Abstract
Carbohydrate modification of proteins includes N-linked and O-linked glycosylation, proteoglycan formation, glycosylphosphatidylinositol anchor synthesis, and O-GlcNAc modification. Each of these modifications requires the sugar nucleotide UDP-GlcNAc, which is produced via the hexosamine biosynthesis pathway. A key step in this pathway is the interconversion of GlcNAc-6-phosphate (GlcNAc-6-P) and GlcNAc-1-P, catalyzed by phosphoglucomutase 3 (Pgm3). In this paper, we describe two hypomorphic alleles of mouse Pgm3 and show there are specific physiological consequences of a graded reduction in Pgm3 activity and global UDP-GlcNAc levels. Whereas mice lacking Pgm3 die prior to implantation, animals with less severe reductions in enzyme activity are sterile, exhibit changes in pancreatic architecture, and are anemic, leukopenic, and thrombocytopenic. These phenotypes are accompanied by specific rather than wholesale changes in protein glycosylation, suggesting that while universally required, the functions of certain proteins and, as a consequence, certain cell types are especially sensitive to reductions in Pgm3 activity.
Glycosylation is the process by which proteins and lipids are modified by the addition of glycans from a cellular pool of sugar nucleotides. One of these nucleotides, UDP-N-acetylglucosamine (UDP-GlcNAc), is vital for multiple forms of glycosylation, being incorporated into N-glycans, O-glycans, proteoglycans, and glycosylphosphatidylinositol (GPI)-anchored proteins (23) as well as being the donor for the reversible O-GlcNAc modification of proteins (32).
While UDP-GlcNAc is fundamental for glycosylation, its synthesis has been studied largely in Saccharomyces cerevisiae, and relatively little is known about the process in higher organisms. UDP-GlcNAc is synthesized de novo from glucose via the hexosamine biosynthesis pathway and is also produced via salvage pathways. In yeast, one gene involved in the de novo synthesis of UDP-GlcNAc is phosphoacetylglucosamine mutase 1 (Agm1). Agm1 encodes an essential yeast protein that catalyzes the conversion of GlcNAc-6-phosphate (GlcNAc-6-P) to GlcNAc-1-P, a key step in the hexosamine biosynthesis pathway (9, 16). The human homologue has been studied under two different names: phosphoglucomutase 3 (Pgm3) and Agm1. Pgm3 was initially discovered through genetic studies of polymorphisms in human populations, which revealed multiple phosphoglucomutase loci (17). For over 30 years, the major interest in Pgm3 was in its use as a genetic marker in forensic and genetic fields, with little attention given to the biochemical or cellular role of this gene. Independent study in other fields led to the cloning of human Agm1 both by complementation of the yeast null mutant (24) and as a gene upregulated in response to erythropoietin (EPO) treatment (22). Only recently was it realized that Pgm3 and Agm1 are the same gene (27). While a targeted deletion of the murine Pgm3/Agm1 homologue has not been described, deletion of EMeg32, which is also required for UDP-GlcNAc synthesis, is embryonic lethal (8), underscoring the importance of this pathway but precluding the study of deficiencies in UDP-GlcNAc synthesis in adult tissues.
In this paper, we describe the generation of mice with two hypomorphic mutations in the gene encoding Pgm3. We report that while perturbations in Pgm3 cause global reductions in UDP-GlcNAc levels, specific rather than wholesale changes to protein glycosylation result. Incremental changes in Pgm3 function produce a phenotype gradient that highlights the fundamental importance of the hexosamine biosynthesis pathway and reveals cellular weak spots for UDP-GlcNAc. While mice with a severe reduction in Pgm3 activity die during embryonic development, mice with partial Pgm3 activity are viable, and defects in these mice are restricted to specific cell types. Males are sterile, and pathological changes are observed in the testis, salivary gland, pancreas, and kidney. In addition, deficiencies are seen in certain hematopoietic cell lineages, namely, red blood cells, lymphocytes, and platelets. These results demonstrate that specific cell types are particularly sensitive to changes in glycosylation and suggest that defects in UDP-GlcNAc synthesis may underlie some of the many human glycosylation diseases for which the genetic defect is currently unknown.
MATERIALS AND METHODS
Isolation of an ENU-induced mutant Pgm3 allele.
Male Mpl−/− C57BL/6 mice (2) were treated with a total dose of 200 to 400 mg/kg of N-ethyl-N-nitrosourea (ENU) (Sigma) divided into one, two, or three weekly injections, as described previously (7). ENU-treated mice were mated with untreated Mpl−/− mice to yield first-generation progeny. First-generation mice were intercrossed to yield second-generation progeny, which were brother/sister mated to produce third-generation (G3) mice. At 7 weeks of age, blood from G3 mice was collected from the retro-orbital plexus into tubes containing K2 EDTA (Sarstedt), and peripheral blood cell values were determined using an Advia 120 automated hematological analyzer (Bayer). Upon isolation in a G3 pedigree, multilineage defect 1 (mld1) was bred to Mpl+/+ C57BL/6 mice. All animals used in the studies described were on an Mpl+/+ C57BL/6 genetic background.
Genetic mapping.
Male C57BL/6 mld1/+ Mpl−/− mice were mated to +/+ Mpl+/+ BALB/c females. Random pairs of F1 mice were mated, a quarter of which produced affected mld1/mld1 F2 mice. DNA from 10 affected F2 mice and 30 unaffected siblings was genotyped for 138 simple sequence length polymorphisms spaced evenly genome wide (10), upon which mld1 was assigned to chromosome 9 between D9Mit130 and D9Mit278. Additional mice with informative recombinations were identified from a further 448 F2 mice, and the candidate interval was refined by analysis of recombinants with publicly available and in-house simple sequence length polymorphism markers designed using the UCSC mouse genome browser (http://genome.uscs.edu).
Mutation identification.
The coding exons and splice junctions of candidate genes were PCR amplified from genomic DNA, and products were treated with ExoSap-IT (USB Corporation). Sequencing was carried out using BigDye Terminator chemistry (Applied Biosystems). Reaction mixtures were purified on Sephadex G-50 columns (Amersham Biosciences) in a MultiScreen HV plate (Millipore) and resolved and analyzed on an Applied Biosystems 3700 capillary electrophoresis machine.
Isolation and characterization of a loss-of-function gene trap allele of Pgm3.
We obtained an embryonic stem (ES) cell line (W037B08, strain 129/SvP) harboring a pT1βgeo gene trap vector insertion within Pgm3 from the German Gene Trap Consortium (14). We identified the genomic insertion site of the gene trap vector by PCR as being in intron 4, 87 bp downstream of Pgm3 exon 4. By using reverse transcription (RT)-PCR, we confirmed the splicing of the vector into the Pgm3 mRNA (5′ primer GCG AGG TCT GAG AGA GTT GG [Pgm3 exon 1] and 3′ primer GAC AGT ATC GGC CTC AGG AAG ATC G [gene trap vector]). W037B08 ES cells were injected into C57BL/6 blastocysts, and the resulting male chimeras were bred to C57BL/6 females. Agouti offspring were tested for transmission of the gene trap insertion by diagnostic PCR by employing oligonucleotides designed to distinguish the Pgm3 wild-type from Pgm3gt trapped alleles (primer sequences are available upon request).
Analysis of Pgm3 mRNA.
RT-PCR was performed on cDNA from spleen, testes, and bone marrow from Pgm3mld1/mld1 and Pgm3+/+ mice using primers located in exons 1 and 7 of Pgm3. RT-PCR products were cloned into pGEM-T Easy (Promega), and DNA was sequenced from at least 30 individual clones per genotype.
Hematopoietic cell analysis.
