Abstract
The mRNA deadenylation process, catalyzed by the CCR4 deadenylase, is known to be the major factor controlling mRNA decay rates in Saccharomyces cerevisiae. We have identified the proline-rich region and RRM1 domains of poly(A) binding protein (PAB1) as necessary for CCR4 deadenylation. Deletion of either of these regions but not other regions of PAB1 significantly reduced PAB1-PAB1 protein interactions, suggesting that PAB1 oligomerization is a required step for deadenylation. Moreover, defects in these two regions inhibited the formation of a novel, circular monomeric PAB1 species that forms in the absence of poly(A). Removal of the PAB1 RRM3 domain, which promoted PAB1 oligomerization and circularization, correspondingly accelerated CCR4 deadenylation. Circular PAB1 was unable to bind poly(A), and PAB1 multimers were severely deficient or unable to bind poly(A), implicating the PAB1 RNA binding surface as critical in making contacts that allow PAB1 self-association. These results support the model that the control of CCR4 deadenylation in vivo occurs in part through the removal of PAB1 from the poly(A) tail following its self-association into multimers and/or a circular species. Known alterations in the P domains of different PAB proteins and factors and conditions that affect PAB1 self-association would, therefore, be expected to be critical to controlling mRNA turnover in the cell.
mRNA degradation is a process involving the interaction and exchange of multiple multisubunit complexes and RNA binding proteins (8). Central to mRNA degradation is the removal of the poly(A) tail (deadenylation) that is controlled by a number of proteins associating with the mRNA in a structure termed the mRNP. Principal among these factors present in the mRNP are the poly(A) binding protein (PAB1), translation initiation and termination factors, the cytoplasmic deadenylases, and the factors that bind to the mRNA and elicit alterations in the mRNA degradative rate. The processes of mRNA degradation and deadenylation and the protein complexes that are involved are highly evolutionarily conserved from Saccharomyces cerevisiae to humans.
The principal pathway for mRNA degradation in yeast proceeds through several steps. First, there is an initial trimming of about 15 to 20 nucleotides (nt) of the poly(A) tail to a length of about 60 to 80 nt that is specific for each mRNA and that appears to be carried out by PAN2/PAN3, presumably a cytoplasmic process (2, 19, 48). This trimming requires PAB1 and the translation termination factors eRF1 and eRF3 (5, 24), and all these factors are known to associate with each other (10, 23, 24, 29). Second, the major part of deadenylation utilizes the CCR4-NOT deadenylase complex (16, 48). CCR4 is the catalytic component of this complex (7, 47) and shortens the poly(A) tail of mRNA to an end point size of about 8 to 12 nt (14). Poly(A) tail shortening down to an oligo(A) form (8 to 12 A's) may lead, in turn, to the reduced ability of PAB1 to bind the poly(A) tail that may alter the translation initiation complex association with the mRNA cap structure (9, 30, 46). Changes in the mRNP structure around the cap apparently can lead to decapping by DCP1/DCP2 (5, 45, 46) and 5′-3′ degradation of the RNA by the exonuclease XRN1 (34). The transition from deadenylation to decapping occurs with apparent sequestering of the oligoadenylated mRNA in P-bodies where decapping takes place (40).
The rate of deadenylation of the mRNA is therefore critical to the stability of most mRNA. Analyses of the relative contributions to total mRNA degradation of deadenylation, decapping, and 5′-3′ degradation clearly indicate that changes in rates of deadenylation lead to the largest changes in mRNA degradation (4). Specific and global analysis of mRNA deadenylation rates (14, 53) indicated that they vary over a 20-fold range. CCR4 has been shown to be necessary for both rapid and slow deadenylation for a number of mRNAs (7, 12, 21, 48, 49, 51, 54), suggesting that it affects deadenylation of most, if not all, mRNAs in yeast. Moreover, the length of the poly(A) tail can alter the translatability of the mRNA.
Several factors have previously been shown to control the deadenylation process. Of the two deadenylases in yeast, defects in PAN2/PAN3 alone do not affect the deadenylation rate, whereas defects in the CCR4-NOT complex, specifically CCR4 and CAF1, slow the deadenylation process (12, 48). The substrate recognition features of CCR4 indicate that its in vivo activity may be directly controlled by its accessibility to a certain length of RNA and that mRNP structures involving the poly(A) tail and its adjacent 3′ untranscribed region (UTR) sequences may limit these interactions and thereby significantly affect CCR4 activity (50, 51).
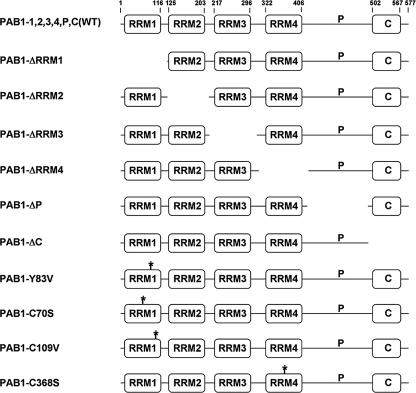
In addition to the deadenylases, a number of components of the poly(A) mRNP structure have been implicated in controlling deadenylation. Notable among these proteins is PAB1. PAB1 consists of four RNA binding motifs (RRM domains) and a C-terminal region comprising the penultimate proline-rich (P) domain and a terminal structured region (C) (Fig. 1). The C region binds PAN3, which is required for PAN2 activity, eRF3, and other proteins (24, 27, 29). The P domain of higher eukaryotic PAB1 is responsible for PAB1-PAB1 interactions (26, 31). The RRM domains of PAB1 consist of four β-strands that form the RNA binding surface backed by two α-helices (18). While RRM1 and RRM2 of PAB1 appear to bind most strongly to poly(A), RRM3 and RRM4 can also make critical contacts and may bind U-rich regions located adjacent to the poly(A) tail (35, 41). The α-helical side of RRM2 contacts eukaryotic initiation factor 46 (eIF4G), which is believed to be important in forming the closed-loop structure between the mRNA cap, eIF4E, eIF4G, PAB1, and the poly(A) tail (38, 44). The functions of the other RRM domains remain less clear, although deletion of RRM4 has been shown recently to reduce by 30% mRNA transport to the cytoplasm (3). RRM1, RRM2, RRM3, and the P and C regions play no apparent role in mRNA transport (3; K. Weis, personal communication). Previous studies concerning the role of PAB1 in deadenylation indicated that PAB1 is inhibitory to CCR4 in vitro (47). In contrast, a pab1 deletion strain has been shown to severely block CCR4 deadenylation in vivo, and it has been suggested that the lack of PAB1 allows nonspecific RNA binding proteins to associate with the poly(A) tail and to block deadenylation (5). A couple of other less comprehensive PAB1 defects have also been shown to block deadenylation (33), supporting a role for PAB1 in promoting deadenylation.
FIG. 1.
PAB1 variants that are discussed in the paper. Residues for each domain are indicated at the top.
