Abstract
Gα12 and Gα13 function as molecular regulators responding to extracellular stimuli. NF-E2-related factor 2 (Nrf2) is involved in a protective adaptive response to oxidative stress. This study investigated the regulation of Nrf2 by Gα12 and Gα13. A deficiency of Gα12, but not of Gα13, enhanced Nrf2 activity and target gene transactivation in embryo fibroblasts. In mice, Gα12 knockout activated Nrf2 and thereby facilitated heme catabolism to bilirubin and its glucuronosyl conjugations. An oligonucleotide microarray demonstrated the transactivation of Nrf2 target genes by Gα12 gene knockout. Gα12 deficiency reduced Jun N-terminal protein kinase (JNK)-dependent Nrf2 ubiquitination required for proteasomal degradation, and so did Gα13 deficiency. The absence of Gα12, but not of Gα13, increased protein kinase C δ (PKC δ) activation and the PKC δ-mediated serine phosphorylation of Nrf2. Gα13 gene knockout or knockdown abrogated the Nrf2 phosphorylation induced by Gα12 deficiency, suggesting that relief from Gα12 repression leads to the Gα13-mediated activation of Nrf2. Constitutive activation of Gα13 promoted Nrf2 activity and target gene induction via Rho-mediated PKC δ activation, corroborating positive regulation by Gα13. In summary, Gα12 and Gα13 transmit a JNK-dependent signal for Nrf2 ubiquitination, whereas Gα13 regulates Rho-PKC δ-mediated Nrf2 phosphorylation, which is negatively balanced by Gα12.
G-protein-coupled receptors (GPCRs) transmit signals through conformational changes upon ligand activation and interaction with heterotrimeric GTP-binding proteins (G proteins), leading to the regulation of physiological processes. G proteins consist of α, β, and γ subunits and are defined by the identity of their α subunits, which can be divided into four families: Gαs, Gαi/o, Gαq, and Gα12. Among these, the two members of the Gα12 family, namely, Gα12 and Gα13, are activated by various stimuli, including thrombin, thromboxane A2, lysophosphatidic acid (LPA), and thyroid-stimulating hormone receptors (9, 10, 31, 35, 39). Gα12 and Gα13 have been shown to regulate a variety of cellular processes, such as actin stress fiber formation, neurite retraction, platelet aggregation, and apoptosis (10, 29, 35, 44).
In spite of their mostly overlapping functions, Gα12 and Gα13 seem to differ in their abilities to couple to different ligands and to recruit different signaling pathways for physiological cellular effectors (7, 9, 12, 34, 35, 41, 42). Gene knockout experiments revealed that Gα13 deficiency resulted in impaired angiogenesis and intrauterine death, whereas Gα12−/− mice remained alive. Thus, it is most likely that Gα12 and Gα13 play a role in the cellular signaling pathways linked to the essential biological processes of cell survival. Nevertheless, the regulatory factors controlled by the distinct signaling pathway that is triggered by Gα12 and Gα13 activation have not yet been identified. Our study has shown that oxidative stress induces actin stress fiber formation for NF-E2-related factor-2 (Nrf2) activation (20). It is therefore possible that Gα12 family members function as molecular switches responding to oxidative stress.
Oxidative stress is characterized by high levels of reactive oxygen species, which have damaging effects on cellular components and trigger defensive responses. Nrf2 is a member of the cap'n'collar family of bZIP transcription factors and is expressed widely in a variety of tissues. The transcriptional activation of antioxidant defense genes through an antioxidant response element (ARE) depends on Nrf2 activity. Genes containing a functional ARE(s) include those encoding hemeoxygenase 1 (HO-1), UDP-glucuronosyl transferase 1A (UGT1A), glutathione S-transferase A1/2, NAD(P)H:quinone reductase, and γ-glutamylcysteine synthetase, which play a crucial role in defense against oxidative stress or electrophilic chemicals (30). We previously reported that glutathione depletion or tert-butylhydroquinone (t-BHQ) treatment enhances the nuclear accumulation and DNA binding activity of Nrf2 (17, 18, 20). Nrf2 activation requires phosphorylation at serine-40 by protein kinase C δ (PKC δ) (13, 33, 43). Despite the well-known function of Nrf2, the upstream regulatory effectors that control Nrf2 have not been identified. We hypothesized that Gα12 and Gα13 function as upstream regulators of Nrf2 activity by responding to extracellular stimuli.
To determine whether the Gα12- and Gα13-mediated signaling pathways regulate Nrf2-dependent gene transactivation, we used mouse embryonic fibroblasts (MEFs) and animals lacking Gα12 and/or Gα13. We investigated whether Gα12 and Gα13 transmit signals to the regulatory processes of Nrf2 and, if so, whether they have a distinct or an overlapping role(s). Here, we report that Gα12 and Gα13 coordinately regulate the ubiquitination of Nrf2, a step required for proteasomal degradation, and that they transmit differential signals for Rho-PKC δ-mediated Nrf2 phosphorylation.
MATERIALS AND METHODS
Materials.
Anti-Nrf2, anti-Keap1, anti-Gα12, anti-Gα13, anti-UGT1A, antihemagglutinin (anti-HA), anti-PKC δ, and antiactin antibodies were supplied from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-HO-1 antibody was purchased from Stressgen (San Diego, CA). Horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G was obtained from Zymed Laboratories (San Francisco, CA). Constitutively activated mutants of Gα12 and Gα13 were kindly provided by N. Dhanasekaran (Temple University, Philadelphia, PA). Small interfering RNAs (siRNAs) for Gα12 and Gα13 and nontargeting scrambled RNA (scRNA) were obtained from Dharmacon (Lafayette, CO). Phorbol-12-myristate-13-acetate (PMA), GF109203X, and rottlerin were purchased from Calbiochem (San Diego, CA). Anti-phosphorylated serine antibody, t-BHQ, and other reagents were supplied by Sigma-Aldrich (St. Louis, MO).
Cell culture.
MEFs were generated from genetically engineered mice (40) that contained gene knockouts for rhodopsin kinase (RK) (39) and Gα12 and Gα13 (46). Cells were cultured as described previously (25).
Subcellular fractionations and immunoblot analyses.
Cells were fractionated and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analyses, as described previously (3, 19). Bands were developed using the ECL chemiluminescence system (Amersham, Buckinghamshire, United Kingdom).
siRNA knockdown.
To knock down Gα12 or Gα13, cells were transfected with siRNA directed against mouse Gα12 and Gα13, or a nontargeting scRNA (100 pmol/ml), by using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Twenty-four hours after transfection, the nuclear extracts prepared from the cells were used for immunoblot assays. Gα12 or Gα13 knockdown was confirmed by immunoblot analysis.
Gel shift assay.
A gel shift assay was performed as described previously (25). A double-stranded DNA probe containing the ARE of the GSTA2 gene was used for the gel shift analysis. The sequence of the ARE-containing oligonucleotide was 5′-GATCATGGCATTGCACTAGGTGACAAAGCA-3′ (core sequences are underlined). In some analyses, the specificity of Nrf2 binding to DNA was determined by competition experiments. SP-1 oligonucleotide (5′-ATTCGATCGGGGCGGGGCGAGC-3′) was used as a negative control.
Knockout animals.
Gα12 homozygous knockout mice, described previously (10), were supplied by M. Simon. Genotypes were determined by PCR assays on tail DNA using specific primers for the respective G proteins and conditions previously described (10).
Animal treatment and hematological analysis.
Hemolysis was induced in 8-week-old mice by intraperitoneal injections of phenylhydrazine hydrochloride (PHZ) (Sigma-Aldrich; 30 mg/kg body weight/day for 2 days) dissolved in phosphate-buffered saline (pH 7.4, adjusted with NaOH). Blood was collected by retro-orbital phlebotomy using heparin-treated capillary tubes. Hematological parameters were analyzed by using the ADVIA 1650 chemistry system (Bayer HealthCare, Tarrytown, NY).
Transient-transfection and reporter gene assays.
Transient transfection was performed with Lipofectamine (Invitrogen, Carlsbad, CA). Briefly, cells were incubated with the plasmid of interest and Lipofectamine reagent for 3 h. Culture medium was replaced with minimal essential medium, and cells were further incubated for 24 h to assess gene expression. In reporter gene assays, we used the GSTA2 promoter-luciferase construct pGL-1651 that contains the ARE (25). Specific base substitutions were made by oligonucleotide-mediated mutagenesis according to the manufacturer's instructions (Stratagene). The serine-40 residue of mouse Nrf2 was mutated to alanine using a mutagenic primer (5′-GTGTTTGACTTTGCTCAGCGACAGAAGGAC-3′). The DNA sequence was verified by using an automatic DNA sequence analyzer. Cells were transiently transfected with pGL-1651 for 3 h in the presence of Lipofectamine reagent. The transfected cells were incubated in Dulbecco's modified Eagle's medium containing 1% fetal bovine serum for 3 h and exposed to t-BHQ (10 or 30 μM) for 18 h at 37°C. The activity of luciferase was measured by adding luciferase assay reagent (Promega, Madison, WI).
DNA microarray.
