 2B). However, it is important to note that treatments were carried out in an asynchronous population of cells. Over the time course, therefore, the apparent normalization of DNA replication as measured by [3H]TdR incorporation could have resulted from continued entry into S phase of cells that had been outside of S phase at the time of CPT treatment.
FIG. 2.
Inhibition of DNA synthesis and preferential delay in cells in mid- and late S phase for several hours after removal of CPT. (A) Experimental protocol for global DNA synthesis measurement. HT29 cells were treated with 1 μM CPT for 30 min. CPT was washed out (W), and cells were grown in drug-free medium for the indicated times. [3H]TdR was incorporated into the DNA for 10 min at the end of each time point and measured by trichloroacetic acid precipitation. (B) [3H]TdR incorporation at the indicated times after CPT removal. Shown are averages ± SEM of three experiments. (C) Experimental protocol for measuring S-phase progression. HT29 cells were pulse-labeled with 50 μM BrdU for 30 min, washed (W), and then treated with 1 μM CPT for 30 min. CPT was removed, and the S-phase population was monitored immediately after treatment (0 h) and at the indicated times after removal of CPT (2, 4, 6, 8, and 16 h). (D) FACS profiles for untreated cells (control: CL). (E) FACS profiles for CPT-treated cells. Panels D and E show the results of one of three similar experiments. (F) Phosphorylation of Chk1 on serine-317 immediately after CPT treatment (0 h) and at the indicated times after removal of the drug (2 to 8 h). (G) Similar samples were run for phosphorylation of Chk2 on threonine-68.
To determine the effect of CPT on the recovery of DNA replication, we focused specifically on the S-phase population of CPT-treated cells. We used pulse-labeling with BrdU to selectively label cells in S phase at the time of CPT treatment. In this way, we were able to follow the recovery of DNA replication in the treated S-phase cells over time. For this analysis, BrdU was incorporated into DNA for 30 min (Fig. 2C); cells were washed and then treated with CPT for 30 min (Fig. 2C, 0 h). CPT was then removed, and cells were grown in drug-free medium for 2 to 16 h. Fluorescence-activated cell sorting (FACS) profiles of BrdU incorporation versus DNA content revealed the progression of untreated cells through the cell cycle. In the untreated control cells (Fig. 2D), the S-phase population moved through S and reached G2/M 4 to 6 h after the initial pulse incorporation of BrdU. The labeled cells continued to proceed through G2/M and entered G1 6 to 8 h later. After 16 h, the labeled cells entered the next S phase.
Figure 2E shows that CPT produced a marked delay in progression through S phase for the BrdU-labeled cells. Cells progressed through S phase very slowly, remaining in mid- to late S phase at 6 to 8 h post-CPT. At 16 h post-CPT, the cells had progressed to G2 without advancing to the next cell cycle as the untreated cells did (Fig. 2, compare panels D and E). These results indicate that CPT produces a delay in S-phase progression, followed by an accumulation of cells in G2 phase.
Induction of the S- and G2/M-phase checkpoints during this experiment was determined by analyzing the ATR-dependent phosphorylation of Chk1 on Ser-317. Figure 2F shows phosphorylation of Chk1 immediately after CPT treatment, a finding consistent with those of previous studies (59). This phosphorylation was sustained up to 8 h after the removal of the drug. We also examined Chk2 activation under similar conditions. Figure 2G shows that Chk2 is also phosphorylated (on Thr-68) immediately after CPT treatment but, in contrast to Chk1-S317, the phosphorylation of Chk2-T68 is a transient event and is not maintained after the removal of the drug. These experiments demonstrate that delayed S-phase progression after CPT treatment is coincident with Chk1 activation.
DNA synthesis inhibition is more intense in mid-/late-S-phase cells than in early-S-phase cells.
S-phase progression appeared to be inhibited more in the latter half of the S phase according to BrdU pulse-labeling experiments (Fig. 2E). This suggested that the cells treated with CPT in early S phase progressed to mid- to late S phase (see the 4- and 6-h time points), where the cells remained delayed for at least 8 h.
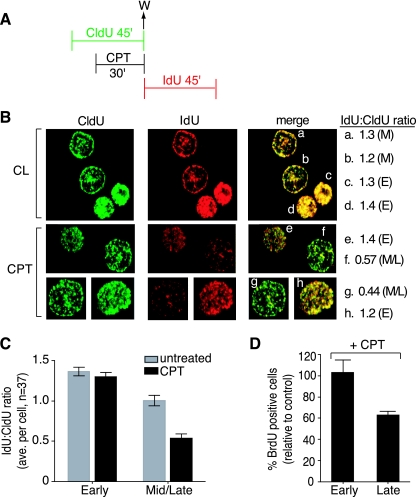
To investigate the possibility of a differential inhibition of DNA synthesis between mid- to late-S-phase cells and early-S-phase cells, the halogenated nucleotides CldU and IdU were incorporated into the DNA according to the protocol shown in Fig. 3A. CPT was added 15 min after the addition of CldU. After a further 30 min, CPT and CldU were washed out, and IdU was incorporated into the DNA for the next 45 min. The cells were then fixed and examined by fluorescence microscopy with antibodies to CldU (green) and IdU (red) (Fig. 3B).
FIG. 3.
Preferential inhibition of DNA replication in mid- to late-S-phase cells. (A) Experimental protocol. W, wash. (B) Confocal microscopy images of representative cells in early (E) or mid- to late (M) S phase. Replication foci were labeled with CldU (green) and IdU (red). The ratio of IdU (mean pixel intensity) to CldU (mean pixel intensity) is shown for each representative cell (a to h) (M, mid-S phase; E, early S phase). (C) IdU/CldU ratio in untreated and CPT-treated cells (37 cells were counted in each group). (D) Percentage of BrdU-positive cells after CPT treatment; early- and late-S-phase populations are expressed as a percentage of the corresponding untreated populations (average ± SEM of three independent experiments).
Representative cells are depicted in Fig. 3B, revealing the different patterns associated with DNA synthesis in different phases of S phase. Early-S-phase cells have a pattern of replication foci distributed throughout the nucleus (Fig. 3B, cells c, d, e, and h). Mid-S-phase cells are characterized by the distribution of replication foci around the periphery of the nucleus and fewer foci within the nucleus itself (Fig. 3B, cells a, b, f, and g). Late-S-phase cells would have a small number of large foci within the nucleus. It should be noted that in the HT29 cells used here there is only a very small population of cells with a late-S-phase pattern at any given time, and these cells (patterns) can be more difficult to distinguish. These late-S-phase cells were scored with the mid-S-phase cells.
