Abstract
E3 ubiquitin ligases play important roles in regulating transforming growth factor β (TGF-β)/Smad signaling. Screening of an E3 ubiquitin ligase small interfering RNA library, using TGF-β induction of a Smad3/Smad4-dependent luciferase reporter as a readout, revealed that Arkadia is an E3 ubiquitin ligase that is absolutely required for this TGF-β response. Knockdown of Arkadia or overexpression of a dominant-negative mutant completely abolishes transcription from Smad3/Smad4-dependent reporters, but not from Smad1/Smad4-dependent reporters or from reporters driven by Smad2/Smad4/FoxH1 complexes. We show that Arkadia specifically activates transcription via Smad3/Smad4 binding sites by inducing degradation of the transcriptional repressor SnoN. Arkadia is essential for TGF-β-induced SnoN degradation, but it has little effect on SnoN levels in the absence of signal. Arkadia interacts with SnoN and induces its ubiquitination irrespective of TGF-β/Activin signaling, but SnoN is efficiently degraded only when it forms a complex with both Arkadia and phosphorylated Smad2 or Smad3. Finally, we describe an esophageal cancer cell line (SEG-1) that we show has lost Arkadia expression and is deficient for SnoN degradation. Reintroduction of wild-type Arkadia restores TGF-β-induced Smad3/Smad4-dependent transcription and SnoN degradation in these cells, raising the possibility that loss of Arkadia function may be relevant in cancer.
The transforming growth factor β (TGF-β) superfamily of ligands comprises TGF-βs, Activin/Nodal family members, bone morphogenetic proteins (BMPs), and growth and differentiation factors (26). These ligands signal through a heterotetrameric complex of two type II receptors and two type I receptors, both serine/threonine kinases. The ligand brings the receptors together, enabling the type II receptor to phosphorylate and activate the type I receptor. The activated type I receptor signals to the nucleus primarily through phosphorylation of receptor-regulated Smads (R-Smads) (12). Broadly speaking, TGF-β and Activin/Nodal ligands induce activation of the R-Smads, Smad2 and Smad3, while the BMP and growth and differentiation factor ligands induce activation of Smad1, -5, and -8. Activated R-Smads form homomeric complexes and heteromeric complexes with Smad4 which accumulate in the nucleus. There they are recruited to promoter elements in conjunction with other transcription factors to regulate transcription both positively and negatively.
Different Smad complexes target different promoter elements. Smad3/Smad4 complexes bind directly to direct or inverted repeats of the GTCT sequence or its reverse complement, AGAC (44), such as those found in the PAI-1 promoter (6) or c-Jun promoter (41). A spliced variant of Smad2 (Smad2Δexon3) also binds as a complex with Smad4 to these same repeated GTCT or AGAC sequences (5, 42). Complexes of Smad4 with Smad1 or Smad5 also bind DNA directly and have recently been shown to recognize a GRCKNCN5GTCT consensus in cooperation with the zinc finger protein Schnurri (43). Such BMP-responsive elements (BREs) are found in the Id1 promoter (20). Full-length Smad2 cannot bind DNA directly; thus, Smad2/Smad4 complexes are recruited to DNA via other transcription factors, the best characterized being members of the FoxH1 family (3) and Mix family (13).
The relatively simple Smad pathway is subject to multiple levels of regulation which allows the pathway to be fine tuned and modulated by other growth factor signaling pathways and the cell cycle (12). The pathway is also regulated by negative-feedback mechanisms which limit the duration of Smad signaling. E3 ubiquitin ligases are emerging as important negative regulators of TGF-β signaling pathways (17). Protein ubiquitination occurs in three stages utilizing E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligase) enzymes (32). E3 ubiquitin ligases are predominantly of two types: those that contain RING fingers and those that contain HECT domains. They interact specifically with the substrate, and they facilitate (RING finger E3s) or catalyze (HECT domain E3s) the transfer of ubiquitin from the E2 enzyme, respectively.
The HECT domain-containing protein Smurf1 (Smad ubiquitination regulatory factor 1) was the first E3 ubiquitin ligase shown to be involved in TGF-β signaling. It binds Smad1 and Smad5 through its WW domain and a PY motif in the Smads and induces ubiquitination and degradation of these Smads (45). A close family member, Smurf2, was then shown to regulate levels of Smad1 and Smad2 (17). Smurf2 may also degrade activated R-Smads, as the association between Smurf2 and Smad2 or Smad3 is promoted by TGF-β signaling (17). Other E3 ubiquitin ligases preferentially degrade phosphorylated R-Smads, such as the multisubunit RING E3 ubiquitin ligase, Skp-1/Cul/Fbox complex which targets phosphorylated Smad3, and the HECT domain E3 ligases, Nedd4-2 and WWP1/Tiul1, which target phosphorylated Smad2. Like the R-Smads, Smad4 is regulated by E3 ligases and Smurf1/2, Nedd4-2, and WWP1/Tiul1, as well as the RING finger protein, Ectodermin/Tif1γ, have all been implicated in Smad4 degradation (8, 29).
The Smurfs also have other targets in the cell. They are recruited via the inhibitory Smads, Smad6 and Smad7, to the activated TGF-β, Activin, and BMP-type I receptors and induce their degradation (9, 18). This provides a negative-feedback mechanism to terminate signaling. These E3 ubiquitin ligases also promote TGF-β signaling by degrading repressors of the pathway. The transcriptional repressors Ski and SnoN interact with activated Smad2 and Smad3 and also Smad4 and have been thought to repress transcription by disrupting formation of active heteromeric Smad complexes, recruiting transcriptional corepressor complexes, and blocking interaction of activated Smads with transcriptional activators (25). SnoN (37) and to a lesser extent, Ski (38) are ubiquitinated and degraded rapidly via the proteasome upon TGF-β stimulation. This requires Smad2 or Smad3, and lysines 440, 446, and 449 of SnoN have been shown to be required for SnoN ubiquitination (1, 36, 39). The E3 ubiquitin ligases so far implicated in this process are Smurf2, which is recruited to SnoN via Smad2 or the anaphase-promoting complex (APC), which is recruited via Smad3 (1, 36, 39).
Unlike most other E3 ubiquitin ligases that modulate the TGF-β signaling pathway, Arkadia, a RING finger E3 ubiquitin ligase encoded by the gene, RNF111, was identified as a protein that enhances a subset of responses mediated by the TGF-β family member Nodal during early mouse and Xenopus embryonic development (11, 31). Subsequent work has indicated that Arkadia binds to Smad7, an inhibitory Smad, and causes its degradation. The lowering of basal levels of Smad7 in this way is thought to enhance both TGF-β and BMP signaling (19). It has recently been shown that Axin acts as a scaffold protein and cooperates with Arkadia to promote degradation of Smad7 (24).
To produce a comprehensive picture of the roles of E3 ubiquitin ligases in the TGF-β signaling pathway, we undertook a small interfering RNA (siRNA) screen of 289 well-annotated human E3 ubiquitin ligases and related proteins from the RefSeq database using a HaCaT cell line containing a stably integrated Smad3/Smad4-dependent luciferase reporter, CAGA12-Luc (6). Strikingly, we found in this screen that only knockdown of Arkadia abolished TGF-β-induced transcription to the same extent as knocking down components of the pathway, like Smad3 and Smad4. Since this would not be expected from the modulatory role ascribed to Arkadia in the literature, we investigated the mechanism of Arkadia function. Our data indicate that Arkadia functions to specifically promote transcription via Smad3/Smad4 binding sites by degrading the transcriptional repressor, SnoN, in response to TGF-β signaling.
MATERIALS AND METHODS
Cells, plasmids, and siRNAs.
HaCaT, 293T, NIH 3T3 and SEG-1 cells were cultured in Dulbecco modified Eagle medium containing 10% fetal calf serum. The HaCaT CAGA12-Luc/TK-Renilla cell line was generated by successive rounds of clonal selection of the CAGA12-Luc plasmid with puromycin and the TK-Renilla plasmid (Promega) with blasticidin. The HaCaT c-JunSBR6-Luc line, which contains a Smad3/Smad4-dependent reporter that contains six repeats of the Smad binding region (SBR) of the c-Jun promoter (c-JunSBR6-Luc reporter), was also generated using blasticidin selection. Plasmids and siRNAs and transfection conditions are described in the supplemental material.
Cell treatments.
Cells were induced at the indicated times with 2 ng/ml TGF-β1 (PreproTech), 20 ng/ml Activin (R&D Systems), or 20 ng/ml BMP4 (R&D Systems). In the case of 293T, cells were treated overnight with 10 μM SBI (SB-431542; Tocris) to inhibit autocrine signaling prior to washing the cells with fresh medium and induction with Activin. For proteasome inhibition, 293T cells were treated for 3 h with 25 μM MG132 (Sigma) in the presence of SBI prior to induction with Activin in the presence of MG132 for another hour.