After 24 h of serum deprivation, cells were treated with t-BHQ for 12 h and then harvested with TRIzol. Total RNAs prepared from cells were used for microarray analyses (15). The 16,000 mouse oligonucleotide arrays were inkjet printed by using Agilent Technologies. These arrays include 13,536 70-mers (Operon Technologies) and 2,304 65-mers (Sigma-Genosys). The aminoallyl method was used for the preparation of fluorescently labeled target samples. Three independent experiments with each dye swap measurement were conducted. Following filtering, normalization, and significance analysis of microarray, the profiles of expression changes induced by t-BHQ were calculated as the average log2 values(treated cells divided by the control cells) that were ≥1, which corresponds to twofold expression changes where P is <0.01. The normalized expression data sets were loaded into the Ingenuity software (http://www.ingenuity.com) to identify the significantly upregulated genes with their corresponding changes. In biological network analysis, the submitted genes that are mapped to Nrf2 (the node labeled NFE2L2) in the Ingenuity Pathways Knowledge Base are focused genes, which are regulated by Nrf2.
Real-time PCR analysis.
Total RNA was isolated from cells using the improved single-step method of thiocyanate-phenol-chloroform RNA extraction, and cDNA was synthesized by reverse transcriptase using an oligo(dT) primer. Then, real-time PCR was performed with a Light Cycler 1.5 apparatus (Roche, Mannheim, Germany) using a Light Cycler DNA master SYBR green-I kit according to the manufacturer's instructions. PCR was performed using the selective primers for the genes encoding UGT1A1 (sense, 5′-CCAAGAGTTTGTCTTTCAAC-3′; antisense, 5′-AGAGGCGTTGACATAGGCTT-3′), UGT1A6 (sense, 5′-ATGCCCTGTGGTGTGATCCT-3′; antisense, 5′-AGAGGCGTTGACATAGGCTT-3′), GSTA4 (sense, 5′-ATGTATGCAGAT GGCACCCAGGACCTG-3′; antisense, 5′-GGACAATCCTGACCACCTCAACATAG-3′), GCLM (sense, 5′-CTGCCTTTGCTAAACAATTT-3′; antisense, 5′-TAGACTTGATGATTCCCCTG-3′), NQO1 (sense, 5′-AGGCTGGTTTGAGAGAGT-3′; antisense, 5′-TCTGCATGCTTTCATCTG-3′), HO-1 (sense, 5′-GCTGTGAACTCTGTCCAATG-3′; antisense, 5′-GTCAGTCAACATGGATGCTT-3′) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (sense, 5′-TCGTGGAGTCTACTGGCGT-3′; antisense, 5′-GCCTGCTTCACCACCTTCT-3′). The thermal profile for SYBR green real-time PCR was as follows: 95°C for 10 min, followed by 45 cycles at 95°C for 10 s, 54°C for 10 s, and 72°C for 15 s. A melting-curve analysis was done after amplification to verify the accuracy of the amplicon.
Immunoprecipitation.
To assess Nrf2 ubiquitination, cells were transfected with the plasmid encoding His-tagged ubiquitin. Cell lysates were incubated with anti-His antibody overnight at 4°C. To determine the serine phosphorylation of Nrf2, a fraction of cell lysates was incubated with a polyclonal rabbit anti-Nrf2 antibody overnight at 4°C. The antigen-antibody complex was immunoprecipitated following incubation for 2 h at 4°C with protein G-agarose. Immune complex was solubilized in 2× Laemmli buffer and boiled for 5 min. In another set of assays, His-tagged ubiquitinated proteins were precipitated from cell lysates using Ni2+ affinity beads (Invitrogen). Samples were resolved and analyzed using 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to a nitrocellulose membrane. The samples were then immunoblotted with the antibody directed against phosphorylated serine or Nrf2.
Statistical analysis.
One-way analysis of variance procedures were used to assess significant differences among treatment groups. For each significant effect of treatment, the Newman-Keuls test was used for comparisons of multiple group means. The criterion for statistical significance was set at a P of <0.05 or <0.01.
RESULTS
Nuclear activation of Nrf2 by Gα12 deficiency.
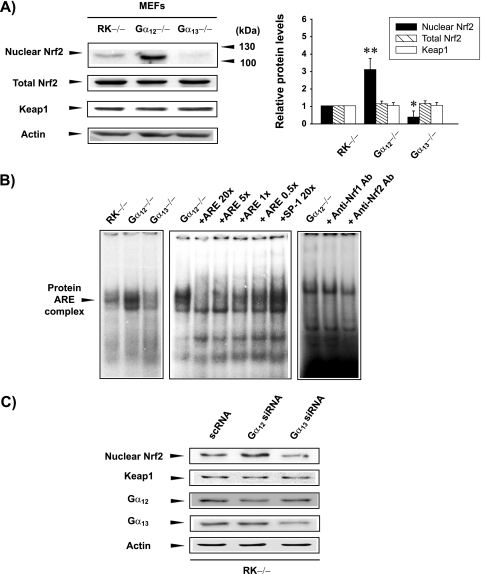
To investigate the regulatory effects of Gα12 and Gα13 on Nrf2, the nuclear accumulation and expression of Nrf2 were monitored in MEFs. In this study, the RK−/− cells were used as a knockout control because the physiological function of RK is restricted to phototransduction. Preliminary experiments showed that Nrf2 responsiveness to a prooxidant in RK−/− cells was identical to that of wild-type (WT) MEFs or hepatocyte-derived cells (HepG2 and H4IIE). We also reported that the Gα12-dependent gene expression pattern in RK−/− cells was identical to that in WT cells (26, 27). Nrf2 levels were minimal in the nuclear fraction of RK−/− cells but high in that of Gα12−/− cells (Fig. 1A). Moreover, the level of nuclear Nrf2 seemed to be lower in Gα13-deficient cells than in the control cells. The absence of Gα12 and Gα13 did not affect the level of Keap1. To determine whether nuclear Nrf2, increased by Gα12 deficiency, is capable of binding to DNA, nuclear extracts prepared from cells were probed with a radiolabeled ARE oligonucleotide (Fig. 1B). An increase in the band intensity of a slowly migrating protein-DNA complex verified that nuclear Nrf2 that accumulated during the absence of Gα12 was active. An excess of the ARE oligonucleotide, but not an excess of SP-1, abolished this band retardation, supporting the specificity of Nrf2 DNA binding. Consistent with the results of gene knockout experiments, siRNA knockdown of Gα12 also resulted in the nuclear accumulation of Nrf2, but knockdown of Gα13 failed to do so. Except for specific knockouts, the expression levels of Nrf2 in the lysates of the cells examined were comparable, suggesting that G proteins might regulate the process of Nrf2 (Fig. 1C).
FIG. 1.
Nrf2 activation by Gα12 deficiency. (A) Nuclear accumulation of Nrf2 by Gα12 deficiency. Nrf2 was immunoblotted in the nuclear or lysate fractions prepared from knockout cells. Keap1 expression was determined in cell lysates. Graph data indicate the means ± standard errors for three separate experiments (asterisks indicate significant differences relative to RK−/− MEFs [*, P < 0.05; **, P < 0.01]). (B) Increase in Nrf2 DNA binding by Gα12 deficiency. The arrowhead in the gel shift analysis indicates a Nrf2 DNA binding complex. Each lane contained 10 μg of nuclear extracts and 5 ng of labeled oligonucleotide. An antibody (Ab) competition experiment was carried out by incubating the samples prepared from Gα12-deficient cells with specific antibodies directed against Nrf1 or Nrf2 (2 μg each). (C) Nuclear accumulation of Nrf2 by Gα12 knockdown. For knockdown experiments, RK−/− cells were transfected with siRNAs directed against Gα12 or Gα13 or nontargeting siRNA. RK−/− cells were used as a knockout control, which is irrelevant to the function of endogenous G protein in MEFs.
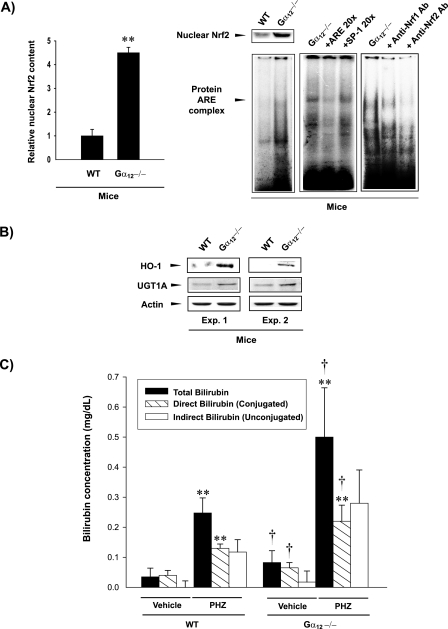
To confirm whether the Gα12 deficiency enhances Nrf2 accumulation and DNA binding activity in vivo, nuclear Nrf2 contents and band intensities of Nrf2 DNA binding complex in the livers of mice carrying targeted mutations of the gene were determined. In Gα12 gene knockout mice, the nuclear localization of hepatic Nrf2 was greatly increased and was accompanied by increased Nrf2 activity compared to that in WT control mice (Fig. 2A). In this series of experiments, we used Gα12 knockout animals because the Gα13 knockout mutation resulted in intrauterine death (40). To assess changes in the representative Nrf2 target genes, we next examined the levels of HO-1 and UGT1A in the livers of the mice. Gα12 gene knockout induced HO-1 and UGT1A in the livers of mice (Fig. 2B), which further supported an in vivo function of Gα12 in Nrf2 regulation. In an effort to assess functional changes for the activation of Nrf2 and its target gene induction, we next determined whether Gα12 deficiency accelerates the metabolism of heme into bilirubin and its glucuronosyl conjugations, which are catalyzed by a series of Nrf2 target genes, including HO-1 and UGT1A (22, 23). As expected, the catabolism of heme into bilirubin and bilirubin glucuronide conjugations were both significantly enhanced by the absence of Gα12 (Fig. 2C). Furthermore, we comparatively evaluated the effects of PHZ on bilirubin production and glucuronosyl conjugations in WT and Gα12 knockout animals because PHZ induces hyperbilirubinemia via chemical hemolysis (Table 1) (36, 38). An intraperitoneal injection of C57BL6 mice with PHZ resulted in marked increases in total and conjugated bilirubin contents in blood (Fig. 2C). As expected, the injection of PHZ into Gα12 knockout mice caused significantly greater increases in total and glucuronidated bilirubin concentrations, indicating that the induction of HO-1 and UGT1A by Gα12 deficiency contributed to the metabolism of a larger amount of heme into bilirubin and the subsequent glucuronosyl conjugations.