Megakaryocytes were manually enumerated in hematoxylin- and eosin-stained sections of sternum and spleen. Clonal cultures of hematopoietic cells were performed as described previously (1). Cultures of 2.5 × 104 adult bone marrow cells or 5 × 104 spleen cells in 1 ml of 0.3% agar in Dulbecco's modified Eagle's medium supplemented with newborn calf serum (20%) were stimulated with 100 ng/ml murine stem cell factor (SCF), 10 ng/ml murine interleukin-3, and 4 U/ml human erythropoietin (EPO) and incubated for 7 days at 37°C in a fully humidified atmosphere of 10% (vol/vol) CO2 in air. Agar cultures were fixed and stained for acetylcholinesterase, Luxol Fast Blue, and hematoxylin, and the cellular composition of each colony was determined at a magnification of ×100 to ×400. CFU-erythroid (CFU-E) and burst-forming units-erythroid (BFU-E) were enumerated using methylcellulose cultures. Spleen (5 × 104) or bone marrow (2.5 × 104) cells were suspended in 1.5% methylcellulose (Fluka) in Iscove's modified Dulbecco's medium supplemented with 20% fetal calf serum. BFU-E were stimulated with 1 μg/ml SCF, 2.5 × 103 U/ml interleukin-3, and 20 U/ml EPO; CFU-E were stimulated with 10 U/ml EPO. Cultures were incubated at 37°C in a fully humidified atmosphere of 5% (vol/vol) CO2 in air for 2 days (CFU-E) or 7 days (BFU-E). Colonies were scored as being erythroid, myeloid, or mixed erythroid at a magnification of ×35, and colonies containing erythroid cells were verified by diaminofluorozine staining.
Myeloablative transplants.
Irradiated C57BL/6/Ly5.1 mice (11 Gy) were intravenously injected with 1 × 106 bone marrow cells from Pgm3mld1/mld1/Ly5.2 mice. In the reciprocal experiment, 1 × 106 bone marrow cells from C57BL/6/Ly5.1 mice were injected into irradiated Pgm3mld1/mld1/Ly5.2 mice. All recipient mice were maintained on oral antibiotics and analyzed at least 16 weeks posttransplantation by flow cytometry and automated blood cell analysis.
Flow cytometry.
Antigens were detected with fluorescent or biotinylated conjugated monoclonal antibodies as follows: monoclonal antibodies to B220 (RA3-6B2) and immunoglobulin M (IgM) (331.12) were purified from hybridoma supernatants and conjugated in the authors' laboratory, and CD43 (S7), IgD (11-26c.2a), Thy-1.2 (53-2.1), and Sca-1 (D7) were obtained from BD Biosciences. Analyses were performed on an LSR flow cytometer (BD Biosciences). Dead cells were excluded based on propidium iodide staining.
Production and analysis of Pgm3 protein.
Each Pgm3 cDNA was cloned into the pET-15b vector, which carries an N-terminal His tag (Novagen). Constructs were transformed into BL21(DE3)(pLysS) bacteria and protein affinity purified using Ni-nitrilotriacetic acid agarose (QIAGEN). Protein levels were measured by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and Western blotting using an anti-His antibody (Penta-His horseradish peroxidase [HRP] conjugate; QIAGEN).
Pgm3 enzyme assay.
Pgm3 activity was assayed as described previously (24). Briefly, 1 μg of recombinant Pgm3 enzyme or 1 mg of lysed tissue was incubated at 30°C for 20 min in a 20-μl reaction mixture containing 50 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 0.1 μM [α-32P]UTP, 100 μM GlcNAc-6-P, 10% (vol/vol) glycerol, and 1 μg ScUap1p (pGEX-2T-ScUAP was kindly provided by Toshiyuki Mio, Chugai Pharmaceutical Co., Japan). Two microliters of each reaction mixture was spotted onto polyethyleneimine-modified cellulose plates, and nucleotide sugars were separated by thin-layer chromatography and visualized by autoradiography.
UDP-N-acetyl amino sugar assays.
Nucleotide sugar levels in tissues were measured essentially as described previously (30). Briefly, tissues were homogenized using 100 mM KCl-50 mM KH2PO4-1 mM EDTA (pH 7.5) (5 ml/g of tissue) in a Dounce homogenizer, centrifuged at 60,000 × g, and deproteinized with perchloric acid. Samples were again centrifuged, and the supernatant was diluted 10-fold in 10 mM KH2PO4 (pH 2.5). The solution was loaded onto a 3-ml strong anion-exchange solid-phase extraction cartridge preequilibrated with 10 mM KH2PO4 (pH 2.5) and washed with 50 mM KH2PO4 (pH 2.5). UDP-sugar samples were eluted using 150 mM KH2PO4 (pH 7.5) and loaded onto a reverse-phase high-performance liquid chromatography (HPLC) (C18, 250 by 4.6 mm) column. The mobile phase was 0.1 M KH2PO4-2 mM tetrabutylammonium phosphate (pH 6.4), and UV detection was done at 262 nm. Elution positions of UDP-Gal, UDP-Glc, UDP-GalNAc, and UDP-GlcNAc were determined using standards.
Lectin affinity purification.
Tissue extracts were precleared by passing them through 0.5 to 5 ml Sepharose CL-4B columns. After adjusting the NaCl concentration to 0.3 M, samples were incubated with 0.4 to 0.8 ml of 50% Griffornia simplificolia II lectin (GS-II)-agarose slurry (EY Laboratories) equilibrated in normal saline containing EDTA-free protease inhibitor cocktail and 1% Triton X-100. GS-II-agarose beads were recovered by pouring mixtures into open Poly-Prep columns (Bio-Rad) and washed with 0.3 M NaCl containing EDTA-free protease inhibitor cocktail and 1% Triton X-100 before elution with six aliquots of a solution containing 200 to 300 μl of 0.1 M N-acetyl-d-galactosamine (Sigma), 0.1% Triton X-100, 0.15 M NaCl, and EDTA-free protease inhibitor cocktail. Columns were incubated in elution buffer for 5 min at room temperature before each eluate was collected and concentrated to ∼40 μl using a Millipore Ultrafree-MC centrifugal filter unit with a regenerated cellulose membrane (molecular mass cutoff of 10 kDa). Concentrated protein samples were mixed with 20 μl of 4× SDS sample buffer containing 0.2 M dithiothreitol and run on a 4 to 12% polyacrylamide gel (Novex).
Protein identification by mass spectrometry.
Protein bands were excised from polyacrylamide gels and digested in situ using trypsin (25). Peptides were separated by capillary chromatography (26) and sequenced using an on-line electrospray ionization ion trap mass spectrometer (LCQ Thermo-Finnigan). Automatically selected tryptic peptide ions were identified using the SEQUEST algorithm incorporated into Finnigan Xcalibur software and a nonredundant protein database produced by the Office of Information Technology of the Ludwig Institute for Cancer Research.
Lectin blotting.
Lectin blotting was performed essentially as described previously (33). Briefly, tissue extracts (30 to 60 μg of protein) were resolved by SDS-PAGE under reducing conditions on a 4 to 12% Novex gel (Invitrogen) and transferred onto a Hybond-C Extra nitrocellulose membrane (Amersham Biosciences). The membrane was blocked with 5% skim milk overnight at 4°C, washed in 20 mM Tris-HCl (pH 7.5)-150 mM NaCl containing 0.1% Tween 20 (TBST), and incubated with 0.5 μg/ml HRP-conjugated p-hydroxphenylacetic acid in TBST for 1 h. After washing in TBST, lectin binding was detected with ECL substrate (Amersham Biosciences).
Western blotting.
Protein lysates were prepared from tissues as described previously (19). Proteins were resolved by SDS-PAGE, transferred onto PVDF-Plus membranes, and blocked for 1 h in 1% bovine serum albumin in phosphate-buffered saline containing 0.1% Tween 20. Primary antibody against O-GlcNAc (clone CTD110.6; Covance) was diluted 1:1,000 in blocking solution and incubated with the membrane for 1 h. Antibody binding was visualized using sheep anti-mouse-Ig-HRP-conjugated secondary antibody (Chemicon).
Erythrocyte ghosts.
Erythrocyte ghosts were prepared as described previously (15). Briefly, blood was collected in heparin, washed three times in phosphate-buffered saline at 4°C, and lysed in 10 volumes of ice-cold lysis buffer (5.0 mmol/liter Na2HPO4 [pH 8.0], 0.5 mmol/liter EGTA, 2.0 mmol/liter phenylmethylsulfonyl fluoride). Membranes were then washed repeatedly in lysis buffer at 4°C to obtain ghosts.
Determination of neutral sugars of serum proteins.