Luciferase assays.
Luciferase assays were performed using the Dual-Luciferase reporter system (Promega) that allows sequential measurement of luciferase and Renilla activity in the same well. Luciferase activities were normalized to Renilla activities. Apart from the siRNA library, which was transfected in duplicate only, all other experiments were performed in quadriplicate and repeated at least three times.
Western blotting, DNA pull-down assays, immunoprecipitations, ubiquitination analysis, and immunofluorescence.
Whole-cell extracts were prepared from six-well plates using radioimmunoprecipitation assay buffer (50 mM Tris [pH 8], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate). Nuclear extracts were prepared as described previously (41). Western blotting was performed using standard procedures. The following antibodies were used: antibodies against Smad4 (B8; Santa Cruz), Smad3 (Abcam), Smad2 (Zymed), Smad2/3 (BD Biosciences), phospho-Smad2 (Cell Signaling Technology), phospho-Smad3 (40), p21 (C19; Santa Cruz), SnoN (H-317; Santa Cruz), PAI-1 (C9; Santa Cruz), Grb2 (BD Biosciences), Arkadia (RNF111 antibody; ABNOVA), Smurf1 (H-60; Santa Cruz), MCM6 (C-20; Santa Cruz), poly(ADP-ribose) polymerase (Roche), hemagglutinin (HA) (Roche), and His (Roche). The Flag antibody was covalently coupled to horseradish peroxidase (Sigma).
DNA pull-down assays were performed as described previously (15). Briefly, for each condition, 5 μg of 5′-biotinylated double-stranded oligonucleotides corresponding to the wild-type SBR of the c-Jun promoter (5′ GGAGGTGCGCGGAGTCAGGCAGACAGACAGACACAGCCAGCCAGCCAGGTCGGCA 3′ [the AGAC motifs are underlined]) or a version mutated in the Smad3/Smad4 binding sites and flanking CCAG repeats (5′ GGAGGTGCGC GGAGTCAGGCATATATATATATACAGCATGCATGCATGGTCGGCA 3′ [mutated motifs underlined]) were bound to 20 μl of Neutravidin-coated beads (Perbio), and DNA pull-down experiments were performed using 200 μg nuclear extract in buffer containing 140 mM NaCl in the presence of 20 μg of nonbiotinylated mutant oligonucleotides to reduce nonspecific binding. After extensive washing, bound proteins were detected by Western blotting.
Immunoprecipitations were performed either using nuclear extract or with whole-cell extract in lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40, 10% glycerol, 1 mM dithiothreitol, 25 mM NaF, 25 mM Na β-glycerophosphate, and protease inhibitors) using 5 μg of the corresponding antibody coupled to protein G plus protein A-Sepharose beads. Flag immunoprecipitations were performed using anti-Flag M2 affinity gel (Sigma). For transfected cells, immunoprecipitations were performed in lysis buffer containing 200 mM NaCl.
For ubiquitination analysis, cells were treated with 50 μM MG132 for 4 h prior to immunoprecipitation of whole-cell extract in lysis buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 1% NP-40, 10% glycerol, 1 mM dithiothreitol, 25 mM NaF, 25 mM Na β-glycerophosphate, protease inhibitors, 50 μM MG132, 0.25 μg/ml ubiquitin-aldehyde) followed by extensive washing with lysis buffer containing 400 mM NaCl. Polyubiquitinated HA-SnoN was detected by Western blotting.
Immunofluorescence was carried out as described previously (33). In all cases, cells were fixed for 10 min in 3.7% formaldehyde at room temperature, except for the detection of SnoN in HaCaT cells (see Fig. 3) where cells were fixed for 5 min in methanol at −20°C.
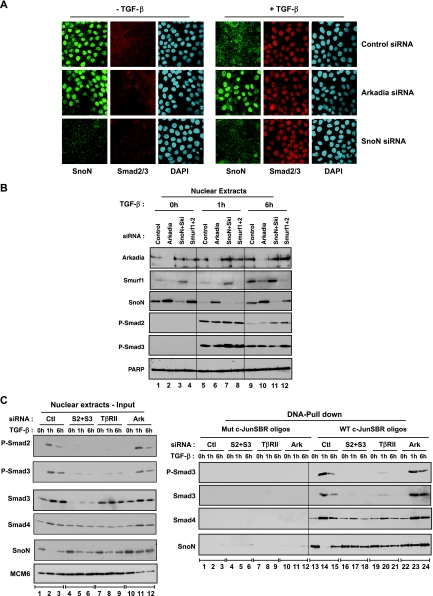
FIG. 3.
TGF-β-induced SnoN degradation requires Arkadia. (A) HaCaT cells were transfected with the indicated siRNA SMARTpools, then treated with TGF-β for 1 h (+ TGF-β) or not treated with TGF-β (− TGF-β), and processed for immunofluorescence using an anti-Smad2/3 antibody and an anti-SnoN antibody. Nuclei were visualized with 4′,6′-diamidino-2-phenylindole (DAPI). (B) HaCaT cells were transfected with the indicated siRNA SMARTpools and then treated with TGF-β for the times indicated or not treated with TGF-β. Nuclear extracts were analyzed directly by Western blotting using antibodies against Arkadia, Smurf1, SnoN, phosphorylated Smad2 (P-Smad2), P-Smad3, and poly(ADP-ribose) polymerase (PARP) as a control. (C) HaCaT cells were transfected with the indicated siRNA SMARTpools and then treated with TGF-β or not treated with TGF-β for the times indicated. Nuclear extracts were either analyzed directly by Western blotting using antibodies against SnoN, Smad3, P-Smad2, P-Smad3, and MCM6 as a control (left blots) or by DNA pull-down assay using the wild-type c-JunSBR oligonucleotide or a version mutated in the Smad3/Smad4 binding sites (right blots). Abbreviations: Ctl, nontargeting control siRNA; S2+S3, Smad2 and Smad3; Ark, Arkadia; Mut and WT c-JunSBR oligos, mutant and wild-type c-JunSBR oligonucleotides, respectively.
RESULTS
Arkadia is an E3 ligase that is absolutely required for TGF-β-induced transcription, and it is specific for transcription via repeated AGAC or GTCT motifs.
To identify the E3 ubiquitin ligases that play important roles in the regulation of the TGF-β pathway, we performed an RNA interference screen using a Smad3/Smad4-dependent luciferase reporter as a readout. We generated a HaCaT cell line that stably expresses CAGA12-Luc, a Smad3/Smad4-dependent reporter containing 12 copies of the CAGAC sites (AGAC motif underlined) from the PAI-1 promoter (6). We refer to this reporter throughout as a Smad3/Smad4-dependent reporter, although it also binds complexes of Smad2Δexon3 with Smad4 (5). The cell line also contains a Renilla reporter driven by the thymidine kinase (TK) promoter to act as an internal control. A library of 289 siRNA SMARTpools targeting known or predicted human E3 ubiquitin ligases was transfected in duplicate into this HaCaT cell line along with nontargeting control siRNAs and the positive-control siRNAs targeting Smad3, Smad4, and TGF-β receptor type II (TβRII). After 72 h, cells were stimulated with TGF-β for 8 h. Luciferase activities were normalized to the appropriate Renilla activities. The duplicate experiments are presented on a logarithmic scale in Fig. 1A. Strikingly, the trend line of all the values yields a slope of approximately 1, indicating that the duplicate experiments were highly reproducible. The list of genes targeted and the normalized values are given in Table S1 and Fig. S1 in the supplemental material.
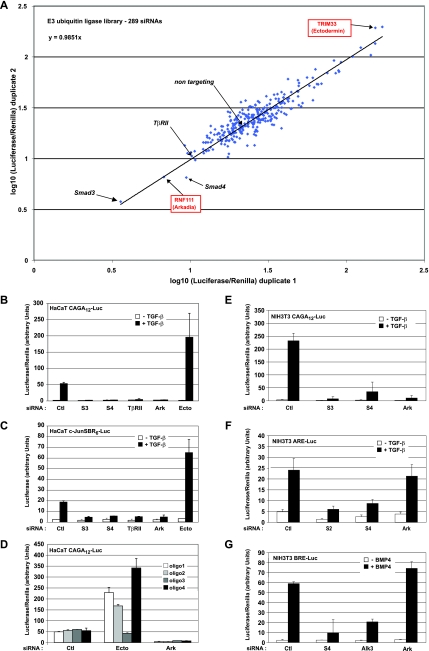
FIG. 1.