FIG. 2.
Nrf2 activation by targeted disruption of the Gα12 gene in mice. (A) Gel shift assay. Nrf2 activation was measured in the livers of Gα12 knockout mice. The specificity of Nrf2 DNA binding was determined by a competition experiment using the unlabeled ARE or SP-1 binding oligonucleotide. An antibody competition experiment was carried out by incubating the samples prepared from Gα12 knockout mice with specific antibodies (2 μg each). Data indicate the means ± standard deviations (three animals) (**, significant compared to WT at a P of <0.01). (B) Induction of HO-1 and UGT1A by targeted disruption of the Gα12 gene in mice. HO-1 and UGT1A were immunoblotted with antibodies in the liver homogenates of the WT or homozygous Gα12 knockout mice (three animals each). Representative blots are shown. (C) Plasma bilirubin levels. Total, direct, and indirect bilirubin contents in the blood of WT or Gα12 knockout mice were determined before and 24 h after PHZ injections (30 mg/kg body weight/day intraperitoneally for 2 days) (seven animals; **, significant compared to the respective parameters of vehicle-treated WT animals at a P of <0.01; †, significant compared to the respective parameters in vehicle- or PHZ-treated WT animals at a P of <0.05).
TABLE 1.
Hemolytic parameters in WT and Gα12 knockout micea
| Mouse genotype and treatment | No. of RBCb (109/ml) | Iron concn (μg/dl) | Body wt (g) | Liver wt/100 g body wt | Spleen wt/100 g body wt |
|---|---|---|---|---|---|
| Wild type | |||||
| Vehicle | 10.67 ± 0.39 | 108 ± 23 | 23.9 ± 2.0 | 3.75 ± 0.42 | 0.23 ± 0.04 |
| PHZ | 6.94 ± 0.73** | 158 ± 52* | 25.9 ± 3.0 | 4.07 ± 0.33 | 0.64 ± 0.23** |
| Gα12−/− | |||||
| Vehicle | 10.11 ± 0.15 | 119 ± 28 | 25.1 ± 1.8 | 3.60 ± 0.29 | 0.26 ± 0.03 |
| PHZ | 6.49 ± 0.88** | 160 ± 27* | 24.0 ± 2.0 | 3.89 ± 0.46 | 0.50 ± 0.11** |
Mice were treated intraperitoneally with PHZ (30 mg/kg body weight/day for 2 days) and subjected to analyses 24 h after treatment. Values are means ± standard deviations. Asterisks represent significant difference from values for vehicle treatment of the same genotype (seven animals; *, P < 0.05; **, P < 0.01).
RBC, red blood cells.
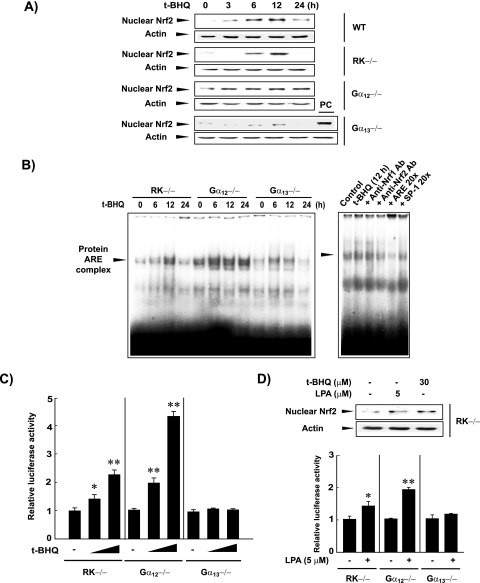
To determine the effects of Gα12 or Gα13 deficiencies on the activation of Nrf2 by a prooxidant, we next examined the effects of t-BHQ on Nrf2 in MEFs. t-BHQ treatment for 6 or 12 h increased nuclear Nrf2 content in WT and RK−/− MEFs (Fig. 3A), which returned to basal levels at 24 h. In Gα12−/− cells, t-BHQ treatment began to enhance nuclear Nrf2 content at 3 h, and this was increased at 6 h to maximum levels, which were maintained for at least 24 h. t-BHQ treatment only weakly increased nuclear Nrf2 levels in Gα13-deficient cells. A greater increase in Nrf2 activation by t-BHQ, under conditions of Gα12 deficiency, was also observed in the gel shift assays (Fig. 3B). Competition experiments using specific antibodies and excessive amounts of unlabeled oligonucleotides confirmed the specificity of the band of the slowly migrating complex.
FIG. 3.
Nrf2 activation and target gene promotion by t-BHQ. (A) Effects of t-BHQ treatment on nuclear accumulation of Nrf2. Nrf2 contents were measured in the nuclear fractions of cells treated with vehicle or t-BHQ (30 μM). A positive control (PC) represents t-BHQ-treated RK−/− cells (12 h). (B) Nrf2 DNA binding activities. Gel shift assays were performed using the nuclear extracts of t-BHQ-treated cells. An antibody competition experiment was carried out by incubating the samples prepared from untreated or t-BHQ-treated cells with specific antibodies directed against Nrf1 or Nrf2 (2 μg each). (C) pGL-1651 reporter assays. Luciferase activity was measured in the lysates of pGL-1651-transfected cells treated with t-BHQ (10 or 30 μM, 18 h). Data indicate the means ± standard errors for four separate experiments (asterisks indicate significant differences relative to the respective control [*, P < 0.05; **, P < 0.01]). (D) Effects of LPA treatment on nuclear Nrf2 levels and pGL-1651 reporter activities. Nrf2 contents and pGL-1651 activities were measured in cells treated with vehicle or 5 μM LPA for 12 h and 24 h, respectively. Data indicate the means ± standard errors for three separate experiments (asterisks indicate significant differences relative to the control [*, P < 0.05; **, P < 0.01]).
As part of our functional studies on the regulation of Nrf2 by Gα12 and Gα13, we examined the transactivation of the pGL-1651 GSTA2 luciferase gene, which contains the ARE. In RK−/− cells, t-BHQ treatment increased luciferase expression by pGL-1651 (Fig. 3C). Luciferase expression was promoted by t-BHQ to a greater extent in Gα12−/− cells than in control or Gα13−/− cells. Our data indicate that Gα12 deficiency enhances the nuclear accumulation of Nrf2 that is active for target gene transactivation. In addition, treatment of the RK−/− cells with LPA (5 μM for 12 h), a representative ligand that activates GPCRs coupled to Gα12 and Gα13, led to nuclear accumulation of Nrf2 (Fig. 3D, gels). This is in line with prooxidant production caused by LPA treatment (6). As expected, LPA treatment induced luciferase expression from pGL-1651 in RK−/− cells (Fig. 3D, graph). Gα12 deficiency further promoted the gene induction by LPA, whereas Gα13 deficiency prevented the gene expression.
Microarray profiles.
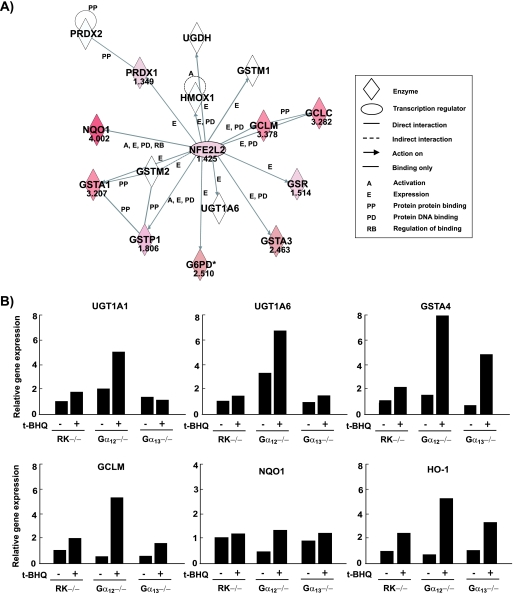
To determine changes in Nrf2-mediated target gene induction, we carried out profiling of t-BHQ-responsive gene expression (12 h) in cells by using oligonucleotide microarrays. The expression of antioxidant and phase II enzymes was enhanced after treating RK−/− cells with t-BHQ. The absence of Gα12 significantly enhanced the ability of t-BHQ to stimulate a battery of Nrf2 target genes compared with that of the control; these genes included Txnrd1, Gsr, Gclm, Gclc, Gsta4, and Gsta2 (Table 2). The smaller changes observed in the relative increases of Gstm1 and UGT1A6 mRNAs by t-BHQ in Gα12−/− cells compared to those in the control may have been due to their higher basal expression (Table 2). Ingenuity pathway analysis in the network validated glutathione and cysteine metabolism as significant pathways potentiated by Gα12 deficiency. Genes not differentially expressed by the absence of Gα12 were omitted from the Ingenuity pathway analysis. All the enhanced genes, including Gsta1, Gsta3, Gsta4, Gstp1, Gclc, Gclm, G6pd, and Gsr, appeared to be regulated by Nrf2 in the map (Fig. 4A).