In this study, we addressed the role of PAB1 in the deadenylation process. Using a set of precise domain deletions in PAB1, we found that the P and RRM1 domains of PAB1 were required for deadenylation. PAB1-PAB1 interactions, as well as the formation of a novel circular species of PAB1, were blocked by deletions in these two domains, suggesting that PAB1 self-association is critical to allowing CCR4 deadenylation. Moreover, PAB1 binding to poly(A) was dramatically decreased or eliminated by its self-association. These results indicate that PAB1 removal from the poly(A) tail following its self-association promotes, in turn, CCR4 deadenylation. Factors and conditions that control PAB1 self-association would therefore be expected to control mRNA deadenylation rates and the turnover of mRNA in the cell.
MATERIALS AND METHODS
Yeast strains.
Saccharomyces cerevisiae strain AS319 (MATα ade2 ura3 leu2 trp1 his3 pab1::HIS3 pAS77 [PAB1-CEN-URA3]) (25) was used for transforming PAB1 variants expressed under their own promoter on plasmid YC504 (pRS314:PAB1-CEN-TRP1) as indicated in Fig. 1. Plasmid AS77 was subsequently lost from each strain following selection on plates containing 5-fluoroorotic acid (25). Other strains isogenic to AS319 were DB267tL/pAS77 (contains pan3::trp1::LEU2), 1771-1/pAS77 (contains pan2::LEU2), and AS319-1a-uN/pAS77 (contains ccr4::ura3::Neo). Strain AS319 with the RP485 plasmid was used for quantitating rates of deadenylation for MFA2pG (48).
Plasmids.
PAB1 variants were constructed by PCR techniques and were expressed in yeast on a pRS314 vector (YC504) as Flag-tagged versions under control of the PAB1 promoter. The following amino acid residues were removed with each PAB1 deletion: PAB1-ΔRRM1, removed 1 to 124; PAB1-ΔRRM2, removed 124 to 211; PAB1-ΔRRM3, removed 211 to 309; PAB1-ΔRRM4, removed 309 to 416; PAB1-ΔP, removed 416 to 492; and PAB1-ΔC, removed 492 to 577.
RNA analysis.
Steady-state and pulse-chase analyses for GAL1 and MFA2pG mRNA were conducted as previously described (48, 51) by following the shortening of the shortest poly(A) tails. Measuring the rates of shortening of the average poly(A) tail length gave similar results. Rates of deadenylation were determined on only those poly(A) tails that were being distributively deadenylated; that is, those that were being shortened as a synchronous pool. S1 analysis for quantitating CCR4, CAF1, and NOT4 mRNA levels was conducted as described previously (10).
Other procedures.
In vitro gel shift assays were conducted as described previously (13) using PAB1 variants expressed in Escherichia coli from a pET28 vector. Recombinant PAB1 variants were purified to greater than 90% purity using Ni-agarose chromatography (25). Nondenaturing gel electrophoresis was conducted on purified PAB1 proteins to identify the presence of nonspecific RNAs that might interfere with the RNA binding measurements. No such interactions were detected. Bovine serum albumin was used as a standard to determine PAB1 protein abundance, and the Kd values were determined as described previously (13). Yeast PAB1 variants were purified by Flag immunoprecipitation as previously described (7) and analyzed by nondenaturing polyacrylamide gel electrophoresis (PAGE) as described previously (17), except that the bisacrylamide/acrylamide ratio was 1:60. Prior to nondenaturing PAGE, the PAB1 samples were incubated with 0.1 mg/ml RNase A at 23°C for 45 min to remove any endogenous mRNA attached to the PAB1. Treatment with RNase I instead of RNase A gave the same results. CAF40 and CCR4 immunoprecipitations were conducted as described previously (6). The rates of in vivo translation were determined as described previously (39). Far-Western analysis was conducted on PAB1 transferred to nitrocellulose filters by incubation with 32P-labeled 7N + 23A RNA substrate (50). Cross-linking with the bis-maleimide cross-linker, bis-maleimidohexane (BMH), was conducted according to the manufacturer's directions (Pierce) for 1 h. The mock solution had 100 mM dimethyl sulfoxide as previously described (43). In vivo BMH cross-linking was conducted as described previously (43).
RESULTS
The RRM1 and proline-rich regions of PAB1 are required for CCR4 deadenylation.
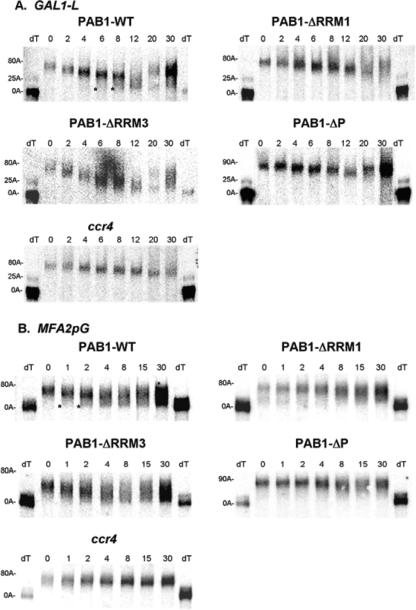
To address the roles of PAB1 in mRNA deadenylation, we deleted each of the known domains in the protein. This systematic approach would allow us to determine if individual regions of PAB1 displayed particular functions. RRM domains 1 through 4, the proline-rich penultimate region (PAB1-ΔP), and the globular, terminal C domain (PAB1-ΔC) were individually deleted (25) (Fig. 1). Each PAB1 variant was transformed into yeast and allowed for viability following loss of the wild-type PAB1. All PAB1 variants were expressed in yeast to comparable levels (data not shown). These six PAB1 variants and wild-type PAB1 (each containing a Flag tag) were analyzed for in vitro poly(A) binding (Table 1) and for their effects in vivo on the rate of deadenylation of MFA2pG and GAL1 mRNA using pulse-chase analysis (Fig. 2A and B; Table 1; data not shown) (48, 51). MFA2pG mRNA, under GAL1 control, was chosen for analysis because of its extensive characterization and because it is rapidly turned over (14). Two GAL1 mRNA species are produced in vivo that result from differential poly(A) site usage and differ by 110 nt in their 3′ UTR (11, 32). GAL1-L (long) deadenylates more rapidly than GAL1-S (short) (51). Following a brief induction of MFA2pG and GAL1 mRNA synthesis with the addition of galactose to the medium, mRNA synthesis was shut off with glucose, and the length of MFA2pG and GAL1 mRNA poly(A) tails was followed as a function of time by Northern analysis.
TABLE 1.
PAB1 effects on deadenylation
| PAB1 varianta | Poly(A) binding Kd (nM)b | Deadenylation rate for MFA2pG (A's/min)c | Deadenylation rate for GAL1-L (A's/min)c | Deadenylation rate for GAL1-S (A's/min)c |
|---|---|---|---|---|
| PAB1 (wt) | 11 | 11 | 3.7 | 2.8 |
| PAB1-Y83V | 220 | 2.6 | 2.0 | 1.8 |
| PAB1-ΔRRM1 | 29 | 3.4 | 2.0 | 2.0 |
| PAB1-ΔRRM2 | 35 | 4.2 | 3.2 | 2.4 |
| PAB1-ΔRRM3 | 15 | 18 | 6.3 | 4.6 |
| PAB1-ΔRRM4 | 22 | 11 | 3.9 | 2.9 |
| PAB1-ΔP | 7.4 | 2.1 | 1.5 | 1.1 |
| PAB1-ΔC | 18 | 7.5 | 3.2 | 2.1 |
PAB1 variants are depicted as in Fig. 1.