CPT induced a decrease in IdU incorporation selectively in the mid- to late-S-phase cells (Fig. 3B, CPT IdU, cells f and g) compared to the early-S-phase cells (cells g and h). This difference was especially evident in the merged images, where early-S-phase cells (Fig. 3B, cells e and h) maintained comparable intensity levels for both nucleotides. Mid- to late-S-phase cells treated with CPT (Fig. 3B, cells f and g), however, had much less pronounced IdU staining, indicating a loss of incorporation of IdU into replication foci after CPT treatment. The ratio of IdU to CldU was measured as the ratio of the mean pixel intensity of IdU versus CldU for each cell (numbers on the right in Fig. 3B). Untreated cells gave a ratio of approximately 1:1, regardless of whether the cells were in early or mid- to late S phase. CPT-treated early-S-phase cells maintained this same ratio (Fig. 3B, cells e and h), whereas the mid- to late-S-phase cells had a reduced ratio of half IdU to CldU (Fig. 3B, cells f and g), indicating that mid- to late-S-phase cells were more efficient at inhibiting DNA replication in response to CPT treatment. This analysis was extended to a larger number of cells (n = 37 for each parameter), and again cells in mid- to late S phase had a greater decrease in incorporation after CPT treatment (Fig. 3C).
In addition, flow cytometry analyses of BrdU incorporation were performed. Cells were treated with CPT for 30 min, followed by BrdU incorporation for 30 min. The percentage of early- or late-S-phase BrdU-positive cells after CPT treatment, compared to untreated cells, was calculated and is shown in Fig. 3D. Again, a preferential decrease in DNA replication in late-S-phase cells was observed. Therefore, CPT inhibits DNA synthesis preferentially in late-replicating DNA.
Histone γ-H2AX colocalizes with DNA replication factories (foci) in CPT-treated cells.
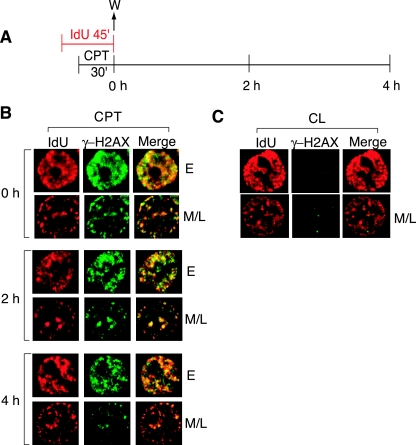
We recently reported that CPT induces the formation of histone γ-H2AX foci selectively in replicating cells (16, 17) and proposed that the γ-H2AX foci correspond to replication-mediated DNA DSBs (16, 17, 59). To determine whether the CPT-induced γ-H2AX foci do correspond to sites of DNA replication, cells were first pulse-labeled with IdU for 45 min, with the last 30 min in the presence of CPT (Fig. 4A). Cells were either fixed immediately (0 h) or 2 and 4 h after IdU and CPT washout (Fig. 4A). The γ-H2AX foci present in the CPT-treated cells coincided with the replication foci marked by IdU incorporation in both early and mid- to late-S-phase cells (Fig. 4B, 0 h). The colocalization of γ-H2AX and IdU foci indicates the presence of DSBs induced by the CPT treatment at sites of ongoing replication. Interestingly, those γ-H2AX foci persisted with the IdU foci for up to 4 h after removal of CPT (Fig. 4B, 2 and 4 h), a finding consistent with the slow repair of the replication-mediated DNA damage after CPT removal (45).
FIG. 4.
γ-H2AX foci coincide with DNA replication foci. (A) Experimental protocol. Cells were fixed at 0, 2, and 4 h after IdU and CPT pulse treatments. (B) Confocal microscopy images of a representative early (E) or mid- to late (M/L) S-phase cell at the indicated times after CPT removal. Cells were stained with anti-IdU (red) and anti-γ-H2AX (green) antibodies. (C) Confocal microscopy images of untreated cells immediately after the IdU pulse-label (corresponding to 0 h in panel A). The images in panels B and C were adjusted for maximum clarity, not to compare intensity. One representative of three similar experiments is shown.
CPT-induced inhibition of DNA replication is due to an intra-S-phase checkpoint.
To determine whether the maintained inhibition of DNA synthesis after the removal of CPT was due to an intra-S-phase checkpoint, we analyzed DNA replication in the presence of the checkpoint inhibitor 7-hydroxystaurosporine (UCN-01) (28, 48, 52, 65) or the specific Chk1 inhibitor CHIR-124 (35, 61). Figure 5A depicts the experimental protocol used. Briefly, cells were pulse-labeled with CldU for 45 min, with CPT added for the last 30 min. CldU and CPT were then washed, and cells were grown in drug-free medium. UCN-01 or CHIR-124 was added after an initial 2-h incubation in drug-free medium to allow time for the establishment of the checkpoint. IdU was then added for 45 min at various times after the removal of CPT, in the absence or presence of UCN-01 or CHIR-124.
FIG. 5.
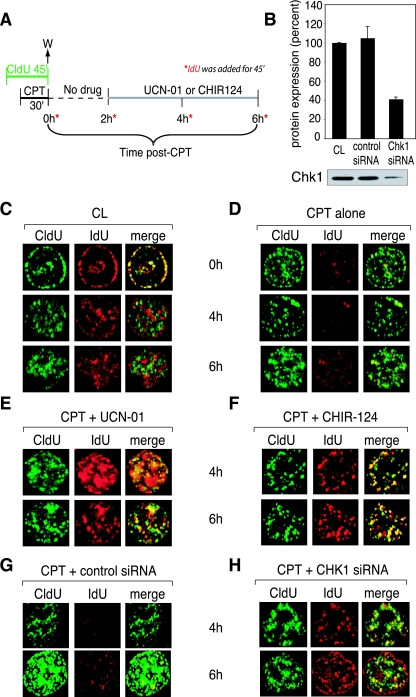
CPT treatment inhibits ongoing replication in preexisting replication foci and blocks the formation of new replication foci by activating an intra-S-phase checkpoint, which is abrogated by UCN-01, CHIR-124, and Chk1 siRNA. (A) Experimental protocol. Drug concentrations: CPT, 1 μM; UCN-01, 0.3 μM; CHIR-124, 0.1 μM. (B) Immunoblot analysis of Chk1 protein expression in nontransfected cells (CL), cells transfected with control siRNA, and cells transfected with Chk1 siRNA. The graph depicts the quantitation of two experiments, with a typical Western blot shown below. (C to H) Confocal microscopy images of representative cells. (C) Control, untreated cells; (D) cells treated with CPT and then grown in drug-free medium for up to 6 h. (E and F) Cells treated with CPT followed by addition of UCN-01 (E) or CHIR-124 (F) 2 h after CPT removal (see protocol in panel A). (G) Cells transfected with nonspecific siRNA. (H) Cells transfected with siRNA against Chk1. Similar results were observed in three independent experiments.