TABLE 2.
Effects of t-BHQ on the representative antioxidant enzyme gene expression
| Gene | Gene product nameb | Relative change in gene expression in cells with indicated genotypea
|
||
|---|---|---|---|---|
| RK−/− | Gα12−/− | Gα13−/− | ||
| Antioxidant enzyme genes | ||||
| Hmox1 | Heme oxygenase (decycling) 1 | 9.90 | 13.11 | 11.59* |
| Txnrd1 | Thioredoxin reductase 1 | 1.50** | 5.97*† | 2.19* |
| Gsr | Glutathione reductase 1 | 1.16 | 2.85*† | 2.13* |
| Cat | Catalase | 1.18 | 2.20* | 1.38** |
| Prdx1 | Peroxiredoxin 1 | 1.27 | NA | 1.56 |
| Xenobiotic metabolism genes | ||||
| Nqo1 | NAD(P)H dehydrogenase quinone 1 | 4.32 | 16.01** | 3.13** |
| Gclm | Glutamate-cysteine ligase modifier subunit | 2.30* | 10.39**‡ | 2.22** |
| Gclc | Glutamate-cysteine ligase catalytic subunit | 2.16* | 9.72**‡ | 3.27**† |
| Gsta4 | GlutathioneS-transferase alpha 4 | 3.64* | 7.27**† | 3.69** |
| Gsta3 | Glutathione S-transferase alpha 3 | NA | 5.51 | 1.13 |
| Gsta2 | GlutathioneS-transferase alpha 2 (Yc2) | 1.10 | 3.43*† | 2.71*‡ |
| Gstp1 | Glutathione S-transferase pi 1 | 1.37* | 2.40 | 1.43* |
| Gss | Glutathione synthetase | 1.39 | 2.31 | 1.37* |
| Gsto1 | Glutathione S-transferase omega 1 | 1.87** | 1.84 | 1.66* |
| Gstm3 | Glutathione S-transferase mu 3 | 1.51 | 1.82* | 1.88** |
| Gstm1 | GlutathioneS-transferase mu 1c | 2.60** | 1.54† | 3.00** |
| UGT1A6 | UDP glycosyltransferase 1 family A6d | 2.98** | 1.40† | 2.55**† |
| Nqo2 | NAD(P)H dehydrogenase quinone 2 | 1.62* | 1.19 | 1.21 |
| Mgst1 | Microsomal glutathione S-transferase 1 | 2.07* | 1.15 | 1.84* |
| Gstm4 | Glutathione S-transferase mu 4 | 1.09 | 1.14 | 0.94† |
| Gstk1 | Glutathione S-transferase kappa 1 | 0.91 | 1.09 | 0.86 |
| Gstm5 | Glutathione S-transferase mu 5 | 1.03 | 0.93 | 1.03 |
| Mgst3 | Microsomal glutathione S-transferase 3 | 1.03 | 0.92 | 1.21 |
| Gstm6 | Glutathione S-transferase mu 6 | 1.43 | 0.84 | 1.29* |
| Gstz1 | Glutathione S-transferase zeta 1 | 0.95 | 0.82 | 0.87 |
| Gstm2 | Glutathione S-transferase mu 2 | 1.52 | 0.66 | 1.25* |
| Gstt3 | Glutathione S-transferase theta 3 | 1.33 | 0.65 | NA |
| Gstt1 | Glutathione S-transferase theta 1 | 0.78** | 0.50 | 0.81 |
Each value indicates the relative change after t-BHQ treatment (30 μM, 12 h). Asterisks indicate significant difference from value for vehicle treatment (*, P < 0.05; **, P < 0.01). Daggers indicate significant difference from RK−/− values (†, P < 0.05; ‡, P < 0.01). NA, data not available.
Genes for products in bold are Nrf2 dependent.
Basal mRNA content was 2.4-fold greater in Gα12−/− cells than in control cells.
Basal mRNA content was 42-fold greater in Gα12−/− cells than in control cells.
FIG. 4.
Microarray profiles and validation. (A) Biological network analysis of Nrf2 (NFE2L2). This network map depicts the relationship between Nrf2 and Nrf2-regulated genes. Bold characters are focused genes whose t-BHQ-inducible expression (30 μM, 12 h) is potentiated by Gα12 deficiency. A greater intensity of red indicates a higher degree of upregulation. The distance and label between nodes indicate the extent and type of relationship of the interactions involved with Nrf2, respectively. The score of each node is displayed as normalized log2 (treated/control) value. (B) Real-time PCR analyses. The expression of representative Nrf2 target genes was analyzed by real-time PCR assays, in which the mRNA level of GAPDH was used as a reference for data normalization. Cells treated with vehicle or t-BHQ (30 μM, 12 h) were subjected to the preparation of mRNA, from which cDNA was synthesized by reverse transcriptase. Changes (n-fold) were calculated by correlation coefficients of crossing point for triplicate PCR results.
We verified these results by real-time PCR analyses. Relative increases in UGT1A1, UGT1A6, GSTA4, GCLM, NQO1, and HO-1 mRNA levels by t-BHQ were all greater in Gα12−/− cells than in control or Gα13−/− cells (Fig. 4B). Diethylmaleate (1 mM, 6 h) or sulforaphane (10 μM, 6 h) treatment caused similar changes in the genes' expression (data not shown). These data were consistent with the results of the microarrays, confirming that Nrf2 activated by the absence of Gα12 was functionally active in increasing its target gene mRNAs. Immunoblot assays confirmed the greater induction of HO-1 and UGT1A proteins by t-BHQ in Gα12−/− cells (see Fig. S1 in the supplemental material).
Gα12- and Gα13-dependent ubiquitination of Nrf2.
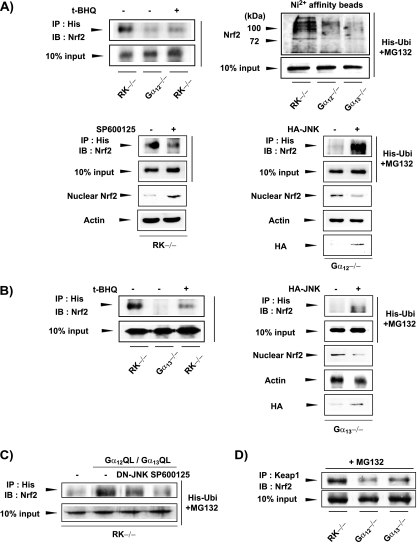
Nrf2 is degraded by the 26S proteasome system after multiple ubiquitinations (37). To understand in more depth the molecular mechanism underlying Nrf2 activation by Gα12 deficiency, we tested the possibility that Gα12 mediates a signal for Nrf2 ubiquitination that would lead to proteasomal degradation. Immunoprecipitation and the immunoblot detection system indicated that the absence of Gα12 basically eliminated Nrf2 ubiquitination compared to that of the control (Fig. 5A, upper left panel). As reported, t-BHQ treatment inhibited Nrf2 ubiquitination in the control cells. Thus, our data suggest that Gα12 contributes to the Nrf2 ubiquitination step. An in vivo ubiquitination assay using a His-tagged ubiquitin construct and Ni2+ affinity beads verified polyubiquitination of Nrf2 in the RK−/− cells (Fig. 5A, upper right panel). Gα12 and Gα13 deficiencies also decreased polyubiquitination of Nrf2. Studies have shown that JNK is involved in the proteasomal degradation of certain transcription factors (1, 11). Previously, we proposed a possible role for Gα12 and Gα13 in the JNK-dependent degradation of phosphorylated IκBα (19). Therefore, we assessed the role of JNK in Nrf2 ubiquitination. The endogenous JNK expression levels were not changed by a deficiency of Gα12 or Gα13 (see Fig. S2A in the supplemental material). In RK−/− cells, the inhibition of JNK using SP600125 reduced the accumulation of ubiquitinated Nrf2 and reciprocally increased nuclear Nrf2 content (Fig. 5A, lower left panel). We then monitored the effect of the ectopic expression of JNK on Nrf2 ubiquitination. When Gα12−/− cells were transfected with a plasmid encoding HA-JNK, the ubiquitination of Nrf2 was substantially increased with a decrease in nuclear Nrf2 levels (Fig. 5A, lower right panel).
FIG. 5.