Poly(A) binding was analyzed in vitro using a gel shift assay with a 7N + 23A radiolabeled RNA (25). These Kd values for PAB1 variants binding to poly(A) agree very well with published values (13, 25). Our values were within 50% of published values except for PAB1-ΔRRM2 (2.1-fold lower) and PAB1-ΔRRM3 (1.9-fold higher). PAB1-ΔP, ΔC was also found to have Kds of binding to poly(A) of 6.0 nM and to be defective in the GAL1 deadenylation rate (data not shown).
Deadenylation rates were determined in strain AS319 with the indicated PAB1 variant expressed from a pRS314 vector (TRP1-PAB1) replacing that of pAS77 by measuring the lengths of the shortest poly(A) tail as a function of time after transcriptional shutoff (48). All rates of deadenylation were determined for the synchronous pool of distributively deadenylated mRNA. GAL1-L (long) and GAL1-S (short) refer to the two GAL1 mRNAs that result from differential poly(A) site usage and differ by about 110 nt in their 3′ UTR ends (11). Poly(A) binding assays are the average of 2 to 3 determinations with standard errors of the means (SEMs) of less than 20%. Deadenylation rates are the averages of two to three determinations, and the SEMs were less than 10%, with the exception of the following: for MFA2pG PAB1-ΔRRM4, it was 25%; for PAB1-ΔP, it was 21%; for PAB1-ΔC, it was 19%; for GAL1-L PAB1-ΔRRM4, it was 12%; and for GAL1-S PAB1-ΔC, it was 12%. The SEMs for the deadenylation rates for wild-type PAB1 were less than 4% for MFA2pG and GAL1-L and less than 2% for GAL1-S. Deadenylation rate values in bold differ significantly from wild type.
FIG. 2.
Multiple PAB1 defects affect CCR4 deadenylation. Pulse-chase analysis was used for determination of rates of mRNA deadenylation. All analyses utilized strain AS319 as indicated in Table 1. Northern analysis of GAL1 and MFA2pG mRNA was conducted following induction of synthesis with galactose for 8 min and shutting off of synthesis at time zero with glucose. Times are in min. The poly(A) tail lengths are indicated on the left and were determined from the known sizes of the deadenylated species: GAL1-L, 290 nt; GAL1-S, 183 nt; MFA2pG, 366 nt; pG, 188 nt; and partially deadenylated GAL1-L, 314 nt, and MFA2pG, 390 nt, as indicated in the figure. dT lanes represent the RNA sample, which had been pretreated with oligo(dT) and RNase H to remove the poly(A) tail prior to Northern analysis. (A) GAL-L; (B) MFA2pG. The asterisks in PAB1-WT for panels A and B indicate the presence of the oligonucleotide poly(A) species present at the same time as the longer poly(A) species. It should be mentioned that the GAL1 and MFA2pG mRNA syntheses are incompletely turned off by glucose, as previously observed (14), and that at the 30-min time point, it is often observed that newly synthesized mRNA with long poly(A) tails becomes present. This new mRNA does not significantly obscure the data for the original pulse of mRNA synthesis.
Several PAB1 deletions significantly affected the deadenylation rate. PAB1-ΔRRM1 and PAB1-ΔP had the most significant decreases in deadenylation (Table 1, Fig. 2). In the wild-type background, the majority of the poly(A) tail was shortened in a synchronous band for the first 4 to 6 min for GAL1-L and 2 min for MFA2pG. This band corresponded to distributive deadenylation by CCR4. However, when the poly(A) tail length reached about 35 to 40 nt for MFA2pG (at about 1 to 2 min for wild type) (Fig. 2B) and GAL1-L (at about 6 to 8 min for wild type) (Fig. 2A), deadenylation became more nonsynchronous: nearly completely deadenylated species (8 to 12 A's) were present at the same time as partially deadenylated species (see also Fig. S1 in the supplemental material). Both PAB1-ΔRRM1 and PAB1-ΔP affected both phases of the deadenylation process for GAL1, reducing the rate of distributive deadenylation (Table 1) and failing to promote the shift to nonsynchronous deadenylation, even though the poly(A) tail lengths reached 30 to 40 A's at about 20 min (Fig. 2A; see also Fig. S1 in the supplemental material). For MFA2pG, the effect of these deletions on the nonsynchronous phase was less clear. These results indicate that these two regions of PAB1 are necessary for CCR4 action throughout the deadenylation process.
PAB1-ΔRRM2 and PAB1-ΔC also affected the rate of CCR4 deadenylation, albeit to lesser extents than did deletion of either RRM1 or the P domain (Table 1). Our PAB1-ΔC data are consistent with previous results that showed that the C-terminal domain of PAB1 interacts with translation termination factor eRF3 and that blocking this interaction results in somewhat slower deadenylation in vivo (23). Removing RRM4 had no significant effect on the deadenylation rate or on the switch to nonsynchronous deadenylation. Importantly, deletion of RRM3, in contrast to the above deletions, accelerated deadenylation for GAL1-L, GAL1-S, and MFA2pG by 1.6- to 1.7-fold (Fig. 2 and Table 1; see also Fig. S1 in the supplemental material). All of the above-described effects on deadenylation were attributable to effects on CCR4, since deleting PAN2 has no effect on the in vivo deadenylation rate (48; data not shown), and in a pan2 background, when CCR4 was the only deadenylase, each of these PAB1 defects still affected deadenylation (data not shown). In addition, no correlation was observed between the ability of PAB1 to bind poly(A) in vitro and its ability to affect deadenylation in vivo (Table 1). This is expected, given the nM Kd of PAB1 binding to RNA, the μM concentration of PAB1 in the cell, and the excess concentration of PAB1 relative to that of total mRNA (20, 53) (see Discussion).
We also addressed whether these PAB1 defects on deadenylation were likely due to secondary effects on in vivo translation, mRNA export, or the concentration of CCR4 in the cell. First, it is unlikely that these defects in deadenylation were due to secondary effects on mRNA export. Previously, it has been shown that deleting RRM1, RRM2, or the P and C terminus of PAB1 does not affect the transport of mRNA into the cytoplasm (3; K. Weis, personal communication), and deleting RRM4, which reduces mRNA export by 30%, did not affect the deadenylation rate. Second, the rate of in vivo protein synthesis for all of our PAB1 variants was monitored, and in most cases there was 10% or less effect on the rate of protein synthesis (36; data not shown). The most severe effect was for PAB1-ΔRRM1, in which the rate of protein synthesis compared to wild-type PAB1 was reduced by 28% (36). However, PAB1-ΔP, which even more severely blocked deadenylation than did PAB1-ΔRRM1, had a negligible effect on the rate of protein synthesis (6%). In contrast, translation initiation defects reduce the rate of translation by greater than 50% (36, 39; data not shown) and accelerate, but do not slow, deadenylation. It is therefore unlikely that the slowing of deadenylation is resulting specifically from defects in translation. In addition, deleting RRM3, which accelerated deadenylation, had no effect on the rate of in vivo translation. Third, except for deleting RRM2, none of the PAB1 deletions affected the abundance of the CCR4-NOT complex in vivo as ascertained by immunoprecipitating it with either CCR4- or CAF40-specific antibodies (data not shown). Deleting RRM2 reduced the abundance of the CCR4-NOT complex in the cell by about fourfold, which therefore appeared to be a primary cause of the reduced rates of deadenylation observed in vivo with this PAB1 variant.