Loss of Arkadia completely and specifically inhibits the Smad3-dependent TGF-β pathway. (A) 289 siRNA SMARTpools were screened in duplicate using the HaCaT CAGA12-Luc/TK-Renilla cell line. Luciferase levels were analyzed and normalized to Renilla levels (Luciferase/Renilla). The two duplicate experiments are represented on a dot plot using a logarithmic (log10) scale which provides an easier representation of the range of values on the same graph. The negative control was a nontargeting siRNA, and positive controls were siRNAs against Smad3, Smad4, and TβRII as indicated. The dots corresponding to the Luciferase/Renilla values for siRNAs that target RNF111 (Arkadia) and TRIM33 (Ectodermin) are also indicated. (B and C) Plots of Luciferase/Renilla values for HaCaT CAGA12-Luc/TK-Renilla cells (B) or luciferase only values for c-JunSBR6-Luc cells (C) transfected with the indicated siRNA SMARTpools and then treated with TGF-β (+ TGF-β) or not treated with TGF-β (− TGF-β). (D) Plots of Luciferase/Renilla values for HaCaT CAGA12-Luc/TK-Renilla cells transfected with the four single siRNAs corresponding to the deconvolution of the nontargeting SMARTpool or those that target Ectodermin or Arkadia. (E to G) NIH 3T3 cells were transfected with the indicated mouse siRNA SMARTpools followed by transfection with plasmids encoding the TK-Renilla reporter and either CAGA12-Luc (E) or ARE-Luc together with the plasmid encoding xFoxH1a (F) or BRE-Luc (G). Cells were treated with TGF-β or BMP4 or not treated with TGF-β or BMP4 as indicated. Luciferase activity was normalized to Renilla activity. Abbreviations in panels B to G: Ctl, nontargeting control siRNA; S3, Smad3; S4, Smad4; Ark, Arkadia; Ecto, Ectodermin; oligo, oligonucleotide.
Most siRNAs had no effect and behaved as the nontargeting control siRNA did. Several siRNAs, however, strongly enhanced the TGF-β-induced level of the CAGA12-Luc reporter (Fig. 1A); these siRNAs included Ectodermin (TRIM33) (8). Intriguingly, only the siRNA that targets Arkadia (RNF111) very strongly inhibited TGF-β-induced CAGA12-Luc activity (Fig. 1A). Knockdown of Arkadia inhibited TGF-β induction of the CAGA12-Luc reporter to approximately the same extent as did siRNAs against major components of the pathway, Smad3, Smad4, or TβRII (Fig. 1A and B). Arkadia knockdown also strongly suppressed TGF-β induction of another Smad3/Smad4-dependent reporter that contains six repeats of the SBR of the c-Jun promoter, c-JunSBR6-Luc reporter (23) (Fig. 1C), strengthening the idea that Arkadia is an activator of Smad3/Smad4-dependent transcription. In contrast, knockdown of Ectodermin enhanced transcription of this reporter, consistent with the results of the screen and the inhibitory role of Ectodermin in the TGF-β pathway (8). Deconvolution of the SMARTpools showed that the four different siRNA oligonucleotides targeting Arkadia have the same effect and that three out of four oligonucleotides that target Ectodermin have the same effect, indicating that these effects are specific to the targeted proteins (Fig. 1D).
To determine whether Arkadia is a general activator of the TGF-β and BMP pathways, we assessed the effect of Arkadia knockdown on other luciferase reporters, which bind different Smad complexes. We used the ARE-Luc reporter, which is derived from the activin responsive element (ARE) of the Xenopus Mix.2 promoter, cotransfected with a plasmid expressing FoxH1 to assay Smad2/Smad4-dependent transcription (33), and the BRE-Luc reporter, which is derived from the BMP-responsive element of the Id1 promoter, to assay Smad1/Smad4-dependent transcription (20). As observed in HaCaT cells, Arkadia is required for Smad3/Smad4-dependent transcription from the CAGA12-Luc reporter in mouse NIH 3T3 cells to the same extent as is Smad3 and Smad4 (Fig. 1E). However, we did not observe any requirement for Arkadia for Smad2/Smad4-dependent transcription or for Smad1/Smad4-dependent transcription, although knockdown of the relevant Smads or, in the case of the BRE-Luc, the relevant type I receptor (Alk3), did inhibit ligand-induced transcriptional activation as expected (Fig. 1F and G).
Therefore, our results indicate that Arkadia is specifically required for TGF-β-induced transcription via Smad3/Smad4 binding sites.
Knockdown of Arkadia has no effect on TGF-β-induced Smad3 phosphorylation or nuclear accumulation.
We next investigated at which level Arkadia is required in the TGF-β pathway. Previous work has suggested that Arkadia regulates the degradation of Smad7 (19). Thus, Arkadia knockdown would be expected to result in increased levels of Smad7 and hence decreased levels of receptors and reduced phosphorylation and nuclear accumulation of Smad2 and Smad3. In contrast, we found that Arkadia knockdown had no effect on TGF-β-induced nuclear accumulation of Smad2 or Smad3 or on Smad3 phosphorylation, when assayed by immunostaining (Fig. 2A). Moreover, knockdown of Arkadia had no effect on the kinetics or levels of phosphorylated Smad3 when assayed by Western blotting (Fig. 2B). As a control, we could show that TβRII knockdown inhibited TGF-β-induced accumulation of Smad2 and -3 and phosphorylation of Smad3 (Fig. 2A and B). This suggests that Arkadia is not functioning through Smad7 in these cells. In fact, neither Smad7 nor its partners, Smurf1 and Smurf2, have a potent basal inhibitory function in HaCaT cells, as demonstrated by the fact that siRNA knockdown of Smad7, Smurf1, or Smurf2 in HaCaT cells had no stimulatory effect on CAGA12-Luc activity (Fig. 2C), although we could demonstrate that the siRNAs were effective (see Fig. 3B; see Fig. S2 in the supplemental material). This is consistent with the very low basal levels of Smad7 mRNA in HaCaT cells (23). Thus, we conclude that Arkadia is unlikely to promote TGF-β-induced transcription through its ability to degrade Smad7 and must act downstream of TGF-β-induced Smad3 phosphorylation and nuclear accumulation. Consistent with this, Arkadia knockdown strongly inhibited the induction of the TGF-β target genes, PAI-1 and p21 (6, 35), which contain repeated AGAC sequences in their promoters (Fig. 2B). In fact, this inhibition was as effective as that observed with TβRII knockdown (Fig. 2B).
FIG. 2.
Arkadia acts downstream of TGF-β-induced Smad phosphorylation and nuclear accumulation. (A) HaCaT cells were transfected with the indicated siRNA SMARTpools, then treated with TGF-β (+ TGF-β) for 1 h or not treated with TGF-β (− TGF-β), and processed for immunofluorescence using an anti-Smad2/3 antibody and an anti-phosphorylated-Smad3 (anti-P-Smad3) antibody. Nuclei were visualized with 4′,6′-diamidino-2-phenylindole (DAPI). (B) HaCaT cells were transfected with the indicated siRNA SMARTpools and then treated with TGF-β or not treated with TGF-β for the indicated times. Whole-cell extracts were analyzed by Western blotting using antibodies against Smad3, P-Smad3, and against the Smad3 target genes PAI-1 and p21. Grb2 was used as a loading control. (C) Plots of Luciferase/Renilla values for HaCaT CAGA12-Luc/TK-Renilla cells transfected with the indicated siRNA SMARTpools and then treated with TGF-β or not treated with TGF-β.
Arkadia is required for TGF-β-induced SnoN degradation.
Since Arkadia has E3 ubiquitin ligase activity, we investigated whether Arkadia could mediate degradation of a transcriptional repressor. SnoN and the related protein Ski, are known to bind to the same repeated GTCT or AGAC elements as activated Smad3/Smad4 complexes do (4, 30), suggesting that they would preferentially inhibit transcription mediated by these elements. Indeed, knockdown of SnoN, and to a lesser extent, Ski, increased TGF-β-induced CAGA12-Luc activity in HaCaT cells (Fig. 2C), indicating that SnoN is the major repressor of the two in HaCaT cells. Knockdown of both SnoN and Ski enhanced this induction further, suggesting that in the absence of SnoN, Ski also acts as a repressor of Smad3/Smad4-dependent transcription.
SnoN is known to be degraded shortly after induction by TGF-β, and this has been proposed to be a prerequisite for TGF-β-induced Smad3/Smad4-dependent transcription (37). As observed by immunofluorescence (Fig. 3A) or Western blotting (Fig. 3B, compare lanes 1 and 5), HaCaT cells have high levels of nuclear SnoN in the absence of a TGF-β signal, but SnoN is completely degraded after 1 h of TGF-β treatment. The disappearance of SnoN from the nucleus is abolished when the proteasome is inhibited by treatment with MG132 and is not associated with an increase in cytoplasmic SnoN (Fig. 3A; see Fig. S3 in the supplemental material). This confirms that TGF-β stimulation readily induces a rapid degradation of SnoN that is mediated by the proteasome. Levels of SnoN are restored 6 h after TGF-β treatment (Fig. 3B, lane 9), consistent with its expression being induced upon prolonged TGF-β signaling (37).