Figure 5C shows representative images for untreated (CL) cells. When IdU was added immediately after CldU, both were colocalized, due to incorporation into the same or adjacent replication foci (Fig. 5C, 0 h CL). As the time period between the pulses with the two nucleotides increased, the foci no longer colocalized, and the pattern of IdU foci became one of cells that had progressed later into S phase (Fig. 5C, CL, 4 and 6 h). Figure 5D represents cells after CPT treatment. Immediately after CPT removal (0 h), incorporation of IdU was decreased in the foci that were present during the CPT treatment, indicating inhibition of DNA replication in those foci. This decrease persisted for several hours after CPT removal (4 to 6 h), which is consistent with the experiment shown in Fig. 2E, where S-phase progression was delayed during the same time period. Moreover, as the time interval between the two nucleotide pulses increased, no new IdU foci were established, indicating an inhibition of DNA replication initiation for several hours after CPT removal.
To determine whether the CPT-induced inhibition of replication was due to checkpoint kinases, UCN-01 or CHIR-124 was added after CPT (see protocol in Fig. 5A). Figure 5E and F show representative images from cells treated with CPT, followed by UCN-01 and CHIR-124 treatment, respectively. To further demonstrate the importance of Chk1, experiments were performed in Chk1-downregulated cells. Figure 5G and H show representative images from cells transfected with a control siRNA (Fig. 5G) or Chk1-targeted siRNA (Fig. 5H). A 60% average decrease in Chk1 protein expression was obtained (Fig. 5B). CPT-treated cells transfected with control siRNA maintained inhibition of IdU similar to that of cells treated with CPT alone (compare Fig. 5G and D, respectively). Treatment with either checkpoint inhibitor or the Chk1 siRNA resulted in the restoration of IdU incorporation at 4 and 6 h post-CPT. New IdU foci were also established in all three cases (Fig. 5E, F, and H). The ability of UCN-01, CHIR-124, and Chk1 siRNA to restore DNA synthesis in preexisting replication foci and to restore the initiation of new replication foci implicates the presence of a CPT-induced, Chk1-dependent checkpoint inhibiting both DNA replication elongation and initiation.
Both origin firing and fork progression are negatively regulated by the intra-S-phase checkpoint.
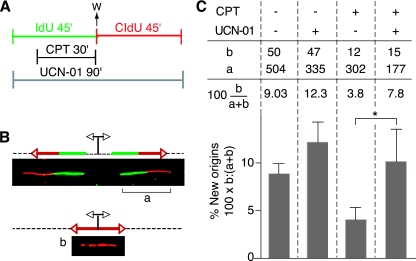
To further examine the checkpoint control on origin activation, we analyzed DNA fiber spreads (23, 31) prepared from CPT-treated cells. To visualize replicons, cells were sequentially pulse-labeled with IdU and CldU for 45 min each, according to the protocol illustrated in Fig. 6A. CPT was added to the cell cultures during the IdU pulse and washed out before adding the CldU pulse. IdU and CldU were detected with specific antibodies, in green and red, respectively. Origins of replication that were activated prior to the IdU pulse generated two bidirectional forks, each appearing as a green or red signal (Fig. 6B, signal a). Conversely, new origins that fired during the CldU pulse and after the CPT treatment resulted in a red signal only (Fig. 6B, signal b). We quantified the frequency of new origins in untreated and CPT-treated cells by dividing the number of red signals (b) by the sum of the red and green/red signals (a + b) (Fig. 6C). The percentage of new origins was 9% in untreated cells. This number dropped to 3.8% when the cells were treated with CPT. To confirm the checkpoint control of this phenomenon (see Fig. 5 and results described above), we treated the cells with UCN-01. The presence of UCN-01 restored the percentage of new origins to 7.8%. It is interesting that treatment of the cells with UCN-01 alone, in the absence of DNA damage, also induced a slight increase in the origin firing compared to that of untreated cells. This is in agreement with the monitoring of origin usage by the checkpoint proteins ATM/ATR previously shown in Xenopus (30, 50, 51) and is consistent with results in mammalian cells demonstrating aberrant firing of late origins after UCN-01 treatment alone (32).
FIG. 6.
Checkpoint control on the initiation of DNA replication after CPT treatment. (A) Experimental protocol. IdU and CldU (100 μM) were added sequentially to cell cultures for 45 min each. IdU was detected with specific antibodies in green, with CldU in red. CPT (2.5 μM) was added for 30 min during the IdU pulse. When indicated, UCN-01 (0.3 μM) was present during both pulses. (B) Schematic drawing and representative images of two replication signals from DNA fibers. At the top, two DNA replication forks moved bidirectionally from an origin (indicated by the diverging black arrows) that was activated before the IdU pulse. Each fork was labeled with both IdU (green) and CldU (red). At the bottom, the replication bubble resulting from an origin that has been activated during the CldU pulse produces a red-only signal. This population of new later origins had been activated after the CPT treatment. (C) Frequencies of new origins activated during the CldU pulse in untreated conditions, with CPT, with UCN-01, and with both CPT and UCN-01. The frequency (as a percentage) was calculated as the number of red signals (b in panel B) divided by the total (a + b) of red (b) plus green/red signals (a in panel B). The table above the histogram shows the actual values and the percentage of new origins for each treatment. Signals were compiled from two independent experiments. The asterisk indicates a significant effect of UCN-01 upon CPT-induced inhibition of replication origin firing (P = 0.01).