PAB1-PAB1 protein interactions are mediated by the P and RRM1 domains, implicating this contact in the control of CCR4 deadenylation.
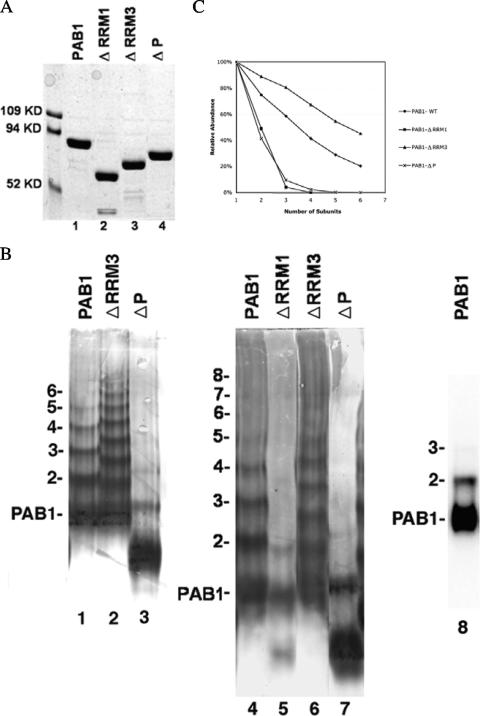
As the P domain is known to be required for interactions among higher eukaryotic PAB1 molecules (26, 31), its effects on deadenylation might be occurring through a required contact among PAB1 molecules. We subsequently tested the model that the effects on the rates of deadenylation caused by deletion of certain PAB1 domains were the result of alterations in PAB1 interactions with itself. We first examined yeast PAB1 self-association following nondenaturing gel electrophoresis. Purified PAB1 was found, as analyzed by sodium dodecyl sulfate (SDS)-PAGE, to migrate as a single band of the expected molecular weight (Fig. 3A), but under nondenaturing gel conditions, PAB1-wt was found to form a series of slower migrating species (Fig. 3B, lane 1). Western analysis with antibody specific to PAB1 identified 6 distinct species of PAB1, although in other experiments up to 13 distinct species could be detected (data not shown). The same results were obtained following nondenaturing PAGE when the PAB1 proteins were identified by Coomassie staining (data not shown). These slower migrating species are indicative of the formation of multimeric PAB1 species, possibly in some type of a concatemer arrangement (see Fig. 6B). Four additional observations support the identification of these species as multimers of PAB1: (i) the different identified species of PAB1 migrate relatively linearly in relation to the expected logarithmic value of their molecular weights (data not shown), (ii) the original PAB1 samples were 95% pure, (iii) PAB1 migrates as a single, cohesive species following SDS-PAGE that is inconsistent with multiple phosphorylation of PAB1 (49), and (iv) other experiments indicate that posttranslational modifications such as acetylation do not result in the multiple species displayed by PAB1 that were observed following nondenaturing gel electrophoresis (15, 17).
FIG. 3.
Yeast PAB1 can form multimers. (A) Coomassie-stained PAB1 proteins following SDS-PAGE. The PAB1 proteins (7 μg each) following purification by Flag immunoprecipitation were subjected to SDS-PAGE and were derived from strain AS319. (B) Western analysis of the PAB1-purified proteins (6 μg each) using Flag antibody following nondenaturing PAGE. PAB1 variants are shown in Fig. 1. Samples were treated with RNase A prior to separation by PAGE. Treatment with RNase I gave the same results. Other experiments indicate that RNase treatment eliminates the mRNA-PAB1 interactions that under nondenaturing conditions partially obscure the individual PAB1 multimeric species. The multimeric species are also not the result of disulfide formation between PAB1 monomers, as treatment with different reducing agents did not affect PAB1 multimerization. In lane 5, the monomeric species for PAB1-ΔRRM1 is the species second from the bottom, and its C-terminal degradation product (37) is the bottom species (see also panel A, lane 2). Numbers refer to the different oligomeric species identified for wild-type PAB1. For lane 8, PAB1 following native PAGE was incubated with radioactive 7N + 23A. Similar results as that observed in lane 8 were obtained for all other deletions in that dimers were deficient in binding poly(A), and trimers and larger oligomers were nearly entirely deficient in binding. (C) Densitometric analysis of the relative abundance of the different oligomeric species. Monomeric PAB1 was set at 1.0. The values represent the averages of the results from two different experiments, and the standard errors of the means were less than 10% in all cases.
FIG. 6.
(A) Model of linear PAB1 bound to poly(A) and circular PAB1. The long linkers between RRM2 and RRM3 (14 amino acid residues) and RRM3 and RRM4 (25 residues) compared to RRM1 and RRM2 (9 residues) can account for the circularization of PAB1. (B) Model of PAB1 oligomerization. BMH can still cross-link PAB1 dimers when the Cys residues in either RRM1 or RRM4 are mutated, suggesting that RRM1 can interact with RRM1 and that RRM4 can interact with RRM4. How the P domain stabilizes oligomer interactions is not clear and not shown.
We analyzed whether PAB1-PAB1 association was important for deadenylation by determining whether deletions in PAB1 that affected deadenylation were defective in PAB1-PAB1 interactions. As shown in Fig. 3B, lanes 3 and 7, PAB1-ΔP failed to form the many multimeric species that PAB1 did and formed the dimeric and trimeric forms in reduced amounts compared to PAB1 (densitometric analysis indicated that PAB1-ΔP dimeric species was reduced twofold compared to that of PAB1, and the trimeric species was reduced by sixfold) (Fig. 3C). Surprisingly, deleting the RRM1 domain also blocked the formation of multimeric species (Fig. 3B, lane 5; summarized in Fig. 3C). As PAB1-ΔP and PAB1-ΔRRM1 had the most severe effects on deadenylation, we interpret these results to suggest that PAB1-PAB1 self-association is one factor required for CCR4 function. Further support for this model is derived from the observation that deleting the RRM3 domain, which accelerates deadenylation in vivo (Table 1 and Fig. 2), consistently resulted in increased multimer formation (Fig. 3B, lanes 2 and 6 compared to wild type, lanes 1 and 4; Fig. 3C).
PAB1 effects on deadenylation correlate with the formation of a novel circular PAB1 species.