Strikingly, knockdown of Arkadia either with the SMARTpool or with two individual siRNAs from the SMARTpool strongly inhibits degradation of SnoN upon TGF-β stimulation (Fig. 3A; Fig. 3B, lanes 5 and 6; Fig. 3C, left blots, lanes 2 and 11; see Fig. S4 in the supplemental material). In contrast, knockdown of Smurf1 and Smurf2, which have previously been implicated in SnoN degradation (1, 36, 39) has no effect (Fig. 3B, compare lanes 4, 8, and 12). This observation is consistent with the fact that knockdown of Smurf1 and -2 has no effect on Smad3/Smad4-dependent transcription (Fig. 2C) and that they did not register as hits in our screen (see Table S1 in the supplemental material). Note that since Smurf1 is also induced by TGF-β, knockdown of Arkadia, which is an activator of the TGF-β pathway, inhibits Smurf1 induction, whereas knockdown of SnoN and Ski, which are repressors, increases Smurf1 induction (Fig. 3B, compare lanes 5 to 7 to lanes 9 to 11). Interestingly, knockdown of Arkadia has only a small effect on the basal level of SnoN in the absence of TGF-β compared to the more dramatic inhibition of signal-induced degradation of SnoN (Fig. 3B; see Fig. S3 and S4 in the supplemental material), indicating that Arkadia might be required primarily for TGF-β-induced SnoN degradation and not for its steady-state level.
We then used DNA pull-down assays (Fig. 3C) to investigate the binding of SnoN to the SBR of the c-Jun promoter, whose activity is affected by Arkadia knockdown. SnoN bound the SBR in the absence of signal, but its binding was lost after 1 h of TGF-β stimulation and then restored after 6 h (Fig. 3C, right blots, lanes 13 to 15). In the basal state, a low level of Smad4 was bound to the SBR, but phosphorylated Smad3 was absent (Fig. 3C, right blots, lanes 13). Strong binding of phosphorylated Smad3 and Smad4 to the SBR was observed at the 1-h time point after TGF-β stimulation, which correlated with the degradation of SnoN (Fig. 3C, right blots, lanes 14). Binding of phosphorylated Smad3 and Smad4 to the SBR then diminished 6 h after TGF-β stimulation, which is consistent with the lower levels of phosphorylated Smad3 and Smad4 at this time point (Fig. 3C, right blots, lanes 15, and left blots, lanes 3). In the absence of Arkadia, strong binding of SnoN to the SBR was still observed after a 1-h TGF-β treatment (Fig. 3C, right blots, compare lanes 14 and 23; see Fig. S5 in the supplemental material). Because excess oligonucleotides are used in these experiments, it is still possible to detect the TGF-β-induced phosphorylated Smad3/Smad4 complex in these conditions. Noticeably, knockdown of Smad4 had no effect on TGF-β-induced SnoN degradation, but it did inhibit the ability of SnoN to bind the SBR in the absence of TGF-β (see Fig. S5 in the supplemental material [left and right panels, compare lanes 1 and 10]). This suggests that SnoN binds the SBR in conjunction with Smad4 in unstimulated cells (16). TGF-β-induced SnoN degradation was not affected when Smad2 or Smad3 were knocked down individually (see Fig. S5 in the supplemental material [left and right panels, lanes 4 to 9]) but was strongly inhibited when both Smad2 and Smad3 were absent or when TβRII was knocked down (Fig. 3C), suggesting that phosphorylated Smad2 and Smad3 act redundantly downstream of TGF-β in mediating SnoN degradation. In all DNA pull-down assays, binding of SnoN, Smad3, and Smad4 to the SBR was specific, as we detected no binding to an oligonucleotide mutated in the Smad binding sites (Fig. 3C, right blots, lanes 1 to 12).
From these experiments, we conclude that TGF-β-induced degradation of SnoN absolutely requires Arkadia together with phosphorylated Smad2 or Smad3.
Arkadia mutated in the RING domain has a dominant-negative effect on Smad3/Smad4-dependent transcription by preventing TGF-β-induced SnoN degradation.
We next determined whether the involvement of Arkadia in TGF-β-induced SnoN degradation could explain the requirement of Arkadia for Smad3/Smad4-dependent transcription. We used 293T cells, as they could be transfected with sufficient efficiency to visualize Arkadia derivatives. We used Activin in these cells to activate both Smad2 and Smad3, as 293T cells respond better to Activin than TGF-β. As expected, Arkadia is a strong transcriptional activator of the Smad3/Smad4-dependent transcription from the CAGA12-Luc reporter when overexpressed (Fig. 4A, top graph), and its activating effect was abolished by adding increasing amounts of SnoN (see Fig. S6 in the supplemental material). In agreement with the results of Arkadia knockdown experiments, we also observed that overexpression of wild-type Arkadia had a much less dramatic effect on Smad2-dependent transcription from the ARE-Luc reporter (Fig. 4A, middle graph) and no effect on Smad1-dependent transcription (Fig. 4A, bottom graph).
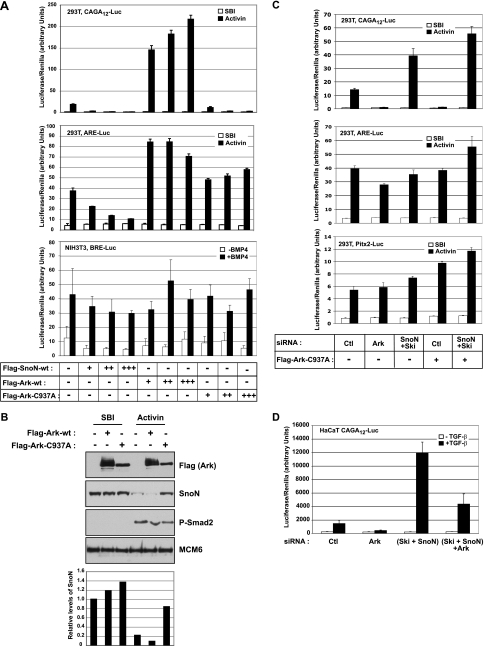
FIG. 4.
Arkadia mutated in its RING domain inhibits Smad3-dependent transcription by preventing TGF-β-induced SnoN degradation. (A) 293T or NIH 3T3 cells were cotransfected with the TK-Renilla reporter and either CAGA12-Luc (top graph) or ARE-Luc together with the plasmid encoding xFoxH1a (middle graph) or with BRE-Luc (bottom graph), with increasing amounts of plasmids encoding Flag-SnoN-wt (Flag-tagged wild-type SnoN), Flag-Ark-wt (Flag-tagged wild-type Arkadia), or Flag-Ark-C937A as indicated (−, none). 293T cells were treated with SB-431542 (SBI) overnight to abolish autocrine signaling before induction or not with Activin. NIH 3T3 cells were treated with BMP4 (+BMP4) or not treated with BMP4 (−BMP4). (B) 293T cells were transfected with plasmids encoding Flag-Ark-wt or Flag-Ark-C937A. Cells were treated with SBI and Activin as described above for panel A. Nuclear extracts were analyzed by Western blotting using antibodies against the Flag tag, SnoN, phosphorylated Smad2 (P-Smad2), and MCM6, as a loading control. Quantification of SnoN levels relative to MCM6 levels is shown below the blots. (C) 293T cells were transfected with the indicated siRNA SMARTpools, followed by transfection with the TK-Renilla reporter and either CAGA12-Luc, ARE-Luc, or Pitx2-Luc together with a plasmid encoding xFoxH1a and in the presence (+) or absence (−) of Flag-Ark-C937A. Cells were treated with SBI and Activin as described above for panel A. (D) HaCaT CAGA12-Luc/TK-Renilla cells were transfected with the indicated siRNA SMARTpools and treated with TGF-β (+TGF-β) or not treated with TGF-β (−TGF-β). Ctl, nontargeting control siRNA; Ark, Arkadia.
Importantly, Arkadia mutated in its RING domain, which is required for its E3 ubiquitin ligase activity (Arkadia C937A), acts dominant negatively on Smad3/Smad4-dependent transcription (Fig. 4A, top graph) and also on ligand-induced SnoN degradation (Fig. 4B). This dominant-negative activity likely results from the ability of this mutant to compete with endogenous Arkadia for SnoN (see below). Thus, expression of the Arkadia C937A mutant or knocking down Arkadia both have the same inhibitory effect on Smad3/Smad4-dependent transcription by preventing degradation of SnoN. Crucially, silencing of SnoN/Ski abolished the dominant-negative effect of Arkadia C937A on Smad3/Smad4-dependent transcription (Fig. 4C) and the inhibitory effect of Arkadia knockdown (Fig. 4D). This indicates that the inhibitory effects of the Arkadia mutant and Arkadia knockdown require endogenous SnoN/Ski.