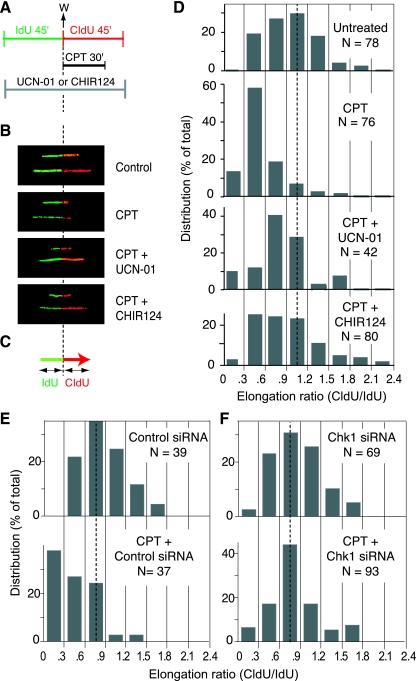
The analysis of individual DNA fibers also allowed us to investigate the presence of a checkpoint control of replication fork progression. Cells were sequentially pulse-labeled by IdU and CldU for 45 min each. CPT was added during the second (CldU) pulse (Fig. 7A). In untreated cells, the elongation of replicons results in adjacent green and red signals of nearly the same length (Fig. 7B, upper panel). After treatment with CPT, the CldU (red) signal was shorter than the IdU (green) signal (Fig. 7B, CPT). The shortening of the red track shows the inhibition of replication fork elongation by CPT. The results were quantified by measuring the lengths of the adjacent red and green signals (Fig. 7C). In untreated conditions the CldU/IdU ratio was ∼1 (Fig. 7D, upper panel). After CPT treatment, the CldU/IdU ratio dropped to ∼0.5 (Fig. 7D). To investigate the putative role of the checkpoint on the fork arrest by CPT, we treated the cells with either UCN-01 or CHIR-124 during both the IdU and CldU pulse (Fig. 7A). Under these conditions, the length of the red track increased (Fig. 7B). The ratio of the red and green signals shifted back closer to 1 (Fig. 7D), indicating a role for Chk1 in inhibiting replication fork progression.
FIG. 7.
Checkpoint control of the elongation of DNA replication after CPT. (A) Experimental protocol. IdU and CldU (100 μM) were added sequentially to the cell cultures for 45 min each. IdU was detected with specific antibodies, in green, with CldU in red. CPT (2.5 μM) was added during the IdU pulse. The Chk1 inhibitors UCN-01 (0.3 μM) or CHIR-124 (0.1 μM) were added during both pulses. (B) Representative replication signals observed on DNA fibers obtained by using the protocol depicted in panel A. In the untreated condition, the length of the green and red tracks should be about equal since the forks progress unperturbed during the two pulses. The red signal is shorter after CPT because of the replication block induced by the drug. The Chk1 inhibitors UCN-01 and CHIR-124 tended to restore the normal length of the red signal. (C) Schematic representation of the measurement of CldU and IdU signals. The CldU/IdU ratio was used to determine elongation. (D) Histograms showing the distribution of elongation ratios calculated in the indicated conditions. In untreated cells, the median ratio is close to 1, as expected. The peak was left-shifted after CPT treatment. The peak tended to be shifted back toward 1 when cells were coincubated with UCN-01 or CHIR-124. The dotted line shows a ratio of 1. Comparison of untreated versus CPT-treated cells, CPT versus CPT+UCN-01-treated cells, and CPT versus CPT+CHIR124-treated cells showed a significant difference (P < 0.001 [Kolmogorov-Smirnov test]). (E) Histograms showing CPT-induced inhibition of DNA replication elongation in cells transfected with a control siRNA. (F) Same analyses as in panel E but in cells transfected with siRNA targeting Chk1. Comparison of control siRNA+CPT versus Chk1 siRNA+CPT showed a significant difference (P < 0.001 [Kolmogorov-Smirnov test]).
Further experiments were performed in cells transfected with a control siRNA (Fig. 7E) or with a Chk1-targeted siRNA (Fig. 7F). CPT-treated cells transfected with control siRNA exhibited a reduced CldU/IdU ratio compared to that of untreated cells (Fig. 7E). CPT-treated cells transfected with Chk1 siRNA showed an attenuation of the CPT-induced replication fork arrest, with a CldU/IdU ratio similar to that of the untreated cells (Fig. 7F), a finding consistent with the results using the drugs shown in Fig. 7D. Together, these experiments support the conclusion of a Chk1-dependent checkpoint inhibiting DNA replication elongation.
Loss of the intra-S-phase checkpoint increases DNA damage.
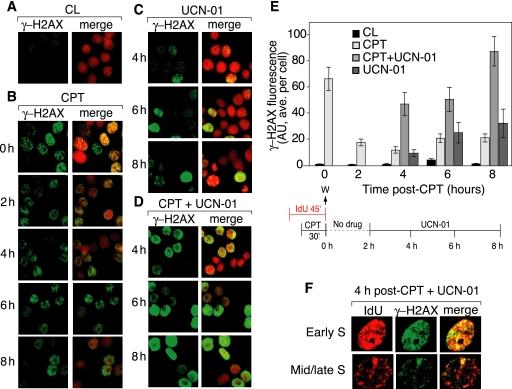
The appearance of a large number of IdU foci colocalizing with preexisting CldU foci in cells treated with UCN-01 (or CHIR-124) for up to 6 h after CPT removal (Fig. 5D and E) prompted us to look for DSBs in those “reactivated” foci by using γ-H2AX immunofluorescence (Fig. 8). In the absence of UCN-01, γ-H2AX foci decreased in intensity after the removal of CPT (Fig. 8B, 2 to 8 h) (17), whereas in cells treated with UCN-01 after CPT removal, γ-H2AX foci increased in intensity (Fig. 8D, 4 to 8 h). Noticeably, a large fraction of the cells showed a diffuse γ-H2AX staining (pan-staining) in the presence of UCN-01. Figure 8E summarizes the average γ-H2AX fluorescence intensities and shows the time-dependent increase in γ-H2AX in the cells treated with UCN-01 after CPT. Figure 8F shows representative cells examined 4 h after CPT treatment in the presence of UCN-01. The UCN-01-induced γ-H2AX foci colocalized with sites of DNA replication in cells both in early and in mid-S phase. These experiments suggest that UCN-01, while restoring DNA replication, induces DNA damage within replication foci.
FIG. 8.