In order to seek independent evidence of PAB1-PAB1 interactions, we conducted in vitro protein cross-linking of the purified PAB1 variants. Chemical cross-linking was conducted with the agent BMH (44), which links cysteine residues that are 13 angstroms apart. Similar results were also obtained with the chemical agents bis-maleimidoethane and 1,4-bis-maleimidobutane, which cross-link Cys residues that are 8.0 and 10.9 angstroms apart, respectively (data not shown). PAB1 has three Cys residues, C70 and C109 in the RRM1 domain and C368 in the RRM4 domain (Fig. 1). All three residues are expected to be on the β-strand-side, exterior surface of PAB1 and thus available for cross-linking to other Cys residues (18). Following treatment of PAB1 with BMH or with a mock solution and separation of the proteins by SDS-PAGE, we observed dimeric and trimeric species of PAB1, consistent with PAB1-PAB1 self-association (Fig. 4A, lane 1). These data confirm that PAB1 can form multimers that can be readily stabilized by the addition of a chemical cross-linker. Moreover, these results indicate that multimer formation involves the β-strand RNA-binding, exterior surface of PAB1, consistent with a recent demonstration that the RRM domain of Nup35 can dimerize through this region (22).
FIG. 4.
(A) BMH cross-linking of PAB1 variants. Equivalent amounts of PAB1 variants were cross-linked in vitro with and without BMH and were detected with anti-Flag antibody following SDS-PAGE. *PAB1 represents an in vivo C-terminal truncation of PAB1 as was previously observed (37). (B) Far-Western analysis of in vitro cross-linked PAB1. Cross-linked PAB1 proteins, as depicted in Fig. 4A, lanes 1 to 12, following SDS-PAGE and transfer to nitrocellulose, were incubated with radioactive 7N + 23A RNA. (C) BMH cross-linking was conducted in vivo (43), after which PAB1 was isolated and identified by Western analysis with anti-Flag antibody. Analysis of the circular PAB1 species indicates that it consists of two related bands. These two species presumably arise from cross-linking of either C70 or C109 to C368 to generate two very similar forms. PAB1-C368S failed to form both species, and PAB1-C70S formed only one species (data not shown).
However, much of the PAB1, following BMH treatment, ran as a novel species of 83 kDa. Since this species is too small to be dimeric PAB1, and since no other small contaminating fragment of PAB1 or other protein was observed in our protein preparations (data not shown), we deduce that this new species of PAB1 results from an internal cross-link between two Cys residues 8 to 13 angstroms apart, most likely between Cys residues located on RRM4 and RRM1. Such an internal cross-link would produce a circular PAB1 (see Fig. 6A for a model) and would be expected to slightly slow the migration of PAB1 following SDS-PAGE. Consistent with this identification of a circular PAB1 that can be cross-linked with BMH, deleting either RRM1 or RRM4, thereby removing one or the other of the donor Cys residues, completely blocked the formation of the circular form (Fig. 4A, lanes 3 and 7). A C368S change also completely blocked the ability of BMH to cross-link the circular form (Fig. 4A, lane 17), as did combining C70S with C109V (lane 15). These results indicate that much of PAB1, in the absence of RNA, exists in a form that involves close proximity of the β-strand surfaces of RRM1 and RRM4, creating a circular species that can be detected following BMH cross-linking. These observations agree with atomic force spectroscopic analysis of PAB1 that showed that PAB1 in the absence of RNA exists essentially as a globular protein, and only in the presence of RNA does it adopt a more linear structure (42), as previously proposed, based on the crystallographic analysis of PAB1 bound to RNA (18) (see also Fig. 6A).
We subsequently examined whether defects in PAB1 that affected deadenylation also affected the formation of the PAB1 circular species as detected with BMH cross-linking. Several observations were made from this analysis. First, deleting the P domain of PAB1 significantly reduced the abundance of the PAB1 circular species that could be formed following BMH cross-linking (Fig. 4A, lane 9, fivefold-less circular form than wild type, lane 1), implying that PAB1 does not readily form internal contacts between RRM4 and RRM1 in the absence of the P domain. Second, deleting RRM3 actually enhanced the abundance of circular forms that could be detected: about 1.6-fold-more circular species compared to monomeric species were observed for PAB1-ΔRRM3 (Fig. 4, lane 5) than for wild type (Fig. 4, lane 1). These correlations suggest that the ability of PAB1 to self-associate to form the circular and multimeric species correlates inversely with its ability to allow CCR4 deadenylation. Third, as we could not determine whether deleting the RRM1 domain affected the formation of the circular species, as such a deletion also would remove the Cys residues, we examined instead the effect of the Y83V alteration on PAB1 circular formation: this mutation was shown to reduce CCR4 deadenylation (Table 1) and is on the exterior RNA binding surface of RRM1 that might be expected to be in close contact with RRM4. PAB1-Y83V was found to reduce by twofold the ability of the circular species to be detected following BMH cross-linking (Fig. 4A, lane 11). The PAB1-Y83V effect on the formation of the circular species was less than was observed for deleting the P domain and, correspondingly, had less of an effect on deadenylation. This result suggests that the Y83V alteration blocks PAB1 circularization and reduces CCR4 deadenylation. However, PAB1-Y83V did not affect the ability of PAB1 to form oligomers (Fig. 4A; data not shown).
PAB1 self-association competes with PAB1 binding to poly(A).
The ability of PAB1 to form a circular species in which the β-strand surfaces of RRM1 and RRM4 domains are in close contact (at most 8 angstroms apart) would be expected to prohibit PAB1 from binding poly(A). Similarly, PAB1 multimer association would be expected to reduce PAB1 binding to RNA, as we have shown that multimer association occurs at least in part between the β-strand surfaces of two PAB1 molecules, as observed following BMH cross-linking (Fig. 4A, lane 1). These observations support the model that PAB1 regulation of mRNA deadenylation occurs through PAB1 binding to poly(A), which serves as an impediment to CCR4 progression (47). The ability of PAB1 to self-associate would conversely aid in removing PAB1 from the RNA, thereby allowing deadenylation to occur. We first tested this model by assaying whether multimeric forms of PAB1 could bind poly(A) as well as monomeric PAB1. Following separation of PAB1 by native gel electrophoresis (Fig. 3B, lane 1), we incubated the resultant forms of PAB1 with radioactive 7N + 23A substrate. Monomeric PAB1 was capable of binding the poly(A) substrate well, but the dimeric PAB1 species was reduced fivefold in its ability to bind RNA (Fig. 3B, lane 8). The trimeric species was much more substantially reduced (at least 20-fold) in binding the poly(A) RNA and binding of the larger multimers to RNA was not detected, even after very long exposures. Similar results were obtained for the other PAB1 variants (data not shown). These data confirm that the multimerization of PAB1 occurs primarily through the RNA binding surfaces of PAB1 (see model, Fig. 6B), and that multimerization would compete with PAB1 contacts to its poly(A) substrate.
We similarly assayed the ability of the BMH cross-linked species to bind our poly(A) substrate. As shown in Fig. 4B, lane 2, monomeric, non-cross-linked PAB1 was capable of binding poly(A), but the circular, cross-linked form of PAB1 (Fig. 4B, lane 1) was nearly deficient in binding poly(A); following extremely long exposures, at least a 50-fold reduction in binding to RNA was observed (data not shown). The dimeric cross-linked PAB1 was also substantially reduced in binding poly(A), although some binding was observed following long exposures (data not shown). These data confirm that BMH cross-linking in the circular species bridges the RNA binding surfaces of RRM1 and RRM4, thereby prohibiting contact to poly(A).