Effects of Gα12 and Gα13 deficiencies or JNK transfection on Nrf2 ubiquitination. (A) Role of Gα12 on Nrf2 ubiquitination. Cells transfected with the plasmid encoding His-tagged ubiquitin (His-Ubi) were incubated with 10 μM MG132 (a proteasomal inhibitor) for 6 h. When cells were treated with t-BHQ (30 μM, 12 h) or SP600125 (10 μM, 6 h), MG132 was added 1 h earlier. Ubiquitinated proteins were immunoprecipitated (IP) with anti-His antibody in lysates and subjected to immunoblottings (IB) for Nrf2. The nuclear Nrf2 levels were comparatively assessed. The effects of HA-JNK were similarly measured. In addition, whole-cell extracts were incubated overnight with Ni2+ affinity beads. After being washed with lysis buffer, the beads were boiled in the sample buffer and the eluate was immunoblotted for Nrf2 (upper right panel). (B) Role of Gα13 in Nrf2 ubiquitination. Assays were performed as described for panel A. (C) JNK-dependent ubiquitination of Nrf2 increased by activated mutants of Gα12 and Gα13. (D) Interaction of Keap1 and Nrf2. Immunoprecipitated Keap1 was subjected to immunoblotting for Nrf2.
We performed similar experiments using Gα13−/− cells and unexpectedly found that Gα13 deficiency also inhibited Nrf2 ubiquitination (Fig. 5B). Transfection of Gα13−/− cells with HA-JNK allowed Nrf2 ubiquitination, which was less pronounced than that seen in transfected Gα12−/− cells (Fig. 5B). Also, we found that constitutive activation of Gα12 (Gα12Q229Lor Gα12QL) and Gα13 (Gα13Q226L or Gα13QL) notably enhanced Nrf2 ubiquitination in RK−/− cells, which was prevented by either transfection with a dominant negative mutant of JNK (DN-JNK) or chemical inhibition of JNK (Fig. 5C). In this experiment, the cells were simultaneously transfected with Gα12QL and Gα13QL because both are involved in the ubiquitination step. Collectively, these findings indicate that the signaling pathways transmitted by both Gα12 and Gα13 regulate Nrf2 ubiquitination in conjunction with JNK, thereby presumably controlling Nrf2 degradation. Keap1 is required for the formation of the proteasomal complex that degrades Nrf2. Next, we determined the interaction between Keap1 and Nrf2. Keap1 binding to Nrf2 was decreased notably by Gα12 deficiency and moderately by Gα13 deficiency (Fig. 5D), supporting the concept that the G proteins regulate the ubiquitination and degradation of Nrf2. Given the reciprocity of the changes in the levels of ubiquitinated and activated Nrf2, we sought to investigate the activating process of Nrf2 that is possibly linked to ubiquitin-mediated proteasomal degradation.
Differential activation of Nrf2 by Gα12 and Gα13.
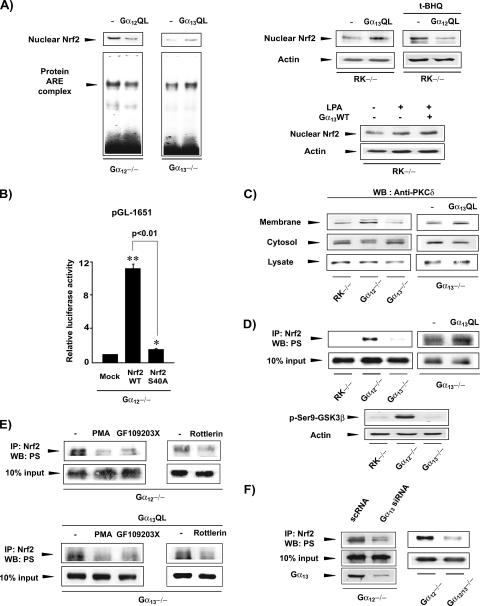
Our finding of Nrf2 activation by Gα12 deficiency suggested that Gα12 represses Nrf2 activation such that a lack of Gα12 facilitates an activating signal. Hence, we reasoned that Gα12 negatively regulates the Nrf2-activating signal transmitted from Gα13. Accordingly, we examined the effect of the transfection of an activated mutant form of Gα12 on Nrf2 activation in cells lacking G protein. The transfection of Gα12QL into Gα12−/− cells decreased nuclear Nrf2 content and DNA binding activity (Fig. 6A), whereas the transfection of Gα13QL into Gα13−/− cells notably increased both. In the rescue experiments using RK−/− cells, Gα13QL increased the basal nuclear Nrf2 content, whereas Gα12QL transfection decreased t-BHQ-inducible nuclear accumulation of Nrf2 (Fig. 6A). Gα13WT transfection in combination with LPA enhanced nuclear Nrf2 accumulation.
FIG. 6.
Regulation of Nrf2 by Gα12QL or Gα13QL. (A) Nuclear contents and DNA binding activities of Nrf2. Immunoblot and gel shift assays were performed with the nuclear extracts prepared from cells transfected with empty vector, Gα12QL, or Gα13QL. Cells were treated with 30 μM t-BHQ or 5 μM LPA for 12 h. (B) pGL-1651 reporter assays. Luciferase activity was measured in the lysates of Gα12-deficient cells transfected with pGL-1651 with or without the plasmid encoding Nrf2 or mutant Nrf2 (serine-40 replaced with alanine). Data indicate the means ± standard errors for four separate experiments (asterisks indicate significant differences relative to the respective control [*, P < 0.05; **, P < 0.01]). (C) PKC δ activation. PKC δ was immunoblotted (Western blotted [WB]) in subcellular fractions. (D) Nrf2 phosphorylation. Serine-phosphorylated Nrf2 was immunoblotted with antiserine antibody in Nrf2 immunoprecipitates. Phosphorylation of glycogen synthase kinase 3β was assessed as a positive control for PKC δ activity. IP, immunoprecipitation; PS, phosphoserine. (E) Effects of PKC inhibition on Nrf2 phosphorylation. Serine phosphorylation of Nrf2 was assessed in cells treated with vehicle, PMA (10 μM, 18 h), GF109203X (5 μM, 6 h), or rottlerin (5 μM, 6 h). (F) Role of Gα13 in Nrf2 phosphorylation by Gα12 deficiency. Serine phosphorylation of Nrf2 was determined in the knockdown or knockout cells indicated.
PKC δ is responsible for Nrf2 phosphorylation at a serine residue (serine-40), which is required for Nrf2 activation (11). In Gα12−/− cells, Nrf2 transfection greatly enhanced luciferase expression from pGL-1651. In contrast, transfection with mutant Nrf2 (where serine-40 is replaced with alanine) failed to induce the gene (Fig. 6B). These data support the association of Gα12 and Gα13 with Nrf2 phosphorylation. In an attempt to identify the downstream effector required for Nrf2 phosphorylation, we investigated whether cell signals from Gα12 and Gα13 regulate PKC δ activation. A deficiency of Gα12, but not of Gα13, caused PKC δ activation, as evidenced by its translocation from the cytoplasm to the plasma membrane (Fig. 6C). However, the expression of PKC δ was unchanged in cell lysates. To examine whether Gα13 positively regulates the signal for Nrf2 activation, the effect of Gα13QL transfection on PKC δ activation in Gα13−/− cells was examined−/−. It was observed that Gα13QL transfection increased PKC δ content in the membrane fraction and that this occurred with a commensurate decrease of PKC δ content in the cytoplasm. Our data indicate that either a Gα12 deficiency or Gα13 activation promotes PKC δ activation.
To determine whether PKC δ activates Nrf2 in these cells, we examined the serine phosphorylation of Nrf2. The absence of Gα12 distinctly increased Nrf2 phosphorylation at the serine residue, whereas the absence of Gα13 failed to do so (Fig. 6D, upper panel). Glycogen synthase kinase 3β, another substrate of PKC δ, was also phosphorylated by Gα12 deficiency but not by Gα13 deficiency (Fig. 6D, lower panel).
Consistent with PKC δ activation, Gα13QL transfection increased Nrf2 phosphorylation. To link PKC δ activation and Nrf2 phosphorylation, we investigated Nrf2 phosphorylation after PKC inhibition using PMA and GF109203X (a selective PKC inhibitor), both of which abolished Nrf2 phosphorylation enhanced by Gα12 deficiency (Fig. 6E). Moreover, rottlerin (a specific PKC δ inhibitor) had the same effect. Gα13QL-induced Nrf2 phosphorylation was also blocked by the chemical inhibition of PKC activity.
We wondered whether Nrf2 activation by Gα12 deficiency resulted from an activating signal transmitted from Gα13 due to relief from Gα12-mediated inhibition. To determine whether Nrf2 activation that is increased by Gα12 deficiency is mediated by Gα13, we examined the effects of Gα13 knockout or knockdown on PKC-dependent Nrf2 phosphorylation. Serine phosphorylation of Nrf2 in Gα12−/− cells was substantially reduced by siRNA knockdown of Gα13 (Fig. 6F). Consistently, the level of Nrf2 phosphorylation was much lower in Gα12 and Gα13 (Gα12/13) double-knockout cells than in Gα12 knockout cells. Our data support the conclusion that a signal transmitted from Gα13 regulates PKC δ-dependent Nrf2 phosphorylation and that this is counter balanced by Gα12.
Rho-mediated Nrf2 activation by Gα13QL during Gα12 deficiency.