Loss of the intra-S-phase checkpoint by UCN-01 increases DNA damage measured as γ-H2AX foci. (A to D) Representative images of γ-H2AX foci (green) alone (left panels) and with propidium iodide staining (red; right panels). (A) Control (CL) untreated HT29 cells; (B) γ-H2AX immunofluorescence after CPT removal (protocol shown at the bottom of panel E); (C) γ-H2AX immunofluorescence in cells treated with UCN-01 alone; (D) γ-H2AX immunofluorescence in cells treated with UCN-01 after CPT removal. (E) γ-H2AX fluorescence (average per cell) for the experiment depicted in panels A to D (mean ± SEM). AU, arbitrary units. (F) Colocalization of γ-H2AX foci (green) and IdU (red) in representative early- and mid- to late-S-phase cells. The images represent one of three similar experiments.
DISCUSSION
Elucidation of the intra-S-phase checkpoint and elaboration of new techniques to explore this checkpoint are important for cancer therapeutics, as well as for understanding carcinogenesis, since a large number of anticancer agents target DNA replication (41) and many tumors are defective in cell cycle checkpoints (24). As outlined in the introduction, Top1cc are among the best-characterized cellular lesions that generate replication-mediated DNA DSBs (for a review, see references 39 and 40). Moreover, Top1cc are not only relevant for the anticancer activity of CPTs and non-CPT Top1 inhibitors, but are also relevant for a large number of other cancer-chemotherapeutic DNA-targeted agents, carcinogens, and endogenous DNA lesions (40). CPT has the unique advantage of inducing Top1cc within minutes of addition to cell cultures and of being readily removed from cells by incubating cell cultures in drug-free medium. In which case, more than 90% of the Top1cc reverses within 15 to 30 min (12). Thus, CPT can be used as a sharp molecular tool to trigger replication-mediated DNA damage (58, 59, 66).
The ability of cells to resume both DNA replication and cell cycle progression after a short treatment with CPT has previously been examined using asynchronous cell cultures (18, 49). These experiments allowed for the possibility that cells outside of S phase at the time of drug treatment could enter S phase and replicate normally. Under such conditions it is difficult to distinguish between the recovery of inhibited DNA replication and normal DNA replication of new S-phase cells by [3H]TdR incorporation, as depicted in Fig. 2A. To avoid the complication of additional drug effects that may be introduced by synchronization agents, we used BrdU to prelabel the S-phase population of cells in order to analyze this population over time. In doing so, we determined that the S-phase population affected by CPT is in fact delayed in its progression through S phase (specifically late S phase) for up to 8 h after the removal of the drug and that these cells are not able to progress to G1 even 16 h after the removal of CPT (see Fig. 2D). Moreover, the CldU/IdU sequential pulse-labeling experiments with various time intervals between the CldU and IdU pulses (see protocol in Fig. 5A) showed that cells that were not labeled with CldU during the CPT treatment still incorporated IdU during the second IdU pulse (data not shown), indicating that these cells were not in S phase at the time of drug treatment, since they lacked CldU foci. These experiments suggest that the checkpoint induced by CPT is specific to S-phase cells. We conclude that cells outside of S phase at the time of drug treatment are able to enter S phase and replicate their DNA normally, contributing to the DNA replication levels measured as “recovery” after CPT removal.
The CldU/IdU double-labeling approach on interphase nuclei (see Fig. 5) confirmed the ability of CPT to inhibit DNA replication (see Fig. 2). Moreover, these analyses demonstrated that new initiation events were blocked for several hours after the removal of CPT, as indicated by the complete loss of new replication foci incorporating only IdU (Fig. 5C). The inhibition of elongation is suggested by the decrease in IdU intensity in preexisting replication foci. This conclusion is somewhat ambiguous, however, since one focus can contain several origins of replication that may fire at different times (3, 23, 29). The situation may occur where the initiation of some origins within a focus is inhibited, while elongation from adjacent origins within the focus is undeterred. This would result in the net effect of decreasing the intensity and/or incorporation of IdU, giving the appearance of elongation inhibition within individual foci. To address this question, we utilized the DNA fiber assay, which can measure initiation and elongation on a per-molecule basis (1, 9, 11). These experiments demonstrated that CPT induced not only an inhibition of DNA replication initiation (Fig. 6), but also an inhibition of elongation after CPT removal (Fig. 7).
Addition of UCN-01, a protein kinase inhibitor that inhibits Chk1 (6, 19); CHIR-124, a specific Chk1 kinase inhibitor (35, 61); or siRNA targeting Chk1 abrogated the inhibition of DNA synthesis after CPT treatment. Both initiation and elongation were restored in each case (Fig. 5 to 7), providing clear evidence for a role of the intra-S-phase checkpoint in controlling replication fork progression (Fig. 1E). The results of our experiments are in agreement with those of genetic experiments showing the involvement of Hus1 and PCNA function in a checkpoint regulating elongation (measured by velocity sedimentation analysis) after CPT and ionizing radiation treatment (66).
We also show here for the first time colocalization of γ-H2AX with IdU after CPT treatment. A previous study described the colocalization of γ-H2AX and BrdU after cisplatin (CDDP) treatment in cells deficient in retinoblastoma protein (5), but this had not yet been demonstrated in cells treated with CPT. The colocalization of γ-H2AX with replication foci further supports a collision mechanism for the formation of DSBs in S phase (Fig. 1D) (2, 20, 22, 45, 58). Our experiments also revealed the persistence of γ-H2AX foci at replication foci for several hours after the removal of CPT, suggesting either a difference in DSB repair kinetics in these cells or the presence of irreparable DNA damage.
In cells that had been treated with CPT, IdU incorporation colocalized with CldU foci for several hours, in contrast to untreated cells. This may represent re-replication events (16) and/or an attempt to replicate damaged DNA. We observed an increased amount of DSBs after treatment with UCN-01, which colocalized with replication foci (Fig. 8F). The immediate resumption of DNA replication in UCN-01-treated cells likely increases the number of collisions between replication forks and cleavage complexes. This, along with aberrant origin firing and the collapse of unstable replication forks in UCN-01 treated cells, would contribute to the observed increase in DNA damage. This significant increase in DNA damage corresponds with published data indicating the synergistic cytotoxicity of CPT and UCN-01 in cancer cells (33, 48, 64) compared to the relatively mild toxicity of brief CPT treatment alone (20). This is an indication that ongoing DNA replication progression is necessary for maximizing the cytotoxic effects of CPT.