One hundred micrograms of serum proteins was precipitated by the addition of trichloroacetic acid to a final concentration of 10%. After 1 h of incubation on ice, samples were washed three times with an excess of 10% trichloroacetic acid. The pellet was dissolved in 0.2 N NaOH and then neutralized with 0.2 N HCl. Next, 11.5 μl of 5% phenol was added to 20 μl of sample. After mixing, 70 μl of concentrated sulfuric acid was added. After mixing, the optical density at 480 nm was determined. The concentration of neutral sugar was determined using a standard curve prepared with mannose. 2-Amino benzamide labeling of oligosaccharides was performed as described previously (4).
Intestinal protein leakage.
Twenty-four-hour fecal samples were collected from wild-type and mutant mice. Samples were homogenized in deionized water, lyophilized, and dissolved in 0.15 M sodium chloride. After centrifugation at 1,000 × g for 10 min, alpha-1-antitrypsin (AAT) concentrations in the supernatant were determining by direct competitive enzyme-linked immunosorbent assay.
RESULTS
Isolation of hypomorphic and null alleles of Pgm3.
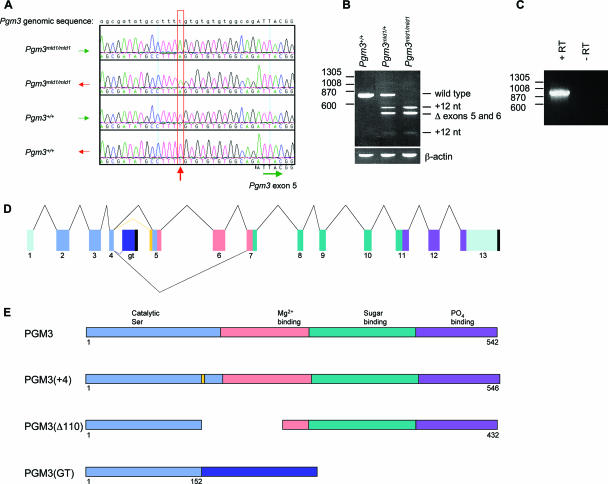
In an N-ethyl-N-nitrosourea mutagenesis screen for recessive mutations causing hematopoietic defects in mice, we isolated a pedigree in which multiple animals exhibited reduced numbers of white blood cells, red blood cells, and platelets (Table 1). The causative mutation in this pedigree, named mld1, was mapped to a 1.86-Mb region of chromosome 9 containing 14 genes via a standard positional cloning approach. Sequencing of the exons and intron/exon boundaries of genes in this interval revealed the presence of a T-to-A transversion in the fourth intron 14 bp upstream of exon 5 of the Pgm3 gene in mld1/mld1 animals but not in six commonly used laboratory mouse strains, including the parental strain C57BL/6 (Fig. 1A). To prove that the mutation in Pgm3 caused the mld1 phenotype, we obtained an independent loss-of-function allele (Pgm3gt) created by the insertion of a gene trap vector into intron 4 (Fig. 1D). Injection of gene trap ES cells into blastocysts resulted in the creation of chimeric mice, which were in turn bred to yield heterozygous Pgm3+/gt mice. A complementation test was then carried out by intercrossing Pgm3+/gt mice and Pgm3+/mld1 mice. Compound heterozygous (Pgm3mld1/gt) mice were found to be anemic, leukopenic, and thrombocytopenic albeit more severely than Pgm3mld1/mld1 animals (Table 1), confirming that mutation of Pgm3 causes the hematological defects observed in the mld1 pedigree.
TABLE 1.
Hematopoietic profile of mice carrying mutations in Pgm3a
| Parameter | Value ± SE
|
||
|---|---|---|---|
| Pgm3+/+ | Pgm3mld1/mld1 | Pgm3mld1/gt | |
| Peripheral blood | |||
| Red blood cells (109/ml) | 10.4 ± 0.5 | 8.0 ± 0.62 | 6.5 ± 2.0 |
| Hematocrit (%) | 53.2 ± 2.4 | 45.8 ± 3.0 | 37.9 ± 10.9 |
| Platelets (106/ml) | 1,159 ± 176 | 620 ± 128 | 858 ± 312 |
| White blood cells (106/ml) | 9.2 ± 1.9 | 3.7 ± 1.3 | 4.8 ± 2.1 |
| Lymphocytes (106/ml) | 8.2 ± 1.8 | 3.5 ± 1.7 | 3.5 ± 1.7 |
| Megakaryocytes (per 100 fields at ×200 magnification) | |||
| Bone marrow | 116 ± 44 | 107 ± 37 | 115 ± 29 |
| Spleen | 2 ± 1 | 33 ± 11 | 39 ± 22 |
| Colony-forming progenitor cells | |||
| Bone marrow (per 2.5 × 104 cells) | |||
| BLAST | 10.3 ± 6.1 | 10 ± 2.8 | ND |
| Granulocyte | 19.5 ± 6.6 | 29.5 ± 2.1 | ND |
| Granulocyte/macrophage | 15.3 ± 4.8 | 15.5 ± 2.1 | ND |
| Macrophage | 13.3 ± 4.9 | 21 ± 11.3 | ND |
| Eosinophil | 0.8 ± 1.0 | 0.5 ± 0.7 | ND |
| Megakaryocyte | 27.8 ± 9.1 | 7 ± 1.4 | ND |
| CFU-E | 30.6 ± 9.4 | 85.9 ± 30.6 | ND |
| BFU-E | 6.4 ± 18.5 | 18.5 ± 5.6 | ND |
| Spleen (per 5.0 × 104 cells) | |||
| BLAST | 0.4 ± 0.5 | 12 ± 0 | ND |
| Granulocyte | 0.6 ± 1.0 | 15 ± 5.7 | ND |
| Granulocyte/macrophage | 0.1 ± 0.3 | 10 ± 8.5 | ND |
| Macrophage | 0.8 ± 1.0 | 12 ± 2.8 | ND |
| Eosinophil | 0 | 0.5 ± 0.7 | ND |
| Megakaryocyte | 4.8 ± 2.2 | 12.5 ± 0.7 | ND |
| CFU-E | 2.5 ± 1.3 | 47.3 ± 7.3 | ND |
| BFU-E | 2.8 ± 1.1 | 20.3 ± 10.3 | ND |
For peripheral blood data, values represent the means ± standard errors for 15 to 52 mice; for all other data, values represent the means ± standard errors for two to four mice. ND, not determined.
FIG. 1.
The Pgm3mld1 and Pgm3gt alleles of Pgm3 lead to aberrant mRNA splicing. (A) The Pgm3mld1 allele contains a T-to-A transversion 14 bp upstream of exon 5. Electropherograms show the genomic DNA sequences from mice of the indicated genotypes. (B) The Pgm3mld1 allele leads to aberrant mRNA splicing. RT-PCR was performed on splenic cDNA using primers located in exons 1 and 7 of Pgm3. An MslI recognition site is present in the 12-nt insertion; hence, PCR products were digested with MslI to distinguish correct splicing from splicing resulting in the 12-nt insertion. Size (bp) is displayed on the left. (C) Confirmation that the gene trap vector splices into Pgm3 mRNA. cDNA was prepared from W037B08 ES cells, and PCR was performed with the 5′ primer in exon 1 of Pgm3 and the 3′ primer in the gene trap vector. The no-reverse-transcriptase (−RT) control confirms that the PCR product is amplified from mRNA rather than from genomic DNA. (D) Schematic of aberrant mRNA splicing of the Pgm3mld1 and Pgm3gt alleles. Pale green, untranslated region; light blue, domain I; pink, domain II; dark green, domain III; purple, domain IV; dark blue, gene trap insertion; yellow, 12-nt insertion; black, poly(A) site. (E) Mutant proteins encoded by the Pgm3mld1 and Pgm3gt alleles. Light blue, domain I; pink, domain II; dark green, domain III; purple, domain IV; dark blue, gene trap insertion; yellow, 12-nt insertion.
The Pgm3mld1 and Pgm3gt alleles lead to aberrant mRNA splicing and encode proteins with reduced enzymatic activity.