In agreement with the loss of Arkadia function experiments, overexpression of the Arkadia mutant also had no inhibitory effect on the Smad2- or Smad1-dependent reporters (Fig. 4A). This indicates that preventing ligand-induced degradation of endogenous SnoN has no effect on these responses. Consistent with this, knockdown of endogenous SnoN/Ski proteins also had no effect on Smad2-dependent transcription from the ARE-Luc reporter or from another Smad2-dependent reporter derived from the Pitx2 gene (34) (Fig. 4C), although it increased Smad3/Smad4-dependent transcription (Fig. 3A and 4C). When overexpressed, SnoN strongly repressed Smad3/Smad4-dependent transcription and had no effect on the Smad1-dependent reporter (Fig. 4A). Overexpression of SnoN had a weaker dose-dependent inhibitory effect on the Smad2-dependent reporter (ARE-Luc) (Fig. 4A), which is likely to result from SnoN titrating phosphorylated Smad2 away from the ARE (37), rather than from direct transcriptional repression.
Taken together, these data indicate that endogenous Ski and SnoN act as specific repressors of transcription mediated via Smad3/Smad4 binding sites, which can be explained by their ability to bind these elements that comprise repeats of AGAC or GTCT motifs (4, 30). We demonstrate that endogenous Arkadia specifically activates transcription mediated via these elements by triggering TGF-β/Activin-induced SnoN (and Ski) degradation.
Phosphorylated Smad2 or Smad3 regulates degradation of SnoN by Arkadia.
We have shown that neither knockdown nor overexpression of Arkadia has much effect on the basal level of SnoN in the absence of ligand induction. We therefore sought to understand how TGF-β/Activin signaling, mediated through either phosphorylated Smad2 or Smad3, can trigger SnoN degradation mediated by Arkadia. For these experiments, we focused mainly on endogenous phosphorylated Smad2, as 293T cells contain very low levels of endogenous Smad3.
We first examined the ability of Arkadia to interact with SnoN and phosphorylated Smad2 using the inactive mutant of Arkadia to prevent degradation of SnoN. In nuclear extracts prepared from 293T cells transfected with Flag-Ark-C937A, Arkadia formed a complex with endogenous SnoN in the absence and presence of Activin and with endogenous phosphorylated Smad2 in the presence of Activin (Fig. 5A). Since nuclear extracts do not contain Smad2 in the absence of ligand, we investigated whether Arkadia specifically binds Smad2 in a ligand-dependent manner by performing coimmunoprecipitations using whole-cell extracts. Arkadia binds endogenous Smad2 only in the presence of Activin signaling, indicating that Smad2 binds to Arkadia only when it is phosphorylated (Fig. 5B). Because of the very low levels of Smad3 in these cells, we could not detect an interaction between endogenous Smad3 and Arkadia. However, when HA-Smad3 or HA-Smad2 was expressed in these cells, we observed that both could interact with Arkadia in a ligand-induced manner (Fig. 5C).
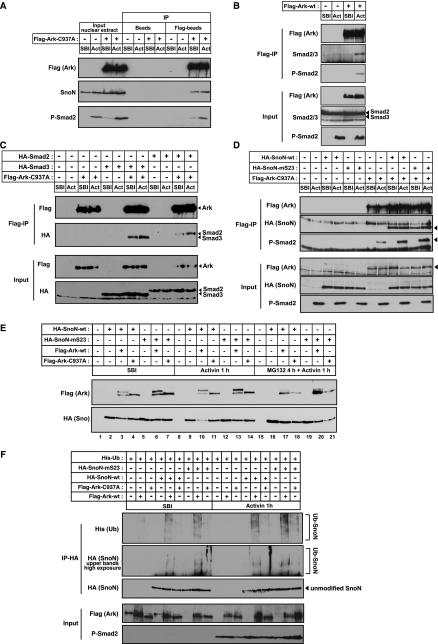
FIG. 5.
Arkadia binds and ubiquitinates SnoN independently of phosphorylated Smad2/3, but Arkadia-mediated SnoN degradation requires phosphorylated Smad2/3. (A) 293T cells were transfected with an empty vector (−) or with Flag-Ark-C937A (+) as indicated. Cells were treated with SB-431542 (SBI) overnight, before induction with Activin (Act) for 1 h. Nuclear extracts were analyzed by Western blotting using anti-Flag, anti-SnoN, or anti-phosphorylated Smad2 (anti-P-Smad2) antibodies, either directly (input) or after immunoprecipitation (IP) with anti-Flag beads (Flag-beads) or empty beads (Beads). (B to D) 293T cells were transfected with Flag-Ark-wt (Flag-tagged wild-type Arkadia) (B) or Flag-Ark-C937, HA-Smad2, or HA-Smad3 as indicated (C) or with HA-SnoN-wt, HA-SnoN-mS23, or Flag-Ark-C937A (D) as indicated. Cells were treated as described above for panel A, but in addition, in panel B, cells were pretreated with MG132 for 4 h prior to Activin induction. Whole-cell extracts were analyzed by Western blotting using the indicated antibodies either directly (Input blots) or after IP with anti-Flag beads (Flag-IP). The arrowheads indicate the bands that correspond to the analyzed proteins. (E) 293T cells were transfected with the indicated plasmids, and cells were treated with SBI overnight and then either treated with MG132 (25 μM) for 3 h and induced with Activin for another hour (MG132 4 h + Activin 1 h) or with Activin only for 1 h (Activin 1 h) or kept in the presence of SBI. Whole-cell extracts were analyzed by Western blotting using anti-Flag and anti-HA antibodies. Wild-type Arkadia migrates as two bands. The top band corresponds to self-ubiquitinated Arkadia. (F) 293T cells were transfected and treated with SBI as described above and then treated with MG132 (50 μM) for 4 h prior to induction with Activin for another hour. Total lysates were immunoprecipitated with an anti-HA antibody. Ubiquitination of HA-SnoN was then assayed by Western blotting the IP with an anti-His antibody and also by overexposure of the top region of the HA blot. Levels of transfected Arkadia and endogenous P-Smad2 were analyzed by Western blotting of the total lysate. Ub, ubiquitinated.
SnoN is able to interact with Arkadia in the absence of an Activin signal; therefore, its binding to Arkadia does not require phosphorylated Smad2 or Smad3. We next investigated whether phosphorylated Smad2 interacted with Arkadia by virtue of its ability to bind SnoN, examining the interaction between Arkadia C937A with wild-type SnoN or a mutant form of SnoN that cannot bind Smad2 or Smad3 (SnoN-mS23) (16). As observed for endogenous SnoN, Arkadia bound overexpressed SnoN both in the presence and absence of signal (Fig. 5D). Interestingly, SnoN-mS23 was still able to interact with Arkadia (Fig. 5D), and endogenous phosphorylated Smad2 was still found in complexes with Arkadia in the presence of this mutated SnoN (Fig. 5D). Altogether, these results indicate that phosphorylated Smad2 does not bind to Arkadia through its ability to interact with SnoN. However, we observed that both overexpression of wild-type SnoN and SnoN-mS23 potentiates the binding of phosphorylated Smad2 to Arkadia (Fig. 5D), which suggests that phosphorylated Smad2 might interact more efficiently with the fraction of Arkadia that is bound to SnoN.
To better understand the role played by phosphorylated Smad2 in Arkadia-mediated degradation of SnoN, we assessed the effects of overexpression of wild-type Arkadia and mutant Arkadia C937A on the stability of wild-type SnoN and mutant SnoN-mS23. Overexpressed SnoN is not degraded in response to Activin, presumably because the endogenous Arkadia is limiting (Fig. 5E, compare lane 2 with lane 9). However, coexpression of wild-type Arkadia with wild-type SnoN strongly reduced SnoN levels upon Activin induction, but not in untreated cells (Fig. 5E, lanes 3 and 10). Since this effect was lost when cells were treated with proteasome inhibitor MG132 (Fig. 5E, lane 17) or when SnoN was coexpressed with the inactive Arkadia mutant (Fig. 5E, compare lane 4 and 11), we conclude that Arkadia induces SnoN degradation in an Activin-dependent manner through its E3 ubiquitin ligase activity. Importantly, wild-type Arkadia cannot degrade the mutant SnoN-mS23, which cannot interact with Smad2 or Smad3 (Fig. 5E, lane 13), despite the fact that SnoN-mS23 can bind Arkadia. This indicates that an interaction of SnoN with phosphorylated Smad2 in the Arkadia/SnoN/phosphorylated Smad2 degradation complex is required for ligand-induced SnoN degradation.