The results described above further suggest that the PAB1 circular form, prior to cross-linking it with BMH, would be in equilibrium with a linear PAB1 capable of binding poly(A). If this were the case, the addition of increased amounts of poly(A) substrate would be predicted to favor the RNA-bound linear form of PAB1 and to reduce the amount able to circularize as detected following BMH treatment. We tested this prediction by incubating PAB1 with increasing amounts of poly(A) substrate prior to stabilizing the circular form following BMH treatment (Fig. 5, lanes 3 to 6). As a control, we also incubated PAB1 with an RNA substrate lacking poly(A) sequences that would be expected to bind poorly to the linear PAB1 species (Fig. 5, lanes 7 to 10). Increasing the concentration of poly(A) substantially decreased the amount of PAB1 capable of being cross-linked by BMH, indicating that there was less circular form available in the presence of poly(A). The non-poly(A) RNA substrate had much less effect on the reduction in the amount of the circular species available for cross-linking. These results indicate that there exists a dynamic interrelationship between PAB1 bound to poly(A) and PAB1 self-associating in a circular structure that can be cross-linked with BMH. It should also be noted (Fig. 5) that the abundance of the cross-linked dimer was also reduced with increasing amounts of poly(A), indicating that there is also an equilibrium between PAB1 bound to RNA and PAB1 interacting with other PAB1 molecules.
FIG. 5.
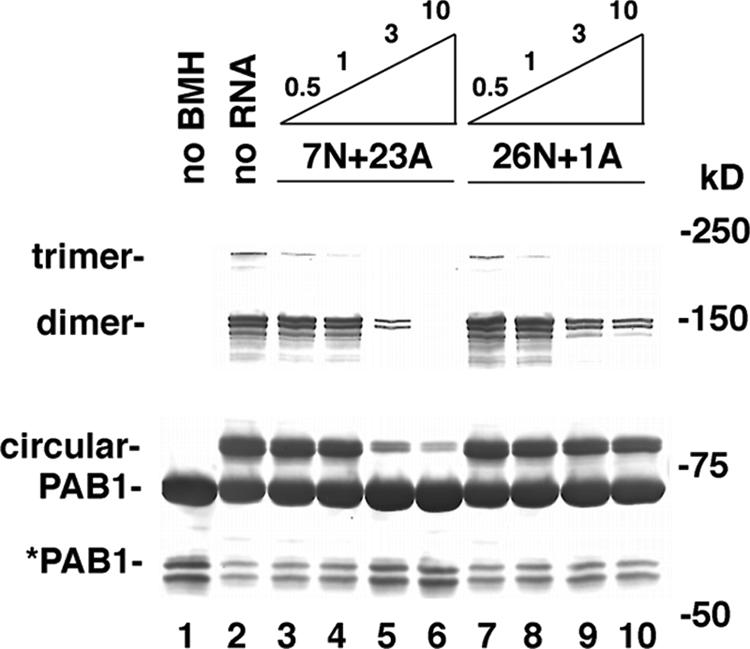
PAB1 bound to poly(A) is in equilibrium with PAB1 associating with itself. Increasing amounts (μM) of the two RNAs as indicated were incubated with PAB1 in the presence of BMH (lanes 3 to 10). Lane 1, no BMH and no RNA; lane 2, BMH without RNA. PAB1 was detected by Western analysis using anti-Flag antibody.
If PAB1 self-association controls its contacts to the poly(A) tail, we should be able to detect in vivo PAB1 present in the circular configuration and/or in the multimer form. To examine PAB1 self-association in vivo, we incubated yeast cells with BMH or with a mock solution, isolated PAB1 protein, and compared the resultant PAB1 species with in vitro BMH cross-linked PAB1. As shown in Fig. 4C, lane 2, in vivo PAB1 following BMH treatment formed an 83-kDa species that was not present in the mock-treated cells and that comigrated with the circular PAB1 species that was detected in vitro (Fig. 4C, lane 3). Also, a 150-kDa PAB1 species that comigrated with the dimeric form of PAB1 was detected (Fig. 4C, lane 2 compared to lane 3). These results suggest that the 83- and 150-kDa species represent self-associated forms of PAB1 that were cross-linked in vivo.
DISCUSSION
Our deletion analysis of PAB1 has established that two of its domains are most critical to CCR4 deadenylation: the RRM1 domain and the P region. Both of these domains were also deficient in forming PAB1 multimers. Removal of the P domain or a point mutation in the RRM1 domain also reduced the formation of a newly described circular species of PAB1. In addition, PAB1 self-association significantly reduced or eliminated its ability to bind poly(A). These correspondences suggest that the ability of PAB1 to allow CCR4 deadenylation of the poly(A) tail depends on its ability to dissociate from the RNA and form multimers and/or the circular form. Interfering with PAB1 self-association, as with the RRM1 or P defects, would inhibit PAB1 removal from the poly(A) tail. Conversely, promoting PAB1 self-association, as with removing the RRM3 domain, would accelerate deadenylation. Several segments of PAB1 are therefore required for promoting deadenylation by virtue of their ability to promote PAB1 self-association.
This model describing the regulation of deadenylation by PAB1 self-association agrees with previous results concerning PAB1 and CCR4 interactions on the poly(A) tail. PAB1 has been shown to inhibit the rate of CCR4 deadenylation in vitro (47). As we have been unable to establish any physical contact between the CCR4-NOT complex and PAB1, whether PAB1 were bound to poly(A) or not, one interpretation of the PAB1 inhibition of CCR4 action in vitro is that PAB1 is simply impeding CCR4 progress or association with the RNA. Our previous observations that CCR4 activity in vitro can be dramatically enhanced by its association with longer stretches of RNA sequence (50, 51) argue that removal of PAB1 from the poly(A) tail should dramatically enhance CCR4 enzymatic activity, shifting it into a processive mode (50). In fact, we and others have observed that during deadenylation in vivo, CCR4 shifts from a slow distributive rate of deadenylation to a rapid, nonsynchronous rate of deadenylation once the poly(A) tail length has been reduced by about 20 or so A's (Fig. 2) (14, 51). Removal of 20 A's would be consistent with the first PAB1 being removed from the poly(A) tail. We suggest that this removal following either circularization or contacts to other PAB1 molecules promotes removal of sequential PAB1 molecules from the RNA. This removal would allow CCR4 to shift into a processive mode, consistent with the nonsynchronous phase of deadenylation that is observed in vivo.
The implication of this model is that specific factors may be engaged in controlling PAB1 self-association. For example, it can be envisaged that 3′ UTR-binding proteins may regulate the mRNA deadenylation process and hence the rate of mRNA turnover through controlling the ability of PAB1 to self-associate. Moreover, the fact that defects in translation initiation factors enhance the deadenylation rate may derive from their direct effects on contacting PAB1. It is therefore significant that RRM1 plays a role in translation (results herein; 25, 36), suggesting it may be contacting translation initiation factors. It may be envisaged that the translation initiation complex may stabilize PAB1 contacts to the poly(A) or through occlusion prevent its self-association, thereby restricting the deadenylation process.