The ENU-induced mutation in intron 4 in the Pgm3mld1 allele and the insertion of the gene trap vector into intron 4 of the Pgm3gt allele were both predicted to alter mRNA splicing. By RT-PCR, Pgm3 mRNA was detected in all major organs from wild-type mice (data not shown). In tissues from Pgm3mld1/mld1 mice, no full-length wild-type mRNA was detectable; instead, approximately equal levels of two aberrantly spliced Pgm3 mRNAs were observed (Fig. 1B). The first of these, Pgm3(+4), contained an in-frame insertion of 12 nucleotides (nt) predicted to result in the insertion of four amino acids (Val-Cys-Val-Ala) in domain 1, which contains key residues required for phosphoryl transfer reactions (Fig. 1E). The second mRNA species, Pgm3(Δ110), lacked exons 5 and 6, resulting from the missplicing of exon 4 to exon 7 (Fig. 1B). This mRNA encodes a protein lacking large parts of domains 1 and 2 (Fig. 1E). In the case of the gene trap allele, PCR analyses confirmed splicing of the vector into the Pgm3 mRNA to create a transcript comprising Pgm3 exons 1 to 4 fused to the βgeo reporter (Fig. 1C). The chimeric message encodes only the first part of domain I of Pgm3 and lacks domains II, III, and IV, which contain regions essential for catalytic activity, including Mg2+, sugar, and PO4 binding loops (Fig. 1E). Any protein translated from this mRNA would be expected to be nonfunctional.
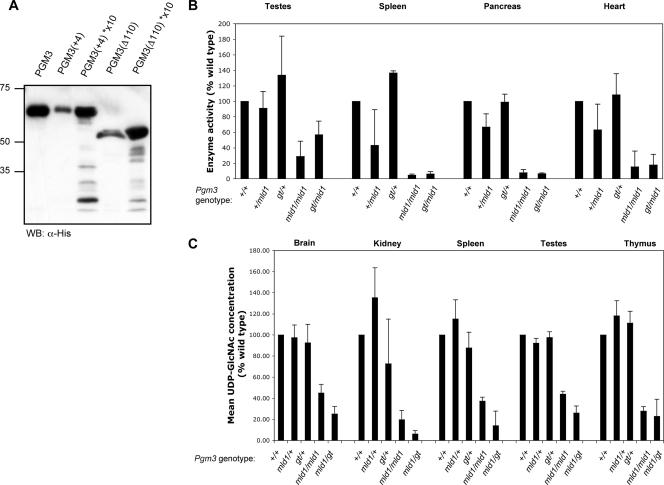
To assess the activity of proteins encoded by the Pgm3mld1 allele, PCR products were derived from the two aberrantly spliced mRNAs and the wild-type mRNA. All three were cloned into vector pET-15b and expressed in bacteria to yield the various Pgm3 proteins preceded by an N-terminal His epitope tag. The level of expression of each of the mutant proteins was lower than that observed from a cDNA encoding the wild-type protein (Fig. 2A). While recombinant wild-type Pgm3 catalyzed the conversion of GlcNAc-6-P to GlcNAc-1-P in vitro, the mutant proteins displayed less than 1% of wild-type enzyme activity [wild-type Pgm3, 346,148 ± 410,416 units activity/μg protein; Pgm3(+4), 238 ± 533 units activity/μg protein; Pgm3(Δ110), 158 ± 317 units activity/μg protein (n = 3)], suggesting that the mld1 mutation leads to a severe reduction in the function of the protein. Consistent with this, we observed significant reductions in Pgm3 activity in tissue extracts from Pgm3mld1/mld1 and Pgm3mld1/gt mice (Fig. 2B).
FIG. 2.
Proteins encoded by hypomorphic (Pgm3mld1) and null (Pgm3gt) alleles of Pgm3 have reduced enzymatic activity and result in decreased levels of UDP-GlcNAc in vivo. (A) Expression and purification of His-tagged Pgm3. Low yields were obtained of the two mutant proteins, Pgm3(+4) and Pgm3(Δ110); lanes with an asterisk (*) contain an approximately 10-fold concentration of the appropriate protein. Molecular mass (kDa) is displayed on the left. WB, Western blot. (B) Pgm3 enzyme assay on tissue extracts. Values represent the means ± standard errors of two tissue samples. (C) UDP-GlcNAc concentration in tissue extracts. Values represent the means ± standard errors of 3 to 11 tissue samples.
Pgm3 is essential for maintenance of the UDP-GlcNAc pool in vivo.
Conversion of GlcNAc-6-P to GlcNAc-1-P, catalyzed by Pgm3, is a key step in the synthesis of UDP-GlcNAc, an essential intermediate in a variety of protein and lipid glycosylation pathways. Consistent with this role, HPLC analysis of tissue extracts from Pgm3mld1/mld1 and Pgm3mld1/gt mice revealed a dramatic reduction in the levels of UDP-GlcNAc, whereas no significant reduction was observed in heterozygous Pgm3+/mld1 and Pgm3+/gt mice (Fig. 2C). In addition, the level of UDP-GalNAc was also reduced in tissue extracts from Pgm3mld1/mld1 and Pgm3mld1/gt mice. No significant alteration was observed in the levels of UDP-Glc or UDP-Gal (data not shown).
Pgm3 is required for early embryonic development.
The effect of reducing Pgm3 function on survival was assessed by genotyping pups derived from a range of crosses involving Pgm3+/gt and Pgm3+/mld1 mice at weaning. Whereas Pgm3mld/1mld1 mice were weaned at the expected Mendelian frequencies (61 observed of 65 expected) and appeared to be ostensibly normal, Pgm3mld1/gt mice were weaned in reduced numbers (18 observed of 47 expected) and displayed an approximately 60% reduction in body weight and reduced viability (data not shown). More dramatically, multiple Pgm3+/gt intercrosses failed to produce any Pgm3gt/gt mice at weaning, suggesting that the more severe reduction in Pgm3 activity and the UDP-GlcNAc level associated with the Pgm3gt allele results in embryonic or perinatal lethality. To determine when Pgm3gt/gt mice were dying, embryos from Pgm3+/gt intercrosses were genotyped at various stages of gestation. While no Pgm3gt/gt embryos were seen at embryonic day 6.5 (E6.5) (zero observed of six expected) or at time points thereafter, they were present at a normal Mendelian frequency at E3.5 (five observed of nine expected), suggesting that Pgm3gt/gt embryos die between E3.5 and E6.5.
Specific intrinsic hematopoietic defects in Pgm3mld1/mld1 and Pgm3mld1/gt mice.
The viability of Pgm3mld1/mld1 and Pgm3mld1/gt mice allowed us to explore the physiological consequences of reducing the activity of this enzyme in adults. Hematological and histological analyses revealed that heterozygous Pgm3+/gt and Pgm3+/mld1 mice were apparently normal (data not shown). In contrast, Pgm3mld1/mld1 and Pgm3mld1/gt mice displayed specific hematological and histological changes. Consistent with the more severe loss of Pgm3 function observed in Pgm3mld1/gt mice, pathological changes in these animals were more pronounced.