To determine whether the role of phosphorylated Smad2 in the complex was to activate Arkadia or to trigger degradation, we investigated the ubiquitination of SnoN by Arkadia. Arkadia, but not the mutant Arkadia C937A, induced polyubiquitination of SnoN and mutant SnoN-mS23 both in the absence and presence of Activin (Fig. 5F).
These results indicate that Arkadia binds and ubiquitinates SnoN in the nucleus in the absence of signal. However, binding of phosphorylated Smad2 (or Smad3) to SnoN and Arkadia upon ligand induction is required for efficient degradation of SnoN.
Arkadia restores Smad3/Smad4-dependent transcriptional activity in the Barrett's-associated esophageal adenocarcinoma cell line SEG-1 that is deficient for TGF-β-induced SnoN degradation.
SnoN is an oncogene that is overexpressed in a variety of tumors (22), and in addition, some esophageal cancer cell lines have lost their ability to degrade SnoN in response to TGF-β (10). The Barrett's-associated esophageal adenocarcinoma cell line SEG-1 (21) has normal levels of Smad4 and Smad2 and, like many other tumor cells, exhibits a low level of Smad3 (Fig. 6A) (22). This cell line responds efficiently to TGF-β as seen by induction of phosphorylation of Smad2 and Smad3 (Fig. 6A). However, the CAGA12-Luc reporter is inactive in these cells (Fig. 6B), and this is associated with an inefficient degradation of SnoN in response to TGF-β (Fig. 6A) (10). Knockdown of SnoN/Ski restored the response to the CAGA12-Luc reporter, indicating that the absence of TGF-β-induced degradation of SnoN (and possibly Ski) is responsible for the lack of a Smad3-dependent transcriptional response in these cells (Fig. 6B). Interestingly, we found that these cells have lost the expression of full-length Arkadia (Fig. 6A), and reexpression of Arkadia was able to restore a strong Smad3-dependent transcriptional response, while it had no effect on the Smad2-dependent transcriptional response through the ARE (Fig. 6C). Moreover, reintroduction of wild-type Arkadia in these cells also fully restores TGF-β-induced degradation of SnoN (Fig. 6D). These results suggest that the absence of a Smad3-dependent response in the SEG-1 cells can be attributed to a defect in the Arkadia-mediated degradation of SnoN upon TGF-β stimulation.
FIG. 6.
Arkadia expression restores the TGF-β-induced Smad3/Smad4 transcriptional response and SnoN degradation in the Barrett's-associated esophageal adenocarcinoma cell line SEG-1. (A) HaCaT and SEG-1 cells were treated with TGF-β for the indicated times (from 0 h to 30 min [30′] to 6 h), and nuclear extracts were analyzed by Western blotting using the indicated antibodies. MCM6 levels acted as a loading control. P-Smad2, phosphorylated Smad2. (B) SEG-1 cells were transfected with the control nontargeting siRNA SMARTpool (Ctl) or with siRNA SMARTpools targeting Ski and SnoN, followed by transfection with CAGA12-Luc and TK-Renilla and treatment with TGF-β (+TGF-β) or not (−TGF-β). (C) SEG-1 cells were transfected with TK-Renilla and either the Smad3-dependent reporter CAGA12-Luc or the Smad2-dependent reporter ARE-Luc together with the plasmid encoding xFoxH1a and in the presence (+) or absence (−) of Flag-Arkwt (Flag-tagged wild-type Arkadia) or Flag-Ark-C937A. Cells were treated with TGF-β (+TGF-β) or not treated with TGF-β (−TGF-β). (D) SEG-1 cells were transfected with plasmids expressing green fluorescent protein (GFP), GFP-Arkwt (GFP-tagged wild-type Arkadia), or GFP-Ark-ΔRING (GFP-tagged Arkadia deleted in the RING domain) for 24 h and then treated with TGF-β for 1 h or not treated with TGF-β. The cells were fixed, the GFP was visualized directly, and SnoN was detected by immunofluorescence. Nuclei were visualized with 4′,6′-diamidino-2-phenylindole (DAPI). White arrows indicate transfected cells.
DISCUSSION
Arkadia is required for the TGF-β-induced transcription via repeated AGAC or GTCT motifs.
The TGF-β superfamily signaling pathways are regulated by E3 ubiquitin ligases, most of which have been demonstrated to act negatively (17). Here we have performed a high-throughput siRNA screen of 289 known or predicted human E3 ubiquitin ligases using the CAGA12-Luc reporter as a transcriptional readout that contains Smad3/Smad4 binding sites (repeated AGAC or GTCT motifs). We have shown that knockdown of only one E3 ubiquitin ligase, Arkadia, inhibits this TGF-β response to the same extent as knockdown of direct components of the pathway, such as Smad3, Smad4, or TβRII, indicating that Arkadia is a strong positive regulator of TGF-β-induced transcription via repeated AGAC or GTCT motifs. Knockdown of Arkadia also prevented TGF-β-induced transcription of another luciferase reporter gene (c-JunSBR6-Luc) driven by these repeated motifs and also endogenous target genes PAI-1 and p21. Knockdown of Arkadia had no effect on Smad1/Smad4-dependent transcription via the BRE or on transcription driven by FoxH1/Smad2/Smad4 complexes via the ARE. Similarly, overexpression of wild-type and mutant Arkadia also affected only transcription from the Smad3/Smad4 binding sites. We therefore conclude that Arkadia is specifically required for transcriptional activation via repeated AGAC or GTCT elements that bind Smad3/Smad4 complexes (44), and we have demonstrated this requirement for Arkadia in the epithelial cell line HaCaT, 293T cells, and the fibroblast cell line NIH 3T3.
It is important to note that these repeated AGAC or GTCT elements also bind complexes of Smad2Δexon3 with Smad4 (5), which means that Arkadia will also be important for a subset of Smad2-mediated transcriptional responses in cells expressing this spliced isoform of Smad2. Although it exists at low levels in mammalian tissue culture cells, Smad2Δexon3 is strongly expressed in embryonic stem (ES) cells and in mouse embryos throughout development (7). Therefore, in mouse embryos, Arkadia will be important not only for Smad3/Smad4-dependent transcription but also for transcription mediated by Smad2Δexon3/Smad4 complexes. This explains why Arkadia−/− embryos do not phenocopy Smad3−/− embryos. In mice lacking Smad3, Smad2Δexon3 compensates; thus, the embryos are normal (7, 14). In mice lacking Arkadia, both Smad3 and Smad2Δexon3 function is compromised and although the embryos form anterior visceral endoderm normally, they lack primitive streak derivatives and anterior definitive endoderm, which leads to anterior patterning defects (11, 27). The Arkadia−/− phenotype is not as severe as the Smad2−/− phenotype (11, 27), as this additionally removes the function of full-length Smad2 and results in embryos that do not form anterior visceral endoderm and therefore lack anterior-posterior polarity and that are highly disorganized by embryonic day 8.5 (7, 14).
Our present work also allows us to explain why Arkadia regulates only a subset of TGF-β/Activin/Nodal transcriptional responses, i.e., those that are mediated via repeated AGAC or GTCT elements. We have shown that Arkadia is required for TGF-β/Activin-induced degradation of the transcriptional repressor SnoN, which specifically binds the same repeated AGAC or GTCT elements as Smad3/Smad4 complexes do (4). Thus, SnoN is a specific transcriptional repressor of promoters driven by these elements. We have validated in SnoN knockdown experiments that endogenous SnoN acts as a specific repressor of reporters driven by these elements, whereas it has no effect on Smad2-dependent reporters driven by AREs or on Smad1-dependent transcription via BREs. Moreover, failure to degrade endogenous SnoN when Arkadia is knocked down or when the Arkadia mutant is overexpressed has no effect on Smad2-dependent promoters. Since SnoN is able to bind to both phosphorylated Smad2 and phosphorylated Smad3 (16), its overexpression can titrate Smad2 from promoters. Therefore, it has a dose-dependent inhibitory effect on Smad2-dependent reporters when overexpressed. The endogenous level of SnoN, however, is not sufficient to inhibit Smad2-dependent transcription in this manner.
Model for the mechanism of Arkadia action.
A proposed model of the mechanism whereby Arkadia activates Smad3/Smad4-dependent transcription is shown in Fig. 7. SnoN binds repeated AGAC or GTCT elements together with Smad4 in unstimulated cells and recruits transcriptional corepressors, such as N-CoR or mSin3A (25). We have shown that in the absence of a signal, SnoN is also complexed with Arkadia in the nucleus. Since we have found no evidence that Arkadia forms complexes on DNA (our unpublished data), we assume that these complexes are in the nucleoplasm. It is likely that SnoN exists in a dynamic equilibrium between its DNA-bound form and non-DNA-bound form, as has been shown for other transcriptional regulators (28). In unstimulated cells, Arkadia has little effect on SnoN levels, since in the absence of phosphorylated Smad2 or Smad3, Arkadia cannot trigger efficient SnoN degradation, even though it can polyubiquitinate SnoN. Upon TGF-β/Activin stimulation, phosphorylated Smad2 or Smad3 forms a complex with Arkadia and SnoN, which leads to degradation of SnoN.