We have taken advantage of the CldU and IdU thymidine analogs to determine the effects of CPT on DNA replication. CldU and IdU have been used in previous studies to measure DNA synthesis during and after replication block (15, 70). The CldU/IdU pulse-labeling experiments in the present study, however, are the first to analyze an agent other than a direct replication inhibitor such as APH. APH blocks replication fork progression by interfering directly with DNA polymerase activity. In contrast, CPT generates Top1-linked DNA single-strand breaks that are converted to DSBs when the forks encounter the Top1cc. Therefore, a new question can be addressed by using these techniques with CPT—namely, whether there is an active checkpoint control on fork progression. The number of CPT-Top1cc interfering with fork progression directly would be small at the dose and time used, meaning that any inhibition of elongation measured is due to a checkpoint response. The use of checkpoint-inhibiting agents in this assay confirmed these results, since elongation inhibition was no longer observed with these drugs (Fig. 7).
The protocol used here has proven informative in determining inhibition of DNA replication at both the levels of initiation and elongation and can also be used to monitor the replication response at different times of S phase. CldU/IdU pulse-labeling experiments and BrdU incorporation experiments revealed selectivity of DNA replication inhibition in late-S-phase cells compared to cells in early S phase. The heterochromatic nature of late-replicating DNA (23, 37) may increase its susceptibility to inhibition, since it is already in a compact and inactive conformation. Chromatin in early S phase is more open and perhaps may require more proteins and/or time to efficiently inhibit DNA replication. It may also be more critical to inhibit synthesis immediately in late-S-phase cells due to their temporal proximity to mitosis. Early-S-phase cells would have more time to repair DNA damage before entering mitosis, and a delayed inhibition of synthesis may then be tolerated. Analysis of replication timing and chromatin structure as related to the intra-S-phase checkpoint is an as-yet-unexplored topic that could provide more insight into how these processes are regulated.
Acknowledgments
We thank Kenneth W. Bair, Chiron Corp., for providing CHIR-124 and unpublished information regarding the specific inhibition of Chk1 by CHIR-124. We also thank Olivier Sordet, Andrew Jobson, and Ashutosh Rao for technical assistance and critical reading of the manuscript.
Our research is supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 21 May 2007.
REFERENCES
- 1.Anglana, M., F. Apiou, A. Bensimon, and M. Debatisse. 2003. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 114:385-394. [DOI] [PubMed] [Google Scholar]
- 2.Avemann, K., R. Knippers, T. Koller, and J. M. Sogo. 1988. Camptothecin, a specific inhibitor of type I DNA topoisomerase, induces DNA breakage at replication forks. Mol. Cell. Biol. 8:3026-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berezney, R., D. D. Dubey, and J. A. Huberman. 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108:471-484. [DOI] [PubMed] [Google Scholar]
- 4.Bjornsti, M. A., P. Benedetti, G. A. Viglianti, and J. C. Wang. 1989. Expression of human DNA topoisomerase I in yeast cells lacking yeast DNA topoisomerase I: restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 49:6318-6323. [PubMed] [Google Scholar]
- 5.Bosco, E. E., C. N. Mayhew, R. F. Hennigan, J. Sage, T. Jacks, and E. S. Knudsen. 2004. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res. 32:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busby, E. C., D. F. Leistritz, R. T. Abraham, L. M. Karnitz, and J. N. Sarkaria. 2000. The radiosensitizing agent 7-hydroxystaurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 60:2108-2112. [PubMed] [Google Scholar]
- 7.Champoux, J. J. 2001. DNA topoisomerase I-mediated nicking of circular duplex DNA. Methods Mol. Biol. 95:81-87. [DOI] [PubMed] [Google Scholar]
- 8.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 9.Chastain, P. D., II, T. P. Heffernan, K. R. Nevis, L. Lin, W. K. Kaufmann, D. G. Kaufman, and M. Cordeiro-Stone. 2006. Checkpoint regulation of replication dynamics in UV-irradiated human cells. Cell Cycle 5:2160-2167. [DOI] [PubMed] [Google Scholar]
- 10.Cliby, W. A., K. A. Lewis, K. K. Lilly, and S. H. Kaufmann. 2002. S phase and G2 arrests induced by topoisomerase I poisons are dependent on ATR kinase function. J. Biol. Chem. 277:1599-1606. [DOI] [PubMed] [Google Scholar]
- 11.Conti, C., S. Caburet, and A. Bensimon. 2001. Targeting the molecular mechanism of DNA replication. Drug Discov. Today 6:786-792. [DOI] [PubMed] [Google Scholar]
- 12.Covey, J. M., C. Jaxel, K. W. Kohn, and Y. Pommier. 1989. Protein-linked DNA strand breaks induced in mammalian cells by camptothecin, an inhibitor of topoisomerase I. Cancer Res. 49:5016-5022. [PubMed] [Google Scholar]
- 13.Dimitrova, D. S., and R. Berezney. 2002. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized, and transformed mammalian cells. J. Cell Sci. 115:4037-4051. [DOI] [PubMed] [Google Scholar]
- 14.Dimitrova, D. S., and D. M. Gilbert. 1999. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell 4:983-993. [DOI] [PubMed] [Google Scholar]
- 15.Feijoo, C., C. Hall-Jackson, R. Wu, D. Jenkins, J. Leitch, D. M. Gilbert, and C. Smythe. 2001. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J. Cell Biol. 154:913-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuta, T., R. L. Hayward, L. H. Meng, H. Takemura, G. J. Aune, W. M. Bonner, M. I. Aladjem, K. W. Kohn, and Y. Pommier. 2006. p21(CDKN1A) allows the repair of replication-mediated DNA double-strand breaks induced by topoisomerase I and is inactivated by the checkpoint kinase inhibitor 7-hydroxystaurosporine. Oncogene 25:2839-2849. [DOI] [PubMed] [Google Scholar]