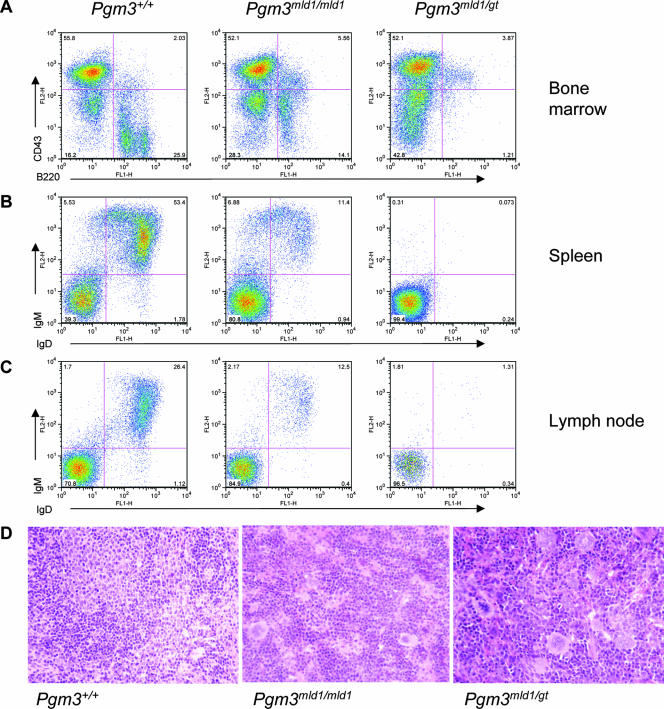
Despite the general importance of protein glycosylation in cell function, emphasized by the death of Pgm3gt/gt mice early in embryogenesis, the effect of the Pgm3mld1 and Pgm3gt alleles on blood cell numbers was lineage dependent (Table 1). While neutrophilic and eosinophilic granulocytes and monocytes were present in normal numbers, a modest reduction in the numbers of red blood cells and platelets was observed, and B lymphocytes were severely affected (Fig. 3A to C). Pgm3mld1/mld1 mice exhibited a two- to threefold increase in the frequency of B220+ CD43+ pro-B cells; however, the cellularity of Pgm3mld1/mld1 bone marrow was approximately 70% of that of wild-type mice, and hence, the number of pro-B cells was slightly elevated. Pgm3mld1/gt mice showed an approximately twofold increase in the frequency of B220+ CD43+ pro-B cells; however, the cellularity of Pgm3mld1/mld1 bone marrow is approximately 50% of that of wild-type mice, and hence, the number of pro-B cells was not significantly different from that of wild-type mice. The frequency and number of B cells at all later stages of maturation were significantly reduced in both Pgm3mld1/mld1 and Pgm3mld1/gt mice. Consistent with this, a reduction in the B-cell number was observed in the peritoneal cavity and peripheral lymphoid organs and was accompanied by histological changes including the absence of lymphoid follicles (Fig. 3D). Although T lymphopoiesis was relatively normal in most Pgm3mld1/mld1 mice, some Pgm3mld1/mld1 mice and all Pgm3mld1/gt mice displayed a moderate to severe reduction in the number of T cells in the spleen and an increased CD4-to-CD8 ratio in the lymph nodes (data not shown).
FIG. 3.
Mice with hypomorphic alleles of Pgm3 exhibit a profound B lymphopenia associated with a defect at the pro-B-cell-to-pre-B-cell transition. (A) Bone marrow cells from 60- to 80-day-old Pgm3+/+, Pgm3mld1/mld1, and Pgm3mld1/gt mice were analyzed by FACS for the expression of B220 and CD43. Data shown are representative of data from at least five age-matched mice. (B) Splenocytes from 60- to 80-day-old Pgm3+/+, Pgm3mld1/mld1, and Pgm3mld1/gt mice were analyzed by FACS for the expression of B220 and IgM. Data shown are representative of data from at least five age-matched mice. (C) Lymph node cells from 60- to 80-day-old Pgm3+/+, Pgm3mld1/mld1, and Pgm3mld1/gt mice were analyzed by FACS for expression of IgD and IgM. Data shown are representative of data from at least five age-matched mice. (D) Spleens of Pgm3mld1/mld1 and Pgm3mld1/gt mice display expansion of the red pulp and an absence of lymphoid follicles.
The anemia observed in Pgm3mld1/mld1 and Pgm3mld1/gt mice was relatively mild (Table 1) and accompanied by compensatory extramedullary hematopoiesis characterized by splenomegaly (Pgm3mld1/mld1 spleen weight was 166 ± 34 mg compared to 87 ± 13 mg for Pgm3+/+) involving the expansion of the red pulp and a dramatic increase in the numbers of BFU-E, CFU-E, and morphologically recognizable erythroid elements. Bone marrow erythropoiesis remained relatively normal. The cause of anemia remains obscure, with both the production and clearance of mature red blood cells not altered significantly in Pgm3mld1/mld1 mice (data not shown). The thrombocytopenia observed in Pgm3mld1/mld1 and Pgm3mld1/gt mice was modest but significant (Table 1) and was accompanied by a normal number of bone marrow megakaryocytes, a reduction in bone marrow megakaryocyte progenitors, and an elevation of splenic megakaryocyte progenitor numbers and megakaryocytes. In vivo labeling of platelets with biotin (3) revealed them to be produced more slowly in Pgm3mld1/mld1 mice but to be cleared at a normal rate, explaining the lower steady-state number in these animals (data not shown).
Since glycoproteins are crucial for the proper function of both blood cells and the stromal elements that generate the hematopoietic microenvironment, we explored the importance of these cell types in the manifestation of the Pgm3 mutant phenotype using reciprocal bone marrow transplants. Irradiated Pgm3+/+ recipients receiving Pgm3mld1/mld1 bone marrow exhibited profound lymphopenia and mild anemia and thrombocytopenia characteristic of the donor (data not shown). In contrast, irradiated Pgm3mld1/mld1 mice receiving Pgm3+/+ bone marrow were rescued from lymphopenia, anemia, and thrombocytopenia (data not shown). These results suggest that defects in protein glycosylation within blood cells themselves, rather than the microenvironment, are responsible for the defects associated with the loss of Pgm3 function.
Reproductive system and other defects in Pgm3mld1/mld1 and Pgm3mld1/gt mice.
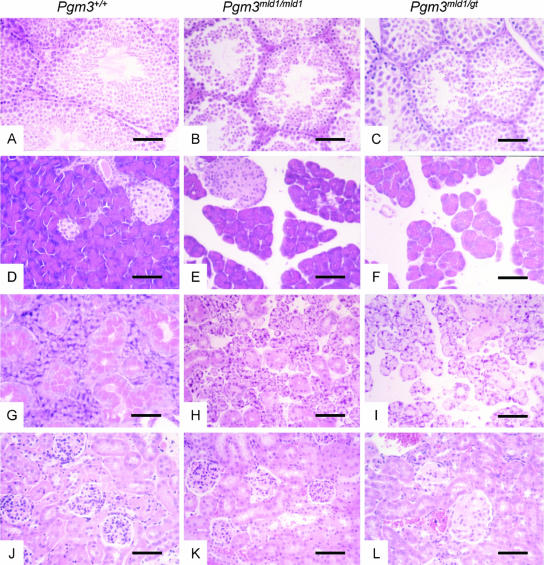
In establishing a colony of Pgm3 mutant mice, we noticed that irrespective of the genotype of the females to which they were mated, male Pgm3mld1/mld1 mice never sired offspring. Histological analyses of the testes showed dramatic defects: although immature spermatogonia were present, all mature spermatocytes were pyknotic (Fig. 4A to C). In contrast, female Pgm3mld1/mld1 mice were able to become pregnant; however, while live pups were born, few survived to weaning. Other histological defects in Pgm3mld1/mld1 mice were observed in the salivary gland and the pancreas (Fig. 4D to I). Pancreatic changes centered on the dispersion of acini associated with the shrinking of the acinar cells; however, no hematopoietic infiltration of the pancreas was observed. These changes did not affect the islets, and no changes in circulating glucose or insulin levels were observed. Pgm3mld1/gt mice but not Pgm3mld1/mld1 mice exhibited glomerulonephritis with amorphous material filling the glomeruli (Fig. 4J to L).
FIG. 4.
Mice with reduced Pgm3 enzyme activity and UDP-GlcNAc levels exhibit defective spermatogenesis, disrupted pancreatic and salivary gland architecture, and glomerulonephritis. Sections were stained with hematoxylin and eosin; the bar represents 60 μm. (A to C) Histopathology of testes. The testes of Pgm3mld1/mld1 (B) and Pgm3mld1/gt (C) mice contain immature spermatogonia, but all mature spermatozoa are pyknotic. (D to F) Histopathology of pancreas. In the pancreas of Pgm3mld1/mld1 (E) and Pgm3mld1/gt (F) mice, acini are reduced in size and dispersed. (G to I) Histopathology of salivary gland. In the salivary gland of Pgm3mld1/mld1 (H) and Pgm3mld1/gt (I) mice, cells are reduced in size and dispersed, with a lesion similar to that observed in the pancreas. (J to L) Kidneys of Pgm3mld1/gt (L) but not Pgm3mld1/mld1 (K) mice display glomerulonephritis with amorphous material filling the glomeruli.
Specific changes in protein glycosylation in tissues affected by Pgm3 mutation.