Another implication of these results is that the proliferation of multiple PABPC genes at the higher eukaryotic level may be involved in controlling PABPC self-association and mRNA turnover. Human iPAB (55) and Xenopus ePAB (embryonic PABPC) (52) differ predominantly from their normal PABPC proteins expressed in their respective organisms in that they contain divergent types of P domains, while their RRM1 to RRM4 domains are more similar to their PABPC counterparts. ePAB is expressed under conditions when the normal PABPC is not expressed and embryonic cells are defective in mRNA deadenylation (52). Similarly, iPAB is highly expressed in T cells under conditions when lymphokine mRNAs are synthesized and particularly stabilized (28). In both of these conditions, it is possible that these divergent P domains reduce P domain contacts that normally aid in PABPC self-association, resulting in slowing of the deadenylation rate. Moreover, humans and other primates possess a PABPC5 gene that completely lacks the P and C domains (1), suggesting that it would be inhibitory to deadenylation for those mRNAs it were to bind. Finally, in yeast, at least 10 to 20% of PAB1 in vivo consists of a proteolytically cleaved species in which the P domain has been removed (37) (Fig. 4 to 6; data not shown). Since there are approximately 200,000 PAB1 molecules per yeast cell (20), there may be as many as 40,000 proteolytically cleaved PAB1 molecules that would be quite capable of binding poly(A) in vivo (Fig. 4B; data not shown) and which would be expected to block deadenylation of the mRNA to which they bind in vivo.
The structure of the PAB1 circular form is likely to involve protein interactions between the β-strand surfaces of RRM4 and RRM1. This model is supported by our observations that the Cys residues in each of these regions are within 8 angstroms of each other, and that mutating either the Cys residue in RRM4 or the pair of Cys residues in RRM1 blocked the formation of the circular species. Based on the crystallographic analysis of a dimeric form of Nup35 (22), the RRM4 and RRM1 surfaces may be contacting each other in an antiparallel fashion. The importance of the P domain for circularization suggests that it may be making contacts to RRM1 to hold the circular form in place. The structure and form of the PAB1-PAB1 multimers is less clear. The number of multimeric species that are observed following native PAGE suggests some type of concatemer arrangement, as depicted in Fig. 6B. The P domain in this case may also be binding RRM1 from an adjacent PAB1 subunit to help stabilize the multimer. Although the P region is also known to contact PBP1, an inhibitor of PAN2/PAN3 (29), it is very unlikely that PBP1 is required for CCR4-NOT function, as defects in PAN2/PAN3 and itself do not affect the deadenylation rate (48) (data not shown). The role of the RRM3 domain in either of these structures is not clear. RRM3 may be making specific protein contacts that restrict self-association. Alternatively, its effects on PAB1 self-association may simply derive from its place as a spacer between the RRM1 and RRM4 domains, in which removing the spacer allows for easier circularization or multimerization.
One obvious expectation from our results would be that mutations that reduce the ability of PAB1 to associate with poly(A) in vitro would be expected to accelerate deadenylation in vivo. However, our analysis of PAB1 mutations indicated that there is no clear correlation between PAB1 association with poly(A) in vitro and its ability to affect deadenylation in vivo (Table 1). This result is consistent with PAB1 being saturating in vivo for poly(A) due to the excess of PAB1 relative to that of mRNA in the cell (4 μM PAB1 to 0.36 μM mRNA) (20, 53) and the high concentration of PAB1 relative to the 11 nM Kd of PAB1 binding to poly(A). Even in mutants where the Kd is increased to 0.2 μM, more than 95% of the poly(A) sites might be expected to be occupied by PAB1. Rather, the presence of PAB1 on the RNA will be dictated by the relative ability of PAB1 to self-associate. These observations suggest that the actual deadenylation rate at a particular mRNA will be governed by the local PAB1-mRNP structure at that mRNA as influenced by other factors in the vicinity.
Supplementary Material
Acknowledgments
We are especially appreciative of the plasmids and strains that were provided to us by D. Mangus, R. Parker, and D. Belostotsky.
This research was supported by Hatch grant H291 and by NIH grant GM78078.
This is Scientific Contribution Number 2285 from the New Hampshire Agricultural Experiment Station.
Footnotes
Published ahead of print on 9 July 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Blanco, P., C. A. Sargent, C. A. Boucher, G. Howell, M. Ross, and N. A. Affara. 2001. A novel poly(A)-binding protein gene (PABPC5) maps to an X-specific subinterval in the Xq21.3/Yp11.2 homology block of the human sex chromosomes. Genomics 74:1-11. [DOI] [PubMed] [Google Scholar]
- 2.Brown, C. E., and A. B. Sachs. 1998. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 18:6548-6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brune, C., S. E. Munchel, N. Fischer, A. V. Podtelejnikov, and K. Weis. 2005. Yeast poly(A)-binding protein Pab1 shuttles between the nucleus and the cytoplasm and functions in mRNA export. RNA 11:517-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, D., and R. Parker. 2001. Computational modeling of eukaryotic mRNA turnover. RNA 7:1192-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caponigro, G., and R. Parker. 1995. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 9:2421-2432. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., J. Rappsilber, Y.-C. Chiang, P. Russell, M. Mann, and C. L. Denis. 2001. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 314:683-694. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., Y. C. Chiang, and C. L. Denis. 2002. CCR4, a 3′-5′ poly (A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21:1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coller, J., and R. Parker. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861-890. [DOI] [PubMed] [Google Scholar]
- 9.Coller, J. M., N. K. Gray, and M. P. Wickens. 1998. mRNA stabilization by poly (A) binding protein is independent of poly (A) and requires translation. Genes Dev. 12:3226-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosson, B., A. Couturier, S. Chabelskaya, D. Kiktev, S. Inge-Vechtomov, M. Philippe, and G. Zhouravleva. 2002. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI+] propagation. Mol. Cell. Biol. 22:3301-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui, Y., and C. L. Denis. 2003. In vivo evidence that defects in transcription elongational factors, RPB2, TFIIS, and SPT5 enhance upstream poly(A) site utilization. Mol. Cell. Biol. 23:7887-7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daugeron, M.-C., F. Mauxion, and B. Seraphin. 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 29:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deardorff, J. A., and A. B. Sachs. 1997. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J. Mol. Biol. 269:67-81. [DOI] [PubMed] [Google Scholar]
- 14.Decker, C. J., and R. Parker. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7:1632-1643. [DOI] [PubMed] [Google Scholar]
- 15.Denis, C. L. 1982. A study of the regulation of the cytoplasmic alcohol dehydrogenases from Saccharomyces cerevisiae. Ph.D. thesis. University of Washington, Seattle.