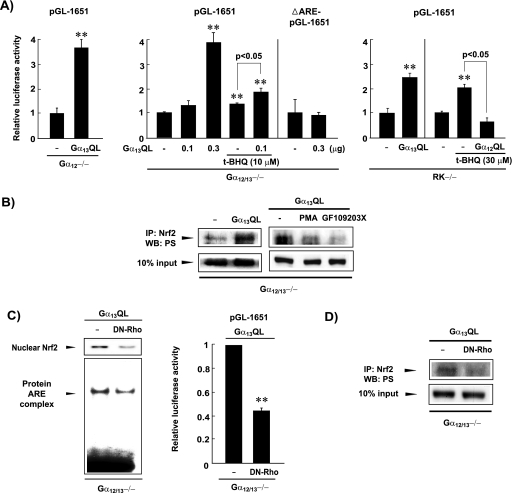
The finding that Gα13 transmits a regulatory signal for Nrf2 activity prompted us to confirm whether Gα13 activation could further increase ARE-mediated gene transactivation during Gα12 deficiency. Gα13QL transfection into Gα12 knockout or Gα12/13 double-knockout cells strongly enhanced luciferase expression from pGL-1651 (Fig. 7A, left). In the Gα12/13−/− cells, Gα13 activation further increased t-BHQ-inducible expression of the Nrf2 target gene. Deletion of the ARE site in pGL-1651 completely abolished reporter gene activity, verifying the essential role of the ARE for Nrf2-mediated gene activation (Fig. 7A, middle panel). In RK−/− cells, Gα13QL also induced pGL-1651 transactivation, whereas Gα12QL prevented the ability of t-BHQ to induce pGL-1651 transactivation (Fig. 7A, right panel). Immunoblot analysis confirmed that Gα13QL transfection into the Gα12/13−/− cells further promoted both PKC δ activation (data not shown) and PKC-mediated Nrf2 phosphorylation, compared with empty-vector transfection (Fig. 7B). Moreover, the strong promotion of Nrf2 target genes by Gα13QL in the double-knockout cells supports the conclusion that Gα13 indeed mediates a positive signal for Nrf2 activation.
FIG. 7.
Rho-mediated Nrf2 activation by Gα13QL during Gα12 deficiency. (A) pGL-1651 reporter activities. Luciferase assays were performed with the lysates of cells transfected with Gα13QL or Gα12QL (0.3 μg plasmid each). To assess the combined effect of Gα13QL and t-BHQ, Gα12/13−/− cells transfected with 0.1 μg Gα13QL were treated with 10 μM t-BHQ. Data represent the means ± standard errors for four separate experiments (**, significant compared to control at a P of <0.01). (B) PKC δ activation by Gα13QL in Gα12/13−/− cells. (C) Effects of DN-Rho on Nrf2 activation and luciferase induction by Gα13QL. (D) Effect of DN-Rho on serine phosphorylation of Nrf2 by Gα13QL. IP, immunoprecipitation; WB, Western blotting.
Rho is a central molecular effector which is involved in a variety of cellular functions and can be regulated by many receptors. Since Gα13 activates Rho (4, 9), we studied the role of Rho in Nrf2 activation by Gα13QL. Transfection with DN-Rho inhibited the ability of Gα13QL to activate Nrf2 or to induce luciferase from pGL-1651 in Gα12/13−/− cells (Fig. 7C). Consistent with this, DN-Rho reversed the serine phosphorylation of Nrf2 by Gα13QL (Fig. 7D). The basal Rho expression levels were very weak but comparable among the MEFs (data not shown). Our data indicate that the Gα13 pathway to Nrf2 activation involves Rho. Taken together, these observations indicate that PKC δ-mediated Nrf2 phosphorylation and its target gene transcription require Rho activity downstream of Gα13.
DISCUSSION
Increases in sensitivity to carcinogen and tumorigenesis in Nrf2-deficient mice support the concept that Nrf2-mediated gene transcription is essential for the prevention of chemical carcinogenesis by cytoprotective agents. Nrf1 and Nrf2 also play critical roles in fetal development and antioxidant defense (32). The main function of Nrf2 is to protect cells from oxidative stress (47). Here, we report the differential role of Gα12 and Gα13 in terms of the regulation of Nrf2. Our knockout and knockdown results revealed the inhibitory role of Gα12 in Nrf2 activity, which was further supported by the finding that Gα12QL reversed Nrf2 activation and target gene induction due to Gα12 deficiency. The observation that HO-1 protein, whose expression depends on Nrf2, was markedly induced in Gα12−/− cells verifies the functional activation of Nrf2 by Gα12 deficiency. Furthermore, hepatic induction of HO-1 and UGT1A in Gα12 knockout mice provided compelling evidence that Gα12 regulates Nrf2 in vivo, demonstrating the physiological importance of Gα12 in Nrf2 activity. Moreover, the finding that the basal and hemolysis-inducible bilirubin production and glucuronosyl conjugations were both elevated during Gα12 deficiency implies that the specific G proteins physiologically contribute to heme catabolism and metabolic excretion of bilirubin via Nrf2 regulation.
Microarray and real-time PCR results demonstrate that Gα12 deficiency markedly enhanced the ability of t-BHQ, an Nrf2 activator, to transactivate Nrf2 target genes. Quantitative real-time PCR assays validated the microarray results that provide only predictive values. In the PCR assays, UGT1A1, 1A6, GCLM, and NQO1 gene transcripts were included because the genes comprised the functional AREs (our customer-made microarrays did not cover all of the genes containing functional AREs). The observation showing strong induction in the genes participating in glutathione metabolism (i.e., Gsta1, Gsta3, Gsta4, Gstp1, Gclc, Gclm, G6pd, and Gsr) (14) supports the significant role of Gα12 in Nrf2 activity. Canonical pathway analysis successfully confirmed glutathione and cysteine metabolism as the major pathways influenced by Gα12 deficiency in combination with t-BHQ treatment. Changes in the levels of UGT1A1, 1A6, and GCLM transcripts correlate well with those in pGL-1651 expression. In this work, t-BHQ was used to promote the expression of target genes because Gα12 deficiency alone affected only a few. Weak activation of Nrf2 by t-BHQ in Gα13−/− cells suggested that Gα13 positively regulates a cell signal for Nrf2 activation. Nevertheless, t-BHQ slightly enhanced Nrf2 activity in Gα13−/− cells, suggesting the role of another minor pathway(s) that might stimulate Nrf2 during the absence of Gα13. The lack of a repressing effect on certain genes (i.e., the GSTA4 and HO-1 genes) by Gα13 deficiency might be due to possible changes in other transcription factors.
Nrf2 and Keap1 are sulfhydryl-containing sensor molecules that respond to oxidative stress (47). Moreover, the Kelch repeat of Keap1 interacts with Neh2 of Nrf2 (13, 37). Initially, it was believed that Keap1 binds with and sequesters Nrf2 in the cytoplasm for repression (16). However, it is now accepted that Keap1 degrades Nrf2 in the resting state and that a decrease in Keap1 activity by prooxidant leads to Nrf2 activation. In the present study, Keap1 expression was unchanged by Gα12 and Gα13 deficiencies, suggesting that the nuclear accumulation of Nrf2 during the absence of Gα12 is not due to changes in Keap1 expression. The actin architecture provides scaffolding for Keap1 and thus controls Nrf2 function (21). Lysine residues within Nrf2 constitute the acceptor sites for Keap1-targeted Nrf2 ubiquitination or stabilization (28). According to our additional studies, nuclear Keap1 content is slightly reduced in the absence of Gα12. Moreover, Keap1 binding to Nrf2 was decreased by deficiencies in Gα12 and Gα13 (Fig. 5D). Therefore, there remains a possibility that Keap1 activity might change in association with a signal transmitted by Gα12.
JNK activation increases E3 ligase activity, enhancing the ubiquitination of target proteins, such as c-Jun and c-FLIP (5, 8). JNK-mediated phosphorylation then enhances c-Jun degradation by allowing its recognition by E3 ligase protein complex (8). Here, we show that Gα12 deficiency reduces JNK-dependent Nrf2 ubiquitination, indicating that Gα12 deficiency disrupts the ubiquitin-mediated proteasomal degradation of Nrf2. Our findings that JNK inhibition resulted in the same inhibitory effect on Nrf2 ubiquitination as Gα12 deficiency and that JNK transfection into Gα12−/− cells restored ubiquitination support the hypothesis that Gα12 regulates the JNK-dependent ubiquitination process. We recently reported that Gα12 deficiency does not allow JNK to be activated in response to sphingosine 1-phosphate (26), consequently decreasing IκBα ubiquitination. Moreover, the finding that Gα13 deficiency also inhibited Nrf2 ubiquitination supports the concept that both Gα12 and Gα13 are necessary for this JNK-dependent process.
Nrf2 activation by t-BHQ may be associated with JNK activation (24). In RK−/− cells, the chemical inhibition of JNK abolished t-BHQ's activation of Nrf2 (see Fig. S2B in the supplemental material). However, our findings showing decreases by JNK overexpression in the increased nuclear Nrf2 content in Gα12−/− or Gα13−/− cells suggest that JNK may not mediate Nrf2 phosphorylation but may regulate the ubiquitination process. Thus, it is unlikely that JNK is directly involved in Nrf2 phosphorylation caused by Gα12 deficiency. It has been shown that Nrf2 undergoes 26S-mediated proteosomal degradation but is stabilized by t-BHQ treatment (37). The present study confirms the inhibitory effect of t-BHQ on Nrf2 ubiquitination. On the basis of the data taken together, it can be speculated that t-BHQ activates Gα13 with relief from the inhibition by Gα12, thereby leading to Nrf2 activation and target gene induction.