UDP-GlcNAc is required for a diverse range of protein modifications, and we therefore sought to identify changes in protein glycosylation that might explain the phenotypes observed in tissues affected by a mutation of Pgm3. Proteins in extracts from a range of tissues and erythrocyte ghosts (Fig. 5F and data not shown) were resolved by SDS-PAGE and visualized by Coomassie staining. Surprisingly, there were few obvious differences in the molecular weights of proteins from Pgm3+/+ and those of proteins from Pgm3mld1/mld1 mice, suggesting that the glycosylation of many proteins is not grossly affected by reductions in UDP-GlcNAc levels. Likewise, fluorescence-activated cell sorter (FACS) analysis of hematopoietic cells revealed no difference in the expression of a range of GPI-anchored proteins including Sca-1 and Thy-1.2 (data not shown).
FIG. 5.
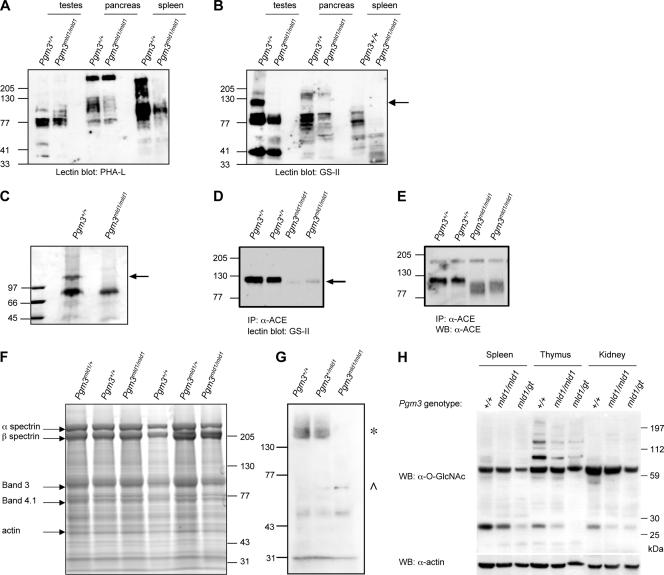
Specific defects in glycosylation resulting from reductions in UDP-GlcNAc levels. (A and B) Membrane lysates were prepared from various tissues, separated by SDS-PAGE, and subjected to lectin blotting with P. vulgaris lectin (PHA-L) (A) or GS-II (B). Most glycoproteins were relatively unaffected by the reduced UDP-GlcNAc levels in Pgm3mld1/mld1 mice; an exception was a ∼110-kDa glycoprotein in the testes (arrow). (C) Affinity purification of a ∼110-kDa glycoprotein from the testes of Pgm3+/+ but not Pgm3mld1/mld1 mice. Membrane lysates from testes were subjected to affinity purification using GS-II-conjugated agarose. Eluted proteins were separated on an SDS-PAGE gel and visualized by Coomassie blue staining. The indicated band (arrow) was excised from the gel and identified by mass spectrometry as testicular ACE. (D) Lysates from Pgm3+/+ and Pgm3mld1/mld1 testes were immunoprecipitated (IP) with an anti-ACE antibody that recognizes both somatic and testicular isoforms of ACE. Immunoprecipitates were separated by SDS-PAGE and subjected to lectin blotting with GS-II. The arrow indicates a band at the expected size of ACE, which is hypoglycosylated in Pgm3 mutants. (E) The blot in D was stripped and reprobed with anti-ACE antibody to demonstrate that testicular ACE is expressed in the Pgm3mld1/mld1 testes but aberrantly glycosylated. (F) Red cell ghosts were prepared, and proteins were separated by SDS-PAGE. Coomassie blue staining demonstrated that major erythrocyte membrane proteins, including glycoproteins such as band 3, have normal molecular weights in Pgm3mld1/mld1 red blood cells. (G) Red cell ghost preparations from F were subjected to Western blotting (WB) with anti-O-GlcNAc antibody. One band (*) is present in Pgm3+/+ and Pgm3mld1/+ but absent in Pgm3mld1/mld1 samples; another band (^) is present in Pgm3mld1/+ and Pgm3mld1/mld1 samples but absent in Pgm3+/+ samples. (H) Lysates were prepared, separated by SDS-PAGE, and subjected to Western blotting with anti-O-GlcNAc antibody.
To examine more specific aspects of glycosylation, extracts of testis, spleen, and pancreas (organs affected in Pgm3mld1/mld1 mice) were electrophoresed on an SDS-polyacrylamide gel, and the proteins were transferred onto a membrane, which was then probed with HRP-conjugated Phaseolus vulgaris lectin, which recognizes complex N-glycans, and GS-II, which binds to the GlcNAc terminal on oligosaccharides (Fig. 5A and B). Although most glycoproteins appeared to be relatively unaffected by reductions in Pgm3 activity and UPD-GlcNAc levels, specific glycoproteins were observed in wild-type cells that were not present in tissues from Pgm3mld1/mld1 mice; most prominent was a species with an apparent molecular weight of 110,000 that was present in the testes. This glycoprotein was purified on GS-II-agarose, excised, and shown by mass spectrometry to be the testis-specific isoform of angiotensin-converting enzyme (ACE) (Fig. 5C). Immunoprecipitation of ACE and blotting with anti-ACE or -GS-II confirmed that although this protein was expressed in Pgm3mld1/mld1 mice, its molecular weight was reduced, and its glycosylation was aberrant (Fig. 5D and E).
Since red blood cells are known to express O-GlcNAc-modified proteins, such as band 4.1, we subjected erythrocyte membrane proteins to Western blotting with an antibody directed at O-GlcNAc. Pgm3mld1/mld1 samples lacked a band present in Pgm3+/+ samples but also showed the presence of an additional band not present in Pgm3+/+ extracts (Fig. 5G) and evident at intermediate levels in Pgm3mld1/+ samples. Analysis of O-GlcNAc modification in tissue lysates revealed a reduced level of O-GlcNAc-modified proteins in Pgm3mld1/mld1 mice and a more dramatic decrease in Pgm3mld1/gt mice (Fig. 5H). Collectively, these results support the proposal that O-GlcNAc modification is regulated by the UDP-GlcNAc concentration (21) and suggest that defects in O-GlcNAc modification may be at least partially responsible for the graded effects of reduced Pgm3 activity.
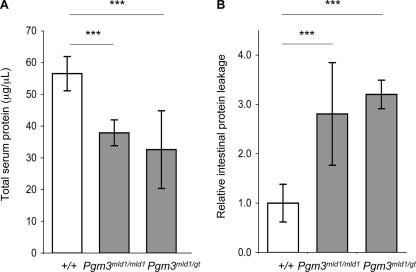
We next analyzed the effects of reduced UDP-GlcNAc biosynthesis on structures of N-glycans from total serum glycoproteins. Glycans were cleaved with PNGase F, labeled with 2-aminobenzamide, and separated by anion-exchange HPLC, but no significant differences were found between wild-type and mutant mice (Pgm3mld1/mld1 and Pgm3mld1/gt) (data not shown). The amount of protein-bound neutral monosaccharide and the relative proportion of albumin were identical in wild-type and mutant sera (data not shown), ruling out preferential deficit of either glycoproteins or albumin. However, the total protein concentration in sera from mutant mice was significantly lower than that for wild-type mice (Fig. 6A). To account for this difference, we analyzed urine for total protein and feces for the accumulation of degradation-resistant AAT, a common metric for the enteric loss of plasma proteins (5). AAT is an endogenous protease inhibitor that is resistant to degradation by endogenous and bacterial proteases in the intestine and, thus, can be detected in the feces and is an established marker for intestinal protein leakage. There was no proteinuria (<1 μg/ml), but AAT in mutant feces was more than threefold higher than that in wild-type littermates (Fig. 6B).
FIG. 6.
Intestinal protein leakage in Pgm3 mutant mice. (A) Total serum protein concentration measured by Bradford assay. (B) Levels of degradation-resistant AAT in fecal samples were measured by enzyme-linked immunosorbent assay. Data show relative levels between genotype classes, indicating intestinal protein leakage in Pgm3 mutants (***, P < 0.001).