- 17.Furuta, T., H. Takemura, Z. Y. Liao, G. J. Aune, C. Redon, O. A. Sedelnikova, D. R. Pilch, E. P. Rogakou, A. Celeste, H. T. Chen, A. Nussenzweig, M. I. Aladjem, W. M. Bonner, and Y. Pommier. 2003. Phosphorylation of histone H2AX and activation of Mre11, Rad50, and Nbs1 in response to replication-dependent DNA double-strand breaks induced by mammalian DNA topoisomerase I cleavage complexes. J. Biol. Chem. 278:20303-20312. [DOI] [PubMed] [Google Scholar]
- 18.Goldwasser, F., T. Shimizu, J. Jackman, Y. Hoki, P. M. O'Connor, K. W. Kohn, and Y. Pommier. 1996. Correlations between S and G2 arrest and the cytotoxicity of camptothecin in human colon carcinoma cells. Cancer Res. 56:4430-4437. [PubMed] [Google Scholar]
- 19.Graves, P. R., L. Yu, J. K. Schwarz, J. Gales, E. A. Sausville, P. M. O'Connor, and H. Piwnica-Worms. 2000. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J. Biol. Chem. 275:5600-5605. [DOI] [PubMed] [Google Scholar]
- 20.Holm, C., J. M. Covey, D. Kerrigan, and Y. Pommier. 1989. Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res. 49:6365-6368. [PubMed] [Google Scholar]
- 21.Hsiang, Y. H., R. Hertzberg, S. Hecht, and L. F. Liu. 1985. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260:14873-14878. [PubMed] [Google Scholar]
- 22.Hsiang, Y. H., M. G. Lihou, and L. F. Liu. 1989. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49:5077-5082. [PubMed] [Google Scholar]
- 23.Jackson, D. A., and A. Pombo. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140:1285-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kastan, M. B., and J. Bartek. 2004. Cell-cycle checkpoints and cancer. Nature 432:316-323. [DOI] [PubMed] [Google Scholar]
- 25.Kaufmann, W. K., J. C. Boyer, L. L. Estabrooks, and S. J. Wilson. 1991. Inhibition of replicon initiation in human cells following stabilization of topoisomerase-DNA cleavable complexes. Mol. Cell. Biol. 11:3711-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohn, E. A., N. D. Ruth, M. K. Brown, M. Livingstone, and A. Eastman. 2002. Abrogation of the S phase DNA damage checkpoint results in S phase progression or premature mitosis depending on the concentration of 7-hydroxystaurosporine and the kinetics of Cdc25C activation. J. Biol. Chem. 277:26553-26564. [DOI] [PubMed] [Google Scholar]
- 27.Larner, J. M., H. Lee, R. D. Little, P. A. Dijkwel, C. L. Schildkraut, and J. L. Hamlin. 1999. Radiation down-regulates replication origin activity throughout the S phase in mammalian cells. Nucleic Acids Res. 27:803-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, S. I., M. K. Brown, and A. Eastman. 1999. Comparison of the efficacy of 7-hydroxystaurosporine (UCN-01) and other staurosporine analogs to abrogate cisplatin-induced cell cycle arrest in human breast cancer cell lines. Biochem. Pharmacol. 58:1713-1721. [DOI] [PubMed] [Google Scholar]
- 29.Ma, H., J. Samarabandu, R. S. Devdhar, R. Acharya, P. C. Cheng, C. Meng, and R. Berezney. 1998. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J. Cell Biol. 143:1415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marheineke, K., and O. Hyrien. 2004. Control of replication origin density and firing time in Xenopus egg extracts: role of a caffeine-sensitive, ATR-dependent checkpoint. J. Biol. Chem. 279:28071-28081. [DOI] [PubMed] [Google Scholar]
- 31.Merrick, C. J., D. Jackson, and J. F. Diffley. 2004. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 279:20067-20075. [DOI] [PubMed] [Google Scholar]
- 32.Miao, H., J. A. Seiler, and W. C. Burhans. 2003. Regulation of cellular and SV40 virus origins of replication by Chk1-dependent intrinsic and UVC radiation-induced checkpoints. J. Biol. Chem. 278:4295-4304. [DOI] [PubMed] [Google Scholar]
- 33.Monks, A., E. D. Harris, A. Vaigro-Wolff, C. D. Hose, J. W. Connelly, and E. A. Sausville. 2000. UCN-01 enhances the in vitro toxicity of clinical agents in human tumor cell lines. Investig. New Drugs 18:95-107. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, H., T. Morita, and C. Sato. 1986. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp. Cell Res. 165:291-297. [DOI] [PubMed] [Google Scholar]
- 35.Ni, Z. J., P. Barsanti, N. Brammeier, A. Diebes, D. J. Poon, S. Ng, S. Pecchi, K. Pfister, P. A. Renhowe, S. Ramurthy, A. S. Wagman, D. E. Bussiere, V. Le, Y. Zhou, J. M. Jansen, S. Ma, and T. G. Gesner. 2006. 4-(Aminoalkylamino)-3-benzimidazole-quinolinones as potent CHK-1 inhibitors. Bioorg. Med. Chem. Lett. 16:3121-3124. [DOI] [PubMed] [Google Scholar]
- 36.Nitiss, J., and J. C. Wang. 1988. DNA topoisomerase-targeting antitumor drugs can be studied in yeast. Proc. Natl. Acad. Sci. USA 85:7501-7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Keefe, R. T., S. C. Henderson, and D. L. Spector. 1992. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J. Cell Biol. 116:1095-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Painter, R. B. 1981. Radioresistant DNA synthesis: an intrinsic feature of ataxia telangiectasia. Mutat. Res. 84:183-190. [DOI] [PubMed] [Google Scholar]
- 39.Pommier, Y. 2006. Topoisomerase I inhibitors, camptothecins, and beyond. Nat. Rev. Cancer 6:789-802. [DOI] [PubMed] [Google Scholar]
- 40.Pommier, Y., J. M. Barcelo, V. A. Rao, O. Sordet, A. G. Jobson, L. Thibaut, Z.-H. Miao, J. A. Seiler, H. Zhang, C. Marchand, K. Agama, J. L. Nitiss, and C. Redon. 2006. Repair of topisomerase 1-mediated DNA damage. Prog. Nucleic Acid Res. 81:179-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pommier, Y., and R. B. Diasio. 2006. Pharmacological agents that target DNA replication in DNA replication and human disease. Cold Spring Harbor Laboratory Press, New York, NY.