FIG. 7.
Model of mechanism of Arkadia action. (A) In the nuclei of unstimulated cells, SnoN is complexed to Arkadia (Ark), and SnoN is also bound to repeated AGAC elements (or the reverse complement [GTCT]) with Smad4, forming a transcriptionally repressive complex. In the absence of signal, SnoN is ubiquitinated by Arkadia, but it is not efficiently degraded. Ub, ubiquitin. (B) Upon TGF-β/Activin/Nodal stimulation, phosphorylated Smad2/3 (P-Smad2/3) interacts with Arkadia (Ark) and SnoN, leading to degradation of SnoN via the proteasome. This allows phosphorylated Smad3 (or Smad2Δexon3) complexed with Smad4 to bind the AGAC sites and activate transcription of target genes. (C) One such target gene is SnoN (shown in red), which will act with Smad4 to repress transcription again. For further details, see Discussion.
From knockdown experiments, we have shown that phosphorylated Smad3 acts redundantly with phosphorylated Smad2 in TGF-β-induced SnoN degradation. We readily detected the Arkadia interaction with endogenous phosphorylated Smad2 but were unable to detect an interaction between Arkadia and endogenous phosphorylated Smad3 in 293T cells, as they have very low levels of Smad3. However, we can detect this interaction if Smad3 is overexpressed in these cells. We presume that the phosphorylated Smad2 or Smad3 in the complex with Arkadia and SnoN is also degraded, but this is likely to be a very small pool, as we do not detect an increase in these species when Arkadia is knocked down (Fig. 2 and Fig. 3C). This is in contrast to the situation in ES cells, where knockout of Arkadia has been reported to increase phosphorylated Smad2 levels (27). It may be that in ES cells, a greater percentage of phosphorylated Smad is complexed with SnoN.
Depletion of the nuclear pool of SnoN in response to ligand will result in the loss of repressive SnoN from the repeated AGAC or GTCT promoter elements. This would then allow access of activated Smad3/Smad4 (or Smad2Δexon3/Smad4 complexes) to the promoter elements and hence transcriptional activation of target genes. Therefore, it is important to note that in response to TGF-β/Activin/Nodal stimulation, full-length Smad2 and Smad3 act redundantly with Arkadia to mediate SnoN degradation, but this affects only a subset of TGF-β/Activin/Nodal responses, i.e., those mediated via repeated AGAC or GTCT elements that are repressed by SnoN.
This model provides a mechanism for Arkadia action and explains why it is required only for transcription via Smad3/Smad4 binding sites. It also raises a number of new questions. The first concerns the role of the previously identified ubiquitin-protein ligases implicated in TGF-β-induced SnoN degradation, Smurf2 and the APC (1, 36, 39). Our results, using knockdown or overexpression of a dominant-negative form of Arkadia, demonstrate that Arkadia is absolutely required for ligand-induced SnoN degradation. However, they do not imply that it is sufficient. It is therefore possible that Smurf2 or the APC could also be components of a larger complex containing Arkadia, phosphorylated Smad2/3, and SnoN. In addition, it is clear that inhibiting the proteasome via MG132 has a much bigger effect on basal levels of SnoN than deleting Arkadia (see Fig. S3 in the supplemental material). Thus, basal SnoN levels are likely to be regulated by additional E3 ubiquitin ligases.
Second, what is the role of phosphorylated Smad2 or Smad3 in Arkadia-mediated SnoN degradation? We show that phosphorylated Smad2 or -3 is not required to target Arkadia to SnoN. Similarly, Arkadia can still interact with phosphorylated Smad2, even in the presence of a SnoN mutant that cannot interact with Smad2 or Smad3. Importantly, however, Arkadia does not trigger degradation of this mutant SnoN upon Activin stimulation, indicating that for SnoN to be degraded, it must be in a complex with Arkadia and phosphorylated Smad2 or Smad3, and the proteins must all be able to interact with each other. We find that Arkadia can polyubiquitinate SnoN even in the absence of a signal. This strongly suggests that the role of phosphorylated Smad2/Smad3 in the complex is to trigger degradation via the proteasome. Further work is required to determine the mechanism by which this occurs.
The third question concerns the role of the previously reported interaction between Arkadia and Smad7 (19, 24). We have shown that in HaCaT cells the requirement for Arkadia is specific for TGF-β/Activin-induced transcription via Smad3/Smad4 binding sites and is downstream of ligand-induced R-Smad phosphorylation and nuclear accumulation. This rules out the idea that in these cells Arkadia primarily acts by degrading Smad7, as this would be expected to affect BMP signaling as well as TGF-β/Activin signaling by affecting receptor levels and thus, R-Smad phosphorylation. Moreover, unlike SnoN which is highly expressed in all the cell lines that we have investigated and which we show is the target of Arkadia-mediated degradation, the levels of Smad7 mRNA in unstimulated HaCaT cells are extremely low (23), suggesting that Smad7 does not act as a repressor in these cells in the absence of signaling or at early time points after TGF-β stimulation. However, Smad7 is strongly induced by TGF-β (23); thus, induction of degradation of Smad7 by Arkadia is much more likely to be functionally relevant after prolonged TGF-β signaling or in cells that express high basal levels of Smad7.
Does loss of Arkadia occur in human tumors?
The TGF-β pathway has a known tumor suppressor function, and key components of the pathway, like Smad4, Smad3, and the receptors, are mutated, deleted, or downregulated in human cancers (22, 46). In addition, elevated SnoN expression has been observed in several different tumor types (22, 46). SnoN induces anchorage-independent growth of chicken and quail embryo fibroblasts when overexpressed (2), and it plays a protumorigenic role at early stages of malignant progression (46). Our demonstration that loss of Arkadia function completely abolishes the Smad3-dependent arm of the TGF-β pathway and that Arkadia is required for TGF-β/Activin-induced SnoN degradation raises the interesting possibility that Arkadia could be a new tumor suppressor gene. From our data, it is clear that loss of Arkadia would also result in an increase in SnoN levels. Intriguingly, analysis of the Oncomine database, searching for differential expression of Arkadia in tumor tissue versus healthy tissue, suggests that Arkadia is downregulated in prostate and breast cancers. We have also identified a Barrett's-associated esophageal adenocarcinoma cell line, SEG-1, that has lost Arkadia expression. This is associated with an absence of SnoN degradation upon TGF-β stimulation in those cells and consequently a loss of the CAGA12-Luc reporter gene activity. We have shown that reexpression of Arkadia can restore Smad3/Smad4-dependent transcription and SnoN degradation, indicating that the activity of Arkadia is compromised in these cells. These results raise the possibility that loss of Arkadia in tumors might contribute to tumorigenesis by abolishing the Smad3-dependent response. We are currently searching for mutations in the Arkadia gene in the SEG-1 cells and in a large panel of cell lines derived from a variety of different tumors to determine whether loss-of-function mutations in Arkadia occur naturally in human tumors.
Supplementary Material
Acknowledgments
We thank A. Atfi, R. Fitzgerald, H. Hamada, E. Leof, S. C. Lin, K. Luo, and P. ten Dijke for cell lines, plasmids, and antibodies. We thank J. Downward and E. Sahai and members of the Hill lab for useful discussions and comments on the manuscript.
The work was supported by CR-UK and an FP6 EU Marie Curie Intra European Fellowship (515316) to L.L.