DISCUSSION
While Agm1 has been shown to be essential for viability in yeast, little is known about the physiological role of Agm1/Pgm3 in mammals. Here, we show that mutations in Pgm3 cause a global reduction in UDP-GlcNAc levels and that despite the general importance of glycosylation in all cell types, incremental changes in UDP-GlcNAc levels selectively affect the modification of specific proteins, leading to a graded series of pathological changes.
The phenotypes of adult Pgm3mld1/mld1 and Pgm3mld1/gt mice are strikingly similar to those seen when genes involved in N-glycan processing are disrupted. For example, Mgat2 encodes GlcNAc transferase II (GlcNAcT-II), a glycosyltransferase found in the Golgi compartment, which is essential for the production of complex-type N-glycans. Like Pgm3mld1/mld1 and Pgm3mld1/gt mice, mice deficient for Mgat2 show reduced viability, reduced size, mild anemia, splenomegaly, thrombocytopenia, defective B- and T-cell development, defective spermatogenesis, and glomerulonephritis. They also, however, display dysmorphic features, locomotor retardation, and defects in bone formation (31), phenotypes not observed in Pgm3mld1/mld1 or Pgm3mld1/gt mice. Since GlcNAcT-II catalyzes a reaction that requires UDP-GlcNAc, it is possible that reducing Pgm3 activity and the level of UDP-GlcNAc leads to suboptimal activity of GlcNAcT-II, resulting in a partial phenocopy of the Mgat2-deficient mouse. These findings suggest that disruptions in N-glycosylation pathways are responsible for the majority of phenotypes observed in adult Pgm3mld1/mld1 and Pgm3mld1/gt mice.
While the decreased UDP-GlcNAc levels seen in Pgm3mld1/gt mice are sufficient for survival through embryonic and neonatal development, Pgm3gt/gt mice died between E3.5 and E6.5, suggesting that a minimum level of UDP-GlcNAc is required for early embryonic development. The lethality of Pgm3gt/gt mice cannot be explained by disruptions in N-glycans alone, as embryos can develop normally, implant, and progress to ∼E9.5 in the complete absence of hybrid and complex N-glycans (29). It is therefore likely that the nearly total absence of Pgm3 affects another pathway or perhaps multiple pathways. While the complete lack of O-GlcNAc transferase (Ogt), the enzyme responsible for O-GlcNAc modification, is embryonic lethal (28), there is evidence that Ogt activity is regulated by the UDP-GlcNAc concentration. Mouse embryonic fibroblasts carrying a mutation in the EMeg32 gene exhibit reduced UDP-GlcNAc levels and a significant defect in O-GlcNAc modification (8). In contrast, there was no general impairment of N-glycosylation or GPI anchor biosynthesis and only modest effects on Golgi compartment processing of N-glycans. This suggests that altering the level of a sugar nucleotide can affect downstream glycosylation pathways to different extents, and embryonic fibroblasts from Pgm3gt/gt mice should provide insights into the extent to which the various pathways are disrupted.
Tissues in Pgm3-deficient mice appear to be differentially sensitive to reductions in the level of UDP-GlcNAc, possibly because glycosylation of certain proteins is more sensitive to the level of UDP-GlcNAc; proteins that are most affected may be those that are critical for the functions of the hematopoietic system, testes, pancreas, salivary gland, and kidney. Examination of glycoproteins in Pgm3mld1/mld1 mice, using electrophoresis, lectins, or antibodies to GPI-linked proteins, demonstrated that most glycoproteins were relatively unaffected by reductions in Pgm3 activity and UDP-GlcNAc levels; however, there were striking exceptions. For example, in the testis, we identified aberrant glycosylation of the testis-specific isoform of ACE in Pgm3mld1/mld1 mice. Testicular ACE is a heavily glycosylated integral membrane protein containing both O- and N-glycosylation sites (34). Since no overall decrease in the level of glycosylation was observed in the testis, testicular ACE appears to be particularly sensitive to reductions in the UDP-GlcNAc level. At present, it is unclear how aberrant glycosylation of ACE contributes to the defective spermatogenesis observed in Pgm3mld1/mld1and Pgm3mld1gt mice. Inactivation of Ace in the mouse leads to male hypofertility, and the homologue in Drosophila melanogaster is required for spermiogenesis (18). However, unlike Pgm3mld1/mld1and Pgm3mld1/gt mice, Ace-deficient mice have normal numbers of sperm (12, 13, 20). An exploration of the function of aberrantly glycosylated ACE may resolve these issues.
It has been shown previously that the loss of heparan-sulfate (HS) on enterocytes plays a central role in a severe clinical phenomenon called protein-losing enteropathy (PLE) (6). The specific loss of HS from the basolateral surface of intestinal epithelial cells leads to the loss of plasma proteins through the intestine of PLE patients. HS consists mainly of alternating units of glucuronic acid and GlcNAc, and limiting UDP-GlcNAc might reduce the amount of the HS chain length, leading to PLE. The elevated fecal AAT levels observed in Pgm3-deficient mice support this hypothesis. Since AAT is reasonably stable in the gut, elevated AAT concentrations in the feces are highly indicative of protein loss from the plasma into the intestine. The other possible cause for plasma protein loss, proteinuria, the loss of proteins by the kidney, was excluded by direct measurement. Accordingly, enteric protein loss is likely to be the major contributor to reduced plasma/serum protein levels, since sera from Pgm3-deficient mice contained the appropriate proportion of glycoproteins, with carbohydrate content and spectrum of glycan structures identical to those of wild-type mice.
UDP-GlcNAc is fundamental to many forms of glycosylation, and reducing the level of this sugar nucleotide in the mouse leads to profound disruptions of specific cell types. It is well known that defects in the synthesis or uptake of other sugar nucleotides underlie several human diseases; however, the UDP-GlcNAc synthesis pathway has not been implicated in any disease to date. Given that the genetic basis of many human glycosylation disorders remains unknown, it will be of great interest to determine whether there are patients with hematological and fertility problems caused by partial-loss-of-function mutations in human Pgm3 or in other genes involved in UDP-GlcNAc synthesis.
Acknowledgments
We thank Jason Corbin, Elizabeth Viney, Sandra Mifsud, Ladina Di Rago, Lynne Hartley, Katya Henley, and Adrienne Hilton for their expert technical assistance. We thank Toshiyuki Mio for generously providing reagents.
This work was supported by grants from the Australian National Health and Medical Research Council (program 257500), the National Cancer Institute (CA22556), the Cancer Council of Victoria, MuriGen Pty. Ltd., and the National Institutes of Heath (R01 DK55615 [H.H.F.]). B.T.K. is supported by a Queen Elizabeth II Fellowship from the Australian Research Council. K.T.G. is supported by a Stella Mary Langford Scholarship from the University of Melbourne. J.A. was supported by a postdoctoral fellowship from the Leukemia and Lymphoma Society. D.L.K. was supported by a postdoctoral fellowship from the Canadian Institutes of Health Research. L.B. and C.K. are supported by fellowships from the Deutsche Forschungsgemeinschaft (KR 2916/1-1 to C.K. and BO 2488/1-1 to L.B.).
Footnotes
Published ahead of print on 4 June 2007.