The present study shows that Gα12 deficiency promotes PKC δ activation and the serine phosphorylation of Nrf2. In addition, it was observed that the level of nuclear Nrf2 increases when its ubiquitination decreases due to the absence of Gα12 and vice versa, which suggests that the ubiquitination of Nrf2 is antagonistically coupled with its activation. Furthermore, our data, which show that Gα13 knockdown or knockout eliminates Nrf2 activation enhanced by Gα12 deficiency, support the conclusion that Nrf2 activation during the absence of Gα12 is mediated by Gα13, thus verifying the antagonistic balance between Gα12 and Gα13. Therefore, it is plausible that the downstream signal required for the phosphorylation and nuclear accumulation of Nrf2 might be relieved by Gα12 inhibition.
Our data indicate that the Gα13 pathway regulates Nrf2 phosphorylation via PKC δ activation. The observation that Gα13 deficiency abrogated the increase in Nrf2 activation due to the absence of Gα12 demonstrates the crucial role of Gα13 in Nrf2 activation. The mechanism underlying the Gα12-mediated repression of Nrf2 activity induced by Gα13 is not yet understood. Here, we show that Nrf2 serine phosphorylation was further increased by Gα13QL in Gα12−/− and in Gα12/13−/− cells, verifying that Gα13 activation enhances Nrf2 phosphorylation even in the absence of Gα12. Therefore, we conclude that Gα13 controls Nrf2 activation. Gαq is involved in the phosphorylation of target proteins via PKC, which suggests that, functionally, Gα12 and Gαq overlap to some extent (2, 48). Moreover, because of cross talk between Gα12 and Gαq, there remains a possibility that the inhibition of Gα12 causes the stimulation of PKC in association with Gαq.
External stimuli mediated by GPCRs and cell adhesion activate Rho (45). Rho is located in the cytoplasm as a complex with GDI in the resting state and dissociates from GDI prior to membrane translocation. Moreover, despite the functional overlap between Gα12 and Gα13, only Gα13 stimulated the GDP-GTP exchange of RhoA through p115RhoGEF (12). The present study shows that Gα13 regulates Nrf2 via a Rho-mediated signaling pathway.
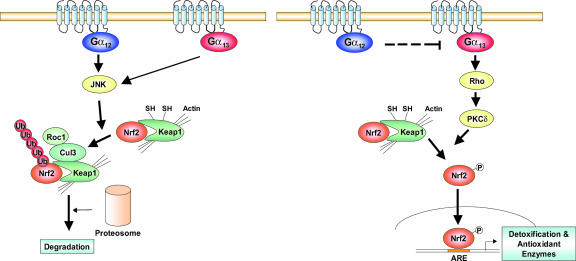
One important result which substantiates our hypothesis that Gα13 regulates Nrf2 via Rho is that DN-Rho transfection into Gα12/13−/− cells completely inhibited the ability of Gα13QL to activate Nrf2 or to promote ARE-mediated gene induction. Moreover, Gα12 has been shown to block Gα13 activation of p115RhoGEF (12), which in conjunction with our results supports the notion that Rho serves as a point of convergence in Nrf2 signaling downstream of Gα12 and Gα13. Based on the present work, we conclude that both Gα12 and Gα13 regulate JNK-dependent Nrf2 ubiquitination but that only Gα13 promotes Nrf2 activity (Fig. 8). Our proposed model is that Nrf2 phosphorylation results from Rho-dependent activation of PKC δ, which is mediated by activating Gα13 and inversely by activating Gα12.
FIG. 8.
Schematic diagrams illustrating the proposed signaling mechanisms by which Gα12 and Gα13 regulate Nrf2 for target gene transactivation.
Supplementary Material
Acknowledgments
This work was supported by Korea Research Foundation grant KRF-2004-015-E00096 (S.G.K.), in part by NIH grant R37GM024236 (M. I. Simon), and by an Ajou University internal research grant (2006; S.C.).
We are grateful to S. Offermanns for helpful discussion regarding this paper.
Footnotes
Published ahead of print on 25 June 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alarcon-Vargas, D., and Z. Ronai. 2004. c-Jun-NH2 kinase (JNK) contributes to the regulation of c-Myc protein stability. J. Biol. Chem. 279:5008-5016. [DOI] [PubMed] [Google Scholar]
- 2.Ally, R. A., K. L. Ives, E. Traube, I. Eltounsi, P. W. Chen, P. J. Cahill, J. F. Battey, M. R. Hellmich, and G. S. Kroog. 2003. Agonist- and protein kinase C-induced phosphorylation have similar functional consequences for gastrin-releasing peptide receptor signaling via Gq. Mol. Pharmacol. 64:890-904. [DOI] [PubMed] [Google Scholar]
- 3.Banan, A., J. Z. Fields, A. Farhadi, D. A. Talmage, L. Zhang, and A. Keshavarzian. 2002. Activation of delta-isoform of protein kinase C is required for oxidant-induced disruption of both the microtubule cytoskeleton and permeability barrier of intestinal epithelia. J. Pharmacol. Exp. Ther. 303:17-28. [DOI] [PubMed] [Google Scholar]
- 4.Buhl, A. M., N. L. Johnson, N. Dhanasekaran, and G. L. Johnson. 1995. G alpha 12 and G alpha 13 stimulate Rho-dependent stress fiber formation and focal adhesion assembly. J. Biol. Chem. 270:24631-24634. [DOI] [PubMed] [Google Scholar]
- 5.Chang, L., H. Kamata, G. Solinas, J. L. Luo, S. Maeda, K. Venuprasad, Y. C. Liu, and M. Karin. 2006. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell 124:601-613. [DOI] [PubMed] [Google Scholar]
- 6.Fischer, O. M., S. Giordano, P. M. Comoglio, and A. Ullrich. 2004. Reactive oxygen species mediate Met receptor transactivation by G protein-coupled receptors and the epidermal growth factor receptor in human carcinoma cells. J. Biol. Chem. 279:28970-28978. [DOI] [PubMed] [Google Scholar]
- 7.Fukuhara, S., H. Chikumi, and J. S. Gutkind. 2001. RGS-containing RhoGEFs: the missing link between transforming G proteins and Rho? Oncogene 20:1661-1668. [DOI] [PubMed] [Google Scholar]
- 8.Gao, M., T. Labuda, Y. Xia, E. Gallagher, D. Fang, Y. C. Liu, and M. Karin. 2004. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306:271-275. [DOI] [PubMed] [Google Scholar]
- 9.Gohla, A., R. Harhammer, and G. Schultz. 1998. The G-protein G13 but not G12 mediates signaling from lysophosphatidic acid receptor via epidermal growth factor receptor to Rho. J. Biol. Chem. 273:4653-4659. [DOI] [PubMed] [Google Scholar]
- 10.Gu, J. L., S. Müller, V. Mancino, S. Offermanns, and M. I. Simon. 2002. Interaction of G alpha(12) with G alpha(13) and G alpha(q) signaling pathways. Proc. Natl. Acad. Sci. USA 99:9352-9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habelhah, H., S. Takahashi, S. G. Cho, T. Kadoya, T. Watanabe, and Z. Ronai. 2004. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. EMBO J. 23:322-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart, M. J., X. Jiang, T. Kozasa, W. Roscoe, W. D. Singer, A. G. Gilman, P. C. Sternweis, and G. Bollag. 1998. Direct stimulation of the guanine nucleotide exchange activity of p115 RhoGEF by Galpha13. Science 280:2112-2114. [DOI] [PubMed] [Google Scholar]
- 13.Huang, H. C., T. Nguyen, and C. B. Pickett. 2002. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 277:42769-42774. [DOI] [PubMed] [Google Scholar]
- 14.Huang, Y., J. Yan, R. Lubet, T. W. Kensler, and T. R. Sutter. 2006. Identification of novel transcriptional networks in response to treatment with the anticarcinogen 3H-1,2-dithiole-3-thione. Physiol. Genomics 24:144-153. [DOI] [PubMed] [Google Scholar]
- 15.Hwang, J. I., S. Choi, I. D. Fraser, M. S. Chang, and M. I. Simon. 2005. Silencing the expression of multiple Gbeta-subunits eliminates signaling mediated by all four families of G proteins. Proc. Natl. Acad. Sci. USA 102:9493-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh, K., N. Wakabayashi, Y. Katoh, T. Ishii, K. Igarashi, J. D. Engel, and M. Yamamoto. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 13:76-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, K. W., M. K. Cho, C. H. Lee, and S. G. Kim. 2001. Activation of phosphatidylinositol 3-kinase and Akt by tert-butylhydroquinone is responsible for antioxidant response element-mediated rGSTA2 induction in H4IIE cells. Mol. Pharmacol. 59:1147-1156. [DOI] [PubMed] [Google Scholar]
- 18.Kang, K. W., S. Y. Choi, and S. G. Kim. 2002. Peroxynitrite activates NF-E2-related factor 2/antioxidant response element through the pathway of phosphatidylinositol 3-kinase: the role of nitric oxide synthase in rat glutathione S-transferase A2 induction. Nitric Oxide 7:244-253. [DOI] [PubMed] [Google Scholar]