Footnotes
Published ahead of print on 25 June 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bonni, S., H. R. Wang, C. G. Causing, P. Kavsak, S. L. Stroschein, K. Luo, and J. L. Wrana. 2001. TGF-β induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat. Cell Biol. 3:587-595. [DOI] [PubMed] [Google Scholar]
- 2.Boyer, P. L., C. Colmenares, E. Stavnezer, and S. H. Hughes. 1993. Sequence and biological activity of chicken snoN cDNA clones. Oncogene 8:457-466. [PubMed] [Google Scholar]
- 3.Chen, X., M. J. Rubock, and M. Whitman. 1996. A transcriptional partner for MAD proteins in TGF-β signalling. Nature 383:691-696. [DOI] [PubMed] [Google Scholar]
- 4.Cohen, S. B., R. Nicol, and E. Stavnezer. 1998. A domain necessary for the transforming activity of SnoN is required for specific DNA binding, transcriptional repression and interaction with TAF(II)110. Oncogene 17:2505-2513. [DOI] [PubMed] [Google Scholar]
- 5.Dennler, S., S. Huet, and J. M. Gauthier. 1999. A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene 18:1643-1648. [DOI] [PubMed] [Google Scholar]
- 6.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J. M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn, N. R., C. H. Koonce, D. C. Anderson, A. Islam, E. K. Bikoff, and E. J. Robertson. 2005. Mice exclusively expressing the short isoform of Smad2 develop normally and are viable and fertile. Genes Dev. 19:152-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupont, S., L. Zacchigna, M. Cordenonsi, S. Soligo, M. Adorno, M. Rugge, and S. Piccolo. 2005. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell 121:87-99. [DOI] [PubMed] [Google Scholar]
- 9.Ebisawa, T., M. Fukuchi, G. Murakami, T. Chiba, K. Tanaka, T. Imamura, and K. Miyazono. 2001. Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 276:12477-12480. [DOI] [PubMed] [Google Scholar]
- 10.Edmiston, J. S., W. A. Yeudall, T. D. Chung, and D. A. Lebman. 2005. Inability of transforming growth factor-β to cause SnoN degradation leads to resistance to transforming growth factor-β-induced growth arrest in esophageal cancer cells. Cancer Res. 65:4782-4788. [DOI] [PubMed] [Google Scholar]
- 11.Episkopou, V., R. Arkell, P. M. Timmons, J. J. Walsh, R. L. Andrew, and D. Swan. 2001. Induction of the mammalian node requires Arkadia function in the extraembryonic lineages. Nature 410:825-830. [DOI] [PubMed] [Google Scholar]
- 12.Feng, X. H., and R. Derynck. 2005. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 21:659-693. [DOI] [PubMed] [Google Scholar]
- 13.Germain, S., M. Howell, G. M. Esslemont, and C. S. Hill. 2000. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 14:435-451. [PMC free article] [PubMed] [Google Scholar]
- 14.Goumans, M. J., and C. Mummery. 2000. Functional analysis of the TGFβ receptor/Smad pathway through gene ablation in mice. Int. J. Dev. Biol. 44:253-265. [PubMed] [Google Scholar]
- 15.Hata, A., J. Seoane, G. Lagna, E. Montalvo, A. Hemmati-Brivanlou, and J. Massague. 2000. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 100:229-240. [DOI] [PubMed] [Google Scholar]
- 16.He, J., S. B. Tegen, A. R. Krawitz, G. S. Martin, and K. Luo. 2003. The transforming activity of Ski and SnoN is dependent on their ability to repress the activity of Smad proteins. J. Biol. Chem. 278:30540-30547. [DOI] [PubMed] [Google Scholar]
- 17.Izzi, L., and L. Attisano. 2006. Ubiquitin-dependent regulation of TGFβ signaling in cancer. Neoplasia 8:677-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kavsak, P., R. K. Rasmussen, C. G. Causing, S. Bonni, H. Zhu, G. H. Thomsen, and J. L. Wrana. 2000. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol. Cell 6:1365-1375. [DOI] [PubMed] [Google Scholar]
- 19.Koinuma, D., M. Shinozaki, A. Komuro, K. Goto, M. Saitoh, A. Hanyu, M. Ebina, T. Nukiwa, K. Miyazawa, T. Imamura, and K. Miyazono. 2003. Arkadia amplifies TGF-β superfamily signalling through degradation of Smad7. EMBO J. 22:6458-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korchynskyi, O., and P. ten Dijke. 2002. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277:4883-4891. [DOI] [PubMed] [Google Scholar]
- 21.Lebman, D. A., J. S. Edmiston, T. D. Chung, and S. R. Snyder. 2002. Heterogeneity in the transforming growth factor β response of esophageal cancer cells. Int. J. Oncol. 20:1241-1246. [PubMed] [Google Scholar]
- 22.Levy, L., and C. S. Hill. 2006. Alterations in components of the TGF-β superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 17:41-58. [DOI] [PubMed] [Google Scholar]
- 23.Levy, L., and C. S. Hill. 2005. Smad4 dependency defines two classes of transforming growth factor β (TGF-β) target genes and distinguishes TGF-β-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol. Cell. Biol. 25:8108-8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, W., H. Rui, J. Wang, S. Lin, Y. He, M. Chen, Q. Li, Z. Ye, S. Zhang, S. C. Chan, Y. G. Chen, J. Han, and S. C. Lin. 2006. Axin is a scaffold protein in TGF-β signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 25:1646-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo, K. 2004. Ski and SnoN: negative regulators of TGF-β signaling. Curr. Opin. Genet. Dev. 14:65-70. [DOI] [PubMed] [Google Scholar]
- 26.Massagué, J., J. Seoane, and D. Wotton. 2005. Smad transcription factors. Genes Dev. 19:2783-2810. [DOI] [PubMed] [Google Scholar]
- 27.Mavrakis, K. J., R. L. Andrew, K. L. Lee, C. Petropoulou, J. E. Dixon, N. Navaratnam, D. P. Norris, and V. Episkopou. 2007. Arkadia enhances Nodal/TGF-β signaling by coupling phospho-Smad2/3 activity and turnover. PLoS Biol. 5:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNally, J. G., W. G. Muller, D. Walker, R. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262-1265. [DOI] [PubMed] [Google Scholar]
- 29.Moren, A., T. Imamura, K. Miyazono, C. H. Heldin, and A. Moustakas. 2005. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J. Biol. Chem. 280:22115-22123. [DOI] [PubMed] [Google Scholar]
- 30.Nicol, R., and E. Stavnezer. 1998. Transcriptional repression by v-Ski and c-Ski mediated by a specific DNA binding site. J. Biol. Chem. 273:3588-3597. [DOI] [PubMed] [Google Scholar]
- 31.Niederlander, C., J. J. Walsh, V. Episkopou, and C. M. Jones. 2001. Arkadia enhances nodal-related signalling to induce mesendoderm. Nature 410:830-834. [DOI] [PubMed] [Google Scholar]
- 32.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 33.Pierreux, C. E., F. J. Nicolás, and C. S. Hill. 2000. Transforming growth factor β-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol. Cell. Biol. 20:9041-9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Randall, R. A., M. Howell, C. S. Page, A. Daly, P. A. Bates, and C. S. Hill. 2004. Recognition of phosphorylated-Smad2-containing complexes by a novel Smad interaction motif. Mol. Cell. Biol. 24:1106-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seoane, J., H. V. Le, L. Shen, S. A. Anderson, and J. Massague. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117:211-223. [DOI] [PubMed] [Google Scholar]
- 36.Stroschein, S. L., S. Bonni, J. L. Wrana, and K. Luo. 2001. Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev. 15:2822-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stroschein, S. L., W. Wang, S. Zhou, Q. Zhou, and K. Luo. 1999. Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science 286:771-774. [DOI] [PubMed] [Google Scholar]
- 38.Sun, Y., X. Liu, E. Ng-Eaton, H. F. Lodish, and R. A. Weinberg. 1999. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor β signaling. Proc. Natl. Acad. Sci. USA 96:12442-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan, Y., X. Liu, and M. W. Kirschner. 2001. The anaphase-promoting complex mediates TGF-β signaling by targeting SnoN for destruction. Mol. Cell 8:1027-1039. [DOI] [PubMed] [Google Scholar]
- 40.Wilkes, M. C., S. J. Murphy, N. Garamszegi, and E. B. Leof. 2003. Cell-type-specific activation of PAK2 by transforming growth factor β independent of Smad2 and Smad3. Mol. Cell. Biol. 23:8878-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong, C., E. M. Rougier-Chapman, J. P. Frederick, M. B. Datto, N. T. Liberati, J. M. Li, and X. F. Wang. 1999. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor β. Mol. Cell. Biol. 19:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yagi, K., D. Goto, T. Hamamoto, S. Takenoshita, M. Kato, and K. Miyazono. 1999. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J. Biol. Chem. 274:703-709. [DOI] [PubMed] [Google Scholar]
- 43.Yao, L. C., I. L. Blitz, D. A. Peiffer, S. Phin, Y. Wang, S. Ogata, K. W. Cho, K. Arora, and R. Warrior. 2006. Schnurri transcription factors from Drosophila and vertebrates can mediate Bmp signaling through a phylogenetically conserved mechanism. Development 133:4025-4034. [DOI] [PubMed] [Google Scholar]
- 44.Zawel, L., J. L. Dai, P. Buckhaults, S. Zhou, K. W. Kinzler, B. Vogelstein, and S. E. Kern. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1:611-617. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, H., P. Kavsak, S. Abdollah, J. L. Wrana, and G. H. Thomsen. 1999. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400:687-693. [DOI] [PubMed] [Google Scholar]
- 46.Zhu, Q., A. R. Krakowski, E. E. Dunham, L. Wang, A. Bandyopadhyay, R. Berdeaux, G. S. Martin, L. Sun, and K. Luo. 2007. Dual role of SnoN in mammalian tumorigenesis. Mol. Cell. Biol. 27:324-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.