- 42.Pommier, Y., C. Redon, V. A. Rao, J. A. Seiler, O. Sordet, H. Takemura, S. Antony, L. Meng, Z. Liao, G. Kohlhagen, H. Zhang, and K. W. Kohn. 2003. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat. Res. 532:173-203. [DOI] [PubMed] [Google Scholar]
- 43.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 45.Ryan, A. J., S. Squires, H. L. Strutt, and R. T. Johnson. 1991. Camptothecin cytotoxicity in mammalian cells is associated with the induction of persistent double strand breaks in replicating DNA. Nucleic Acids Res. 19:3295-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santocanale, C., and J. F. Diffley. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395:615-618. [DOI] [PubMed] [Google Scholar]
- 47.Sedelnikova, O. A., E. P. Rogakou, I. G. Panyutin, and W. M. Bonner. 2002. Quantitative detection of 125IdU-induced DNA double-strand breaks with γ-H2AX antibody. Radiat. Res. 158:486-492. [DOI] [PubMed] [Google Scholar]
- 48.Shao, R. G., C. X. Cao, T. Shimizu, P. M. O'Connor, K. W. Kohn, and Y. Pommier. 1997. Abrogation of an S-phase checkpoint and potentiation of camptothecin cytotoxicity by 7-hydroxystaurosporine (UCN-01) in human cancer cell lines, possibly influenced by p53 function. Cancer Res. 57:4029-4035. [PubMed] [Google Scholar]
- 49.Shao, R. G., C. X. Cao, H. Zhang, K. W. Kohn, M. S. Wold, and Y. Pommier. 1999. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 18:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shechter, D., V. Costanzo, and J. Gautier. 2004. ATR and ATM regulate the timing of DNA replication origin firing. Nat. Cell Biol. 6:648-655. [DOI] [PubMed] [Google Scholar]
- 51.Shechter, D., and J. Gautier. 2005. ATM and ATR check in on origins: a dynamic model for origin selection and activation. Cell Cycle 4:235-238. [PubMed] [Google Scholar]
- 52.Shi, Z., A. Azuma, D. Sampath, Y. X. Li, P. Huang, and W. Plunkett. 2001. S-phase arrest by nucleoside analogues and abrogation of survival without cell cycle progression by 7-hydroxystaurosporine. Cancer Res. 61:1065-1072. [PubMed] [Google Scholar]
- 53.Shiloh, Y. 2003. ATM: ready, set, go. Cell Cycle 2:116-117. [DOI] [PubMed] [Google Scholar]
- 54.Shirahige, K., Y. Hori, K. Shiraishi, M. Yamashita, K. Takahashi, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395:618-621. [DOI] [PubMed] [Google Scholar]
- 55.Snapka, R. M. 1986. Topoisomerase inhibitors can selectively interfere with different stages of simian virus 40 DNA replication. Mol. Cell. Biol. 6:4221-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soe, K., A. Rockstroh, P. Schache, and F. Grosse. 2004. The human topoisomerase I damage response plays a role in apoptosis. DNA Repair 3:387-393. [DOI] [PubMed] [Google Scholar]
- 57.Sordet, O., Q. A. Khan, and Y. Pommier. 2004. Apoptotic topoisomerase I-DNA complexes induced by oxygen radicals and mitochondrial dysfunction. Cell Cycle 3:1095-1097. [PubMed] [Google Scholar]
- 58.Strumberg, D., A. A. Pilon, M. Smith, R. Hickey, L. Malkas, and Y. Pommier. 2000. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 20:3977-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takemura, H., V. A. Rao, O. Sordet, T. Furuta, Z. H. Miao, L. Meng, H. Zhang, and Y. Pommier. 2006. Defective Mre11-dependent activation of Chk2 by ataxia telangiectasia mutated in colorectal carcinoma cells in response to replication-dependent DNA double strand breaks. J. Biol. Chem. 281:30814-30823. [DOI] [PubMed] [Google Scholar]
- 60.Tsao, Y. P., A. Russo, G. Nyamuswa, R. Silber, and L. F. Liu. 1993. Interaction between replication forks and topoisomerase I-DNA cleavable complexes: studies in a cell-free SV40 DNA replication system. Cancer Res. 53:5908-5914. [PubMed] [Google Scholar]
- 61.Tse, A. N., K. G. Rendahl, T. Sheikh, H. Cheema, K. Aardalen, M. Embry, S. Ma, E. J. Moler, Z. J. Ni, D. E. Lopes de Menezes, B. Hibner, T. G. Gesner, and G. K. Schwartz. 2007. CHIR-124, a novel potent inhibitor of Chk1, potentiates the cytotoxicity of topoisomerase I poisons in vitro and in vivo. Clin. Cancer Res. 13:591-602. [DOI] [PubMed] [Google Scholar]
- 62.Tse, A. N., and G. K. Schwartz. 2004. Potentiation of cytotoxicity of topoisomerase I poison by concurrent and sequential treatment with the checkpoint inhibitor UCN-01 involves disparate mechanisms resulting in either p53-independent clonogenic suppression or p53-dependent mitotic catastrophe. Cancer Res. 64:6635-6644. [DOI] [PubMed] [Google Scholar]
- 63.Wang, J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 3:430-440. [DOI] [PubMed] [Google Scholar]
- 64.Wang, J. L., X. Wang, H. Wang, G. Iliakis, and Y. Wang. 2002. CHK1-regulated S-phase checkpoint response reduces camptothecin cytotoxicity. Cell Cycle 1:267-272. [PubMed] [Google Scholar]
- 65.Wang, Q., S. Fan, A. Eastman, P. J. Worland, E. A. Sausville, and P. M. O'Connor. 1996. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J. Natl. Cancer Inst. 88:956-965. [DOI] [PubMed] [Google Scholar]
- 66.Wang, X., J. Guan, B. Hu, R. S. Weiss, G. Iliakis, and Y. Wang. 2004. Involvement of Hus1 in the chain elongation step of DNA replication after exposure to camptothecin or ionizing radiation. Nucleic Acids Res. 32:767-775. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Wang, Y., A. R. Perrault, and G. Iliakis. 1997. Down-regulation of DNA replication in extracts of camptothecin-treated cells: activation of an S-phase checkpoint? Cancer Res. 57:1654-1659. [PubMed] [Google Scholar]
- 68.Xiao, Z., Z. Chen, A. H. Gunasekera, T. J. Sowin, S. H. Rosenberg, S. Fesik, and H. Zhang. 2003. Chk1 mediates S and G2 arrests through Cdc25A degradation in response to DNA-damaging agents. J. Biol. Chem. 278:21767-21773. [DOI] [PubMed] [Google Scholar]
- 69.You, Z., L. Kong, and J. Newport. 2002. The role of single-stranded DNA and polymerase alpha in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 277:27088-27093. [DOI] [PubMed] [Google Scholar]
- 70.Zachos, G., M. D. Rainey, and D. A. Gillespie. 2003. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 22:713-723. [DOI] [PMC free article] [PubMed] [Google Scholar]