REFERENCES
- 1.Alexander, W. S., D. Metcalf, and A. R. Dunn. 1995. Point mutations within a dimer interface homology domain of c-Mpl induce constitutive receptor activity and tumorigenicity. EMBO J. 14:5569-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, W. S., A. W. Roberts, N. A. Nicola, R. Li, and D. Metcalf. 1996. Deficiencies in progenitor cells of multiple hematopoietic lineages and defective megakaryocytopoiesis in mice lacking the thrombopoietic receptor c-Mpl. Blood 87:2162-2170. [PubMed] [Google Scholar]
- 3.Ault, K. A., and C. Knowles. 1995. In vivo biotinylation demonstrates that reticulated platelets are the youngest platelets in circulation. Exp. Hematol. 23:996-1001. [PubMed] [Google Scholar]
- 4.Bigge, J. C., T. P. Patel, J. A. Bruce, P. N. Goulding, S. M. Charles, and R. B. Parekh. 1995. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal. Biochem. 230:229-238. [DOI] [PubMed] [Google Scholar]
- 5.Bode, L., and H. H. Freeze. 2006. Applied glycoproteomics—approaches to study genetic-environmental collisions causing protein-losing enteropathy. Biochim. Biophys. Acta 1760:547-559. [DOI] [PubMed] [Google Scholar]
- 6.Bode, L., S. Murch, and H. H. Freeze. 2006. Heparan sulfate plays a central role in a dynamic in vitro model of protein-losing enteropathy. J. Biol. Chem. 281:7809-7815. [DOI] [PubMed] [Google Scholar]
- 7.Bode, V. C. 1984. Ethylnitrosourea mutagenesis and the isolation of mutant alleles for specific genes located in the T region of mouse chromosome 17. Genetics 108:457-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehmelt, G., A. Wakeham, A. Elia, T. Sasaki, S. Plyte, J. Potter, Y. Yang, E. Tsang, J. Ruland, N. N. Iscove, J. W. Dennis, and T. W. Mak. 2000. Decreased UDP-GlcNAc levels abrogate proliferation control in EMeg32-deficient cells. EMBO J. 19:5092-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boles, E., W. Liebetrau, M. Hofmann, and F. K. Zimmermann. 1994. A family of hexosephosphate mutases in Saccharomyces cerevisiae. Eur. J. Biochem. 220:83-96. [DOI] [PubMed] [Google Scholar]
- 10.Dietrich, W., H. Katz, S. E. Lincoln, H. S. Shin, J. Friedman, N. C. Dracopoli, and E. S. Lander. 1992. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics 131:423-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reference deleted.
- 12.Esther, C. R., Jr., T. E. Howard, E. M. Marino, J. M. Goddard, M. R. Capecchi, and K. E. Bernstein. 1996. Mice lacking angiotensin-converting enzyme have low blood pressure, renal pathology, and reduced male fertility. Lab. Investig. 74:953-965. [PubMed] [Google Scholar]
- 13.Hagaman, J. R., J. S. Moyer, E. S. Bachman, M. Sibony, P. L. Magyar, J. E. Welch, O. Smithies, J. H. Krege, and D. A. O'Brien. 1998. Angiotensin-converting enzyme and male fertility. Proc. Natl. Acad. Sci. USA 95:2552-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen, J., T. Floss, P. Van Sloun, E.-M. Fuechtbauer, F. Vauti, H.-H. Arnold, F. Schnütgen, W. Wurst, H. von Melchner, and P. Ruiz. 2003. A large scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. USA 100:9918-9922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassoun, H., T. Hanada, M. Lutchman, K. E. Sahr, J. Palek, M. Hanspal, and A. H. Chishti. 1998. Complete deficiency of glycophorin A in red blood cells from mice with targeted inactivation of the band 3 (AE1) gene. Blood 91:2146-2151. [PubMed] [Google Scholar]
- 16.Hofmann, M., E. Boles, and F. K. Zimmermann. 1994. Characterization of the essential yeast gene encoding N-acetylglucosamine-phosphate mutase. Eur. J. Biochem. 221:741-747. [DOI] [PubMed] [Google Scholar]
- 17.Hopkinson, D. A., and H. Harris. 1968. A third phosphoglucomutase locus in man. Ann. Hum. Genet. 31:359-367. [DOI] [PubMed] [Google Scholar]
- 18.Hurst, D., C. M. Rylett, R. E. Isaac, and A. D. Shirras. 2003. The Drosophila angiotensin-converting enzyme homologue Ance is required for spermiogenesis. Dev. Biol. 254:238-247. [DOI] [PubMed] [Google Scholar]
- 19.Krebs, D., R. Uren, D. Metcalf, S. Rakar, J. Zhang, R. Starr, D. De Souza, K. Hanzinikolas, J. Eyles, L. Connolly, R. Simpson, N. Nicola, S. Nicholson, M. Baca, D. Hilton, and W. Alexander. 2002. SOCS-6 binds to insulin receptor substrate 4, and mice lacking the SOCS-6 gene exhibit mild growth retardation. Mol. Cell. Biol. 22:4567-4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krege, J. H., S. W. John, L. L. Langenbach, J. B. Hodgin, J. R. Hagaman, E. S. Bachman, J. C. Jennette, D. A. O'Brien, and O. Smithies. 1995. Male-female differences in fertility and blood pressure in ACE-deficient mice. Nature 375:146-148. [DOI] [PubMed] [Google Scholar]
- 21.Kreppel, L. K., and G. W. Hart. 1999. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 274:32015-32022. [DOI] [PubMed] [Google Scholar]
- 22.Li, C., M. Rodriguez, and D. Banerjee. 2000. Cloning and characterization of complementary DNA encoding human N-acetylglucosamine-phosphate mutase protein. Gene 242:97-103. [DOI] [PubMed] [Google Scholar]
- 23.Lowe, J. B., and J. D. Marth. 2003. A genetic approach to mammalian glycan function. Annu. Rev. Biochem. 72:643-691. [DOI] [PubMed] [Google Scholar]
- 24.Mio, T., T. Yamada-Okabe, M. Arisawa, and H. Yamada-Okabe. 2000. Functional cloning and mutational analysis of the human cDNA for phosphoacetylglucosamine mutase: identification of the amino acid residues essential for the catalysis. Biochim. Biophys. Acta 1492:369-376. [DOI] [PubMed] [Google Scholar]
- 25.Moritz, R. L., J. S. Eddes, G. E. Reid, and R. J. Simpson. 1996. S-Pyridylethylation of intact polyacrylamide gels and in situ digestion of electrophoretically separated proteins: a rapid mass spectrometric method for identifying cysteine-containing peptides. Electrophoresis 17:907-917. [DOI] [PubMed] [Google Scholar]
- 26.Moritz, R. L., and R. J. Simpson. 1992. Application of capillary reversed-phase high-performance liquid chromatography to high-sensitivity protein sequence analysis. J. Chromatogr. 599:119-130. [DOI] [PubMed] [Google Scholar]
- 27.Pang, H., Y. Koda, M. Soejima, and H. Kimura. 2002. Identification of human phosphoglucomutase 3 (PGM3) as N-acetylglucosamine-phosphate mutase (AGM1). Ann. Hum. Genet. 66:139-144. [DOI] [PubMed] [Google Scholar]
- 28.Shafi, R., S. P. Iyer, L. G. Ellies, N. O'Donnell, K. W. Marek, D. Chui, G. W. Hart, and J. D. Marth. 2000. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97:5735-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi, S., S. A. Williams, A. Seppo, H. Kurniawan, W. Chen, Z. Ye, J. D. Marth, and P. Stanley. 2004. Inactivation of the Mgat1 gene in oocytes impairs oogenesis, but embryos lacking complex and hybrid N-glycans develop and implant. Mol. Cell. Biol. 24:9920-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Span, P. 2001. Assays for hexosamine pathway intermediates (uridine diphosphate-N-acetyl amino sugars) in small samples of human muscle tissue. Clin. Chem. 47:944-946. [PubMed] [Google Scholar]
- 31.Wang, Y., J. Tan, M. Sutton-Smith, D. Ditto, M. Panico, R. M. Campbell, N. M. Varki, J. M. Long, J. Jaeken, S. R. Levinson, A. Wynshaw-Boris, H. R. Morris, D. Le, A. Dell, H. Schachter, and J. D. Marth. 2001. Modeling human congenital disorder of glycosylation type IIa in the mouse: conservation of asparagine-linked glycan-dependent functions in mammalian physiology and insights into disease pathogenesis. Glycobiology 11:1051-1070. [DOI] [PubMed] [Google Scholar]
- 32.Wells, L., and G. W. Hart. 2003. O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett. 546:154-158. [DOI] [PubMed] [Google Scholar]
- 33.Xia, L., T. Ju, A. Westmuckett, G. An, L. Ivanciu, J. M. McDaniel, F. Lupu, R. D. Cummings, and R. P. McEver. 2004. Defective angiogenesis and fatal embryonic hemorrhage in mice lacking core 1-derived O-glycans. J. Cell Biol. 164:451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, X. C., E. D. Sturrock, Z. Wu, K. Biemann, M. R. Ehlers, and J. F. Riordan. 1997. Identification of N-linked glycosylation sites in human testis angiotensin-converting enzyme and expression of an active deglycosylated form. J. Biol. Chem. 272:3511-3519. [DOI] [PubMed] [Google Scholar]