- 19.Kang, K. W., S. Y. Choi, M. K. Cho, C. H. Lee, and S. G. Kim. 2003. Thrombin induces nitric-oxide synthase via Galpha12/13-coupled protein kinase C-dependent I-kappaBalpha phosphorylation and JNK-mediated I-kappaBalpha degradation. J. Biol. Chem. 278:17368-17378. [DOI] [PubMed] [Google Scholar]
- 20.Kang, K. W., S. J. Lee, J. W. Park, and S. G. Kim. 2002. Phosphatidylinositol 3-kinase regulates nuclear translocation of NF-E2-related factor 2 through actin rearrangement in response to oxidative stress. Mol. Pharmacol. 62:1001-1010. [DOI] [PubMed] [Google Scholar]
- 21.Kang, M. I., A. Kobayashi, N. Wakabayashi, S. G. Kim, and M. Yamamoto. 2004. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. USA 101:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan, M., C. Hammerman, F. F. Rubaltelli, M. T. Vilei, E. Levy-Lahad, P. Renbaum, H. J. Vreman, D. K. Stevenson, and M. Muraca. 2002. Hemolysis and bilirubin conjugation in association with UDP-glucuronosyltransferase 1A1 promoter polymorphism. Hepatology 35:905-911. [DOI] [PubMed] [Google Scholar]
- 23.Kappas, A., C. S. Simionatto, G. S. Drummond, S. Sassa, and K. E. Anderson. 1985. The liver excretes large amounts of heme into bile when heme oxygenase is inhibited competitively by Sn-protoporphyrin. Proc. Natl. Acad. Sci. USA 82:896-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keum, Y. S., Y. H. Han, C. Liew, J. H. Kim, C. Xu, X. Yuan, M. P. Shakarjian, S. Chong, and A. N. Kong. 2006. Induction of heme oxygenase-1 (HO-1) and NAD[P]H: quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA) and its metabolite, tert-butylhydroquinone (tBHQ) in primary-cultured human and rat hepatocytes. Pharm. Res. 23:2586-2594. [DOI] [PubMed] [Google Scholar]
- 25.Ki, S. H., I. J. Cho, D. W. Choi, and S. G. Kim. 2005. Glucocorticoid receptor (GR)-associated SMRT binding to C/EBPβ TAD and Nrf2 Neh4/5: role of SMRT recruited to GR in GSTA2 gene repression. Mol. Cell. Biol. 25:4150-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ki, S. H., M. J. Choi, C. H. Lee, and S. G. Kim. 2007. Galpha12 specifically regulates COX-2 induction by sphingosine 1-phosphate. Role for JNK-dependent ubiquitination and degradation of IkappaBalpha. J. Biol. Chem. 282:1938-1947. [DOI] [PubMed] [Google Scholar]
- 27.Kim, M.-S., S. M. Lee, W. D. Kim, S. H. Ki, M. Aree, C. H. Lee, and S. G. Kim. 2007. Galpha12/13 basally regulates p53 through Mdm4 expression. Mol. Cancer Res. 5:473-484. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi, A., M.-I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kranenburg, O., M. Poland, F. P. Van Horck, D. Drechsel, A. Hall, and W. H. Moolenaar. 1999. Activation of RhoA by lysophosphatidic acid and Galpha12/13 subunits in neuronal cells: induction of neurite retraction. Mol. Biol. Cell 10:1851-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwak, M. K., K. Itoh, M. Yamamoto, T. R. Sutter, and T. W. Kensler. 2001. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1,2-dimethiole-3-thione. Mol. Med. 7:135-145. [PMC free article] [PubMed] [Google Scholar]
- 31.Laugwitz, K. L., A. Allgeier, S. Offermanns, K. Spicher, J. Van Sande, J. E. Dumont, and G. Schultz. 1996. The human thyrotropin receptor: a heptahelical receptor capable of stimulating members of all four G protein families. Proc. Natl. Acad. Sci. USA 93:116-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung, L., M. Kwong, S. Hou, C. Lee, and J. Y. Chan. 2003. Deficiency of the Nrf1 and Nrf2 transcription factors results in early embryonic lethality and severe oxidative stress. J. Biol. Chem. 278:48021-48029. [DOI] [PubMed] [Google Scholar]
- 33.Li, B., X. Wang, N. Rasheed, Y. Hu, S. Boast, T. Ishii, K. Nakayama, K. I. Nakayama, and S. P. Goff. 2004. Distinct roles of c-Abl and Atm in oxidative stress response are mediated by protein kinase Cdelta. Genes Dev. 18:1824-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao, J., H. Yuan, W. Xie, M. I. Simon, and D. Wu. 1998. Specific involvement of G proteins in regulation of serum response factor-mediated gene transcription by different receptors. J. Biol. Chem. 273:27118-27123. [DOI] [PubMed] [Google Scholar]
- 35.Moers, A., B. Nieswandt, S. Massberg, N. Wettschureck, S. Gruner, I. Konrad, V. Schulte, B. Aktas, M. P. Gratacap, M. I. Simon, M. Gawaz, and S. Offermanns. 2003. G13 is an essential mediator of platelet activation in hemostasis and thrombosis. Nat. Med. 9:1418-1422. [DOI] [PubMed] [Google Scholar]
- 36.Nakano, M., K. Ohneda, H. Yamamoto-Mukai, R. Shimizu, O. Ohneda, S. Ohmura, M. Suzuki, S. Tsukamoto, T. Yanagawa, H. Yoshida, Y. Takakuwa, and M. Yamamoto. 2005. Transgenic over-expression of GATA-1 mutant lacking N-finger domain causes hemolytic syndrome in mouse erythroid cells. Genes Cells 10:47-62. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen, T., P. J. Sherratt, H. C. Huang, C. S. Yang, and C. B. Pickett. 2003. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 278:4536-4541. [DOI] [PubMed] [Google Scholar]
- 38.Nicolas, G., C. Chauvet, L. Viatte, J. L. Danan, X. Bigard, I. Devaux, C. Beaumont, A. Kahn, and S. Vaulont. 2002. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J. Clin. Investig. 110:1037-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Offermanns, S., K. L. Laugwitz, K. Spicher, and G. Schultz. 1994. G proteins of the G12 family are activated via thromboxane A2 and thrombin receptors in human platelets. Proc. Natl. Acad. Sci. USA 91:504-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Offermanns, S., V. Mancino, J. P. Revel, and M. I. Simon. 1997. Vascular system defects and impaired cell chemokinesis as a result of Galpha13 deficiency. Science 275:533-536. [DOI] [PubMed] [Google Scholar]
- 41.Radhika, V., J. Hee Ha, M. Jayaraman, S. T. Tsim, and N. Dhanasekaran. 2005. Mitogenic signaling by lysophosphatidic acid (LPA) involves Galpha12. Oncogene 24:4597-4603. [DOI] [PubMed] [Google Scholar]
- 42.Riobo, N. A., and D. R. Manning. 2005. Receptors coupled to heterotrimeric G proteins of the G12 family. Trends Pharmacol. Sci. 26:146-154. [DOI] [PubMed] [Google Scholar]
- 43.Rushworth, S. A., R. M. Ogborne, C. A. Charalambos, and M. A. O'Connell. 2006. Role of PKCδ in curcumin-induced antioxidant response element-mediated gene expression in human monocytes. Biochem. Biophys. Res. Commun. 341:1007-1016. [DOI] [PubMed] [Google Scholar]
- 44.Sah, V. P., T. M. Seasholtz, S. A. Sagi, and J. H. Brown. 2000. The role of Rho in G protein-coupled receptor signal transduction. Annu. Rev. Pharmacol. Toxicol. 40:459-489. [DOI] [PubMed] [Google Scholar]
- 45.Sinnett-Smith, J., J. A. Lunn, D. Leopoldt, and E. Rozengurt. 2001. Y-27632, an inhibitor of Rho-associated kinases, prevents tyrosine phosphorylation of focal adhesion kinase and paxillin induced by bombesin: dissociation from tyrosine phosphorylation of p130(CAS). Exp. Cell Res. 266:292-302. [DOI] [PubMed] [Google Scholar]
- 46.Vogt, S., R. Grosse, G. Schultz, and S. Offermanns. 2003. Receptor-dependent RhoA activation in G12/G13-deficient cells: genetic evidence for an involvement of Gq/G11. J. Biol. Chem. 278:28743-28749. [DOI] [PubMed] [Google Scholar]
- 47.Wakabayashi, N., A. T. Dinkova-Kostova, W. D. Holtzclaw, M. I. Kang, A. Kobayashi, M. Yamamoto, T. W. Kensler, and P. Talalay. 2004. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. USA 101:2040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wittau, N., R. Grosse, F. Kalkbrenner, A. Gohla, G. Schultz, and T. Gudermann. 2000. The galanin receptor type 2 initiates multiple signaling pathways in small cell lung cancer cells by coupling to G(q), G(i) and G(12) proteins. Oncogene 19:4199-4209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.