- 16.Denis, C. L., and J. Chen. 2003. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73:221-250. [DOI] [PubMed] [Google Scholar]
- 17.Denis, C. L., and E. T. Young. 1983. Isolation and characterization of the positive regulatory gene ADR1 from Saccharomyces cerevisiae. Mol. Cell. Biol. 3:360-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deo, R. C., J. B. Bonanno, N. Sonenberg, and S. K. Burley. 1999. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98:835-845. [DOI] [PubMed] [Google Scholar]
- 19.Dheur, S., K. R. Nykamp, N. Viphakone, M. S. Swanson, and L. Minvielle-Sebastia. 2005. Yeast mRNA poly(A) tail length control can be reconstituted in vitro in the absence of Pab1p-dependent Poly(A) nuclease activity. J. Biol. Chem. 280:24532-24538. [DOI] [PubMed] [Google Scholar]
- 20.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 21.Goldstrohm, A. C., D. J. Seay, B. A. Hook, and M. Wickens. 2007. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 282:109-114. [DOI] [PubMed] [Google Scholar]
- 22.Handa, N., M. Kukimoto-Niino, R. Akasaka, S. Kishishita, K. Murayama, T. Terada, M. Inoue, T. Kigawa, S. Kose, N. Imamoto, A. Tanaka, Y. Hayashizaki, M. Shirouzu, and S. Yokoyama. 2006. The crystal structure of mouse Nup35 reveals atypical RNP motifs and novel homodimerization of the RRM domain. J. Mol. Biol. 363:114-124. [DOI] [PubMed] [Google Scholar]
- 23.Hoshino, S., M. Imai, T. Kobayashi, N. Uchida, and T. Katada. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of eRF3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 274:16677-16680. [DOI] [PubMed] [Google Scholar]
- 24.Hosoda, N., T. Kobayashi, N. Uchida, Y. Funakoshi, Y. Kikuchi, S. Hoshino, and T. Katada. 2003. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278:38287-38291. [DOI] [PubMed] [Google Scholar]
- 25.Kessler, S. H., and A. B. Sachs. 1998. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction and eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kühn, U., and T. Pieler. 1996. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J. Mol. Biol. 256:20-30. [DOI] [PubMed] [Google Scholar]
- 27.Khn, U., and E. Wahle. 2004. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 1678:67-84. [DOI] [PubMed] [Google Scholar]
- 28.Lindstein, T., C. H. June, J. A. Ledbetter, G. Stella, and C. B. Thompson. 1989. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science 244:339-343. [DOI] [PubMed] [Google Scholar]
- 29.Mangus, D. A., M. C. Evans, N. S. Agrin, M. Smith, P. Gongidi, and A. Jacobson. 2004. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol. 24:5521-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly (A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4:223-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melo, E. O., R. Dhalia, C. Martins de Sa, N. Standart, and O. P. de Melo Neto. 2003. Identification of a C-terminal poly(A)-binding protein (PABP)-PABP interaction domain. J. Biol. Chem. 278:46357-46368. [DOI] [PubMed] [Google Scholar]
- 32.Miyajima, A., N. Nakayama, I. Miyajima, N. Arai, H. Okayama, and K. Arai. 1984. Analysis of full-length cDNA clones carrying GAL1 of Saccharomyces cerevisiae: a model system for cDNA expression. Nucleic Acids Res. 12:6397-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrissey, J. P., J. A. Deardorff, C. Hebron, and A. B. Sachs. 1999. Decapping of stabilized, polyadenylated mRNA in yeast pab1 mutants. Yeast 15:687-702. [DOI] [PubMed] [Google Scholar]
- 34.Muhlrad, D., C. J. Decker, and R. Parker. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′-3′ digestion of the transcript. Genes Dev. 8:855-866. [DOI] [PubMed] [Google Scholar]
- 35.Mullin, C., K. Duning, A. Barnekow, D. Richter, J. Kremerskothen, and E. Mohr. 2004. Interaction of rat poly(A)-binding protein with poly(A)- and non-poly(A) sequences is preferentially mediated by RNA recognition motifs 3 + 4. FEBS Lett. 576:437-441. [DOI] [PubMed] [Google Scholar]
- 36.Ohn, T., Y.-C. Chiang, D. J. Lee, G. Yao, C. Zhang, and C. L. Denis. 2007. CAF1 plays an important role in mRNA deadenylation separate from its contact to CCR4. Nucleic Acids Res. 35:3002-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sachs, A. B., M. W. Bond, and R. D. Kornberg. 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell 45:827-835. [DOI] [PubMed] [Google Scholar]
- 38.Sachs, A. B., and G. Varani. 2000. Eukaryotic translation initiation: there are (at least) two sides to every story. Nat. Struct. Biol. 7:356-361. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz, D. C., and R. Parker. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:5247-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sladic, R. T., C. A. Lagnado, C. J. Bagley, and G. J. Goodal. 2004. Human PABP binds AU-rich RNA via RNA-binding domains 3 and 4. Eur. J. Biochem. 271:450-457. [DOI] [PubMed] [Google Scholar]
- 42.Smith, B. L., D. R. Gallie, H. Le, and P. K. Hansma. 1997. Visualization of poly(A)-binding protein complex formation with poly(A) RNA using atomic force microscopy. J. Struct. Biol. 119:109-117. [DOI] [PubMed] [Google Scholar]
- 43.Taggart, A. K., and B. F. Pugh. 1996. Dimerization of TFIID when not bound to DNA. Science 272:1331-1333. [DOI] [PubMed] [Google Scholar]
- 44.Tarun, S. Z., Jr., S. E. Wells, J. A. Deardorff, and A. B. Sachs. 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. USA 94:9046-9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tharun, S., W. He, A. E. Mayes, P. Lennertz, J. D. Beggs, and R. Parker. 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404:515-518. [DOI] [PubMed] [Google Scholar]
- 46.Tucker, M., and R. Parker. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69:571-595. [DOI] [PubMed] [Google Scholar]
- 47.Tucker, M., R. R. Staples, M. A. Valencia-Sanchez, D. Muhlrad, and R. Parker. 2002. CCR4p is the catalytic sub-unit of Ccr4p/Pop2p/Notp mRNA deadenylase ecomplex in Saccharomyces cerevisiae. EMBO J. 21:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated proteins, Ccr4p and Caf1p, are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 49.Vallari, R. C., W. J. Cook, D. C. Audino, M. J. Morgan, D. E. Jensen, A. P. Laudano, and C. L. Denis. 1992. Glucose repression of yeast ADH2 occurs through multiple mechanisms, including control of the protein synthesis of its transcriptional activator ADR1. Mol. Cell. Biol. 12:1663-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viswanathan, P., J. Chen, Y.-C. Chiang, and C. L. Denis. 2003. Identification of multiple RNA features that influence CCR4 deadenylation activity. J. Biol. Chem. 278:14949-14955. [DOI] [PubMed] [Google Scholar]
- 51.Viswanathan, P., T. Ohn, Y.-C. Chiang, J. Chen, and C. L. Denis. 2004. Mouse CAF1 can function as a processive deadenylase 3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 279:23988-23995. [DOI] [PubMed] [Google Scholar]
- 52.Voeltz, G. K., J. Ongkasuwan, N. Standart, and J. A. Steitz. 2001. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 15:774-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, Y., C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woolstencroft, R. N., T. H. Beilharz, M. A. Cook, T. Preiss, D. Durocher, and M. Tyers. 2006. Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. J. Cell Sci. 119:5178-5192. [DOI] [PubMed] [Google Scholar]
- 55.Yang, H., C. S. Duckett, and T. Lindsten. 1995. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol. Cell. Biol. 15:6770-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.