Abstract
Smad proteins are critical intracellular signaling mediators for the transforming growth factor β (TGFβ) superfamily. Here, we report that Erbin (for “ErbB2/Her2-interacting protein”), which contains leucine-rich repeats and a PDZ (PSD-95/DLG/ZO-1) domain, interacts specifically with Smad3 and, to a lesser extent, with Smad2 through a novel Smad-interacting domain (SID) adjacent to its PDZ domain. Increased expression of Erbin does not affect the level of TGFβ-induced phosphorylation of Smad2/Smad3, but it physically sequesters Smad2/Smad3 from their association with Smad4 and hence negatively modulates TGFβ-dependent transcriptional responses and cell growth inhibition. An isoform of Erbin encoded by an alternatively spliced transcript in human tissues lacks this SID and fails to inhibit TGFβ responses. Consistently, knockdown of the endogenous Erbin gene with short hairpin RNA enhances TGFβ-induced antiproliferative and transcriptional responses. In addition, Erbin suppresses activin/Smad2-dependent, but not BMP/Smad1-mediated, induction of endogenous gene expression in Xenopus embryos. Therefore, these results define Erbin as a novel negative modulator of Smad2/Smad3 functions and expand the physiological role of Erbin to the regulation of TGFβ signaling.
The transforming growth factor β (TGFβ) superfamily of cytokines includes TGFβ, activin, and bone morphogenetic proteins (BMPs) that control a plethora of physiological processes, such as cell growth, differentiation, and apoptosis. TGFβ and related factors initiate signaling through their binding to heteromeric receptor complexes at the cell surface and subsequent activation of the intracellular Smads (39). Eight Smads exist in mammals, including five receptor-activated Smads (R-Smads), one common mediator Smad (Co-Smad; i.e., Smad4), and two inhibitory Smads (I-Smads). Of the R-Smads, Smad2 and Smad3 transduce signals from TGFβ and activin, while Smad1, Smad5, and Smad8 transduce signals from BMPs. Upon ligand-induced activation of TGFβ receptors, Smad2/Smad3 become phosphorylated by the TGFβ type I receptor (TβRI), dissociate from the receptor, oligomerize with Smad4, and translocate to the nucleus, where they regulate transcription of TGFβ target genes (12, 31). At the end of active signaling, activated Smad2/Smad3 are believed to be dephosphorylated (30) and exported from the nucleus to the cytoplasm in order to sense the TGFβ receptor activity on the cell surface (21, 34, 38, 42). Thus, the strength and duration of TGFβ signaling can be modulated by activation-inactivation cycles and nucleocytoplasmic shuttling of Smads.
Erbin (for “ErbB2/Her2-interacting protein”) is the founding member of the leucine-rich repeat (LRR) and PDZ (PSD-95/DLG/ZO-1) domain (LAP) family, which is characterized by 16 LRRs at the N terminus and 1 to 4 PDZ domains at the C terminus (3, 5, 19). Other members in the LAP family (Densin-180, Let-413, and Scribble) are involved in determining epithelial integrity (1, 3, 5). Through their LRRs and PDZ domains, LAP family proteins are speculated to mediate protein-protein interactions that contribute to the organization of subcellular structures and signal transduction complexes (5, 26, 36).
Erbin originally was identified as a direct and specific binding partner of ErbB2 of the epidermal growth factor receptor (EGFR) family through its C-terminal PDZ domain (4). Erbin localizes to the basolateral side of epithelial cells through its N-terminal LRR domain (4, 29) and functions to restrict ErbB2 to the basolateral membrane (4). Our knowledge of Erbin's functional role in cells has been expanded rapidly by identification of new interaction partners. Via its PDZ domain, Erbin interacts with proteins involved in cell-cell or cell-matrix interaction (19, 22, 23, 24, 28, 35). Via its LRR domain, Erbin interacts with scaffold protein Sur-8 and disrupts the Sur-8-Ras-Raf interaction, thereby inhibiting extracellular signal-regulated kinase (ERK) activation (8, 20, 26, 36). Similarly, the LRR domain also mediates the interaction of Erbin with Nod2, an intracellular sensor of a specific bacterial cell wall component, indicating that Erbin may regulate inflammatory responses via Nod2-dependent activation of NF-κB and cytokine secretion (32).
In this study, we uncover a novel function of Erbin as a modulator of TGFβ signaling. We show that Erbin inhibits TGFβ signaling through a physical interaction with and functional antagonism to TGFβ-specific Smad2 and Smad3. A novel Smad-interacting domain (SID) was identified in a functionally unassigned region immediately upstream of the conserved PDZ domain in Erbin. SID is necessary and sufficient to mediate Erbin's inhibitory effect on TGFβ signaling. Furthermore, Erbin blocked activin-responsive, but not BMP-responsive, endogenous gene expression in early Xenopus embryos. Taking the results together, our studies define Erbin as a novel modulator of TGFβ signaling through its direct interaction with Smad2/Smad3 and suggest that Erbin may regulate cell fate determination by inhibiting TGFβ signaling.
MATERIALS AND METHODS
Expression plasmids.
Mammalian expression plasmids for epitope-tagged (hemagglutinin [HA] or FLAG) Smad and Gal4-Smad proteins (13, 14), Smad2/Smad3 mutants that either lack C-terminal serine phosphorylation [Smad2/Smad3(2SA)] or harbor phosphorylation-mimetic residues [Smad2/Smad3(2SD)] (30), Myc-tagged full-length human Erbin, and Erbin mutants (LRR, 965-1371, and ΔPDZ) (20) were described previously. HA-tagged Smad3-LC was generated by PCR using primers spanning amino acids (aa) 143 to 426 of human Smad3 and was cloned into EcoRI and SalI sites of the expression vector pXF3H (derived from pRK5 [Genentech]). Other Myc-tagged Erbin deletion mutants, including Erbin(392-986), Erbin(962-1084), Erbin(1071-1181), Erbin(1172-1282), Erbin(1229-1279), and Erbin(1172-1234), were generated by PCR using primers spanning the indicated region and were cloned into EcoRI and SalI sites of the expression vector pRK5M. To generate the Myc-tagged Densin-180 expression construct, three fragments were amplified from a cDNA library, a 1.7-kb ClaI-EcoRV fragment, a 1.5-kb EcoRV-SpeI fragment, and a 1.3-kb SpeI-NotI fragment, and they were subcloned in pBluescript KS(−). The resulting full-length Densin-180 subsequently was subcloned at the NotI site in pEF6. Myc-Erbin-v7 was constructed based on the human ErbB2 interacting protein (ERBB2IP), transcript variant 7 sequence (NCBI database accession no. NM_001006600) by using primers specifically skipping the amino acids (residues 1212 to 1280) from the pRK5M-Erbin construct.
Antibodies.
The following commercial antibodies were used in this study: anti-Flag (M2; Sigma), anti-HA (12CA5; Roche), anti-Myc (9E10; Santa Cruz Biotechnology), anti-HA and anti-Myc affinity gels (Sigma), anti-Smad2 and anti-Smad3 (Zymed), anti-Smad2/Smad3 (E-20; Santa Cruz Biotechnology), and anti-phospho-Smad2 (anti-P-Smad2) (Cell Signaling Technology). Anti-phospho-Smad3 (anti-P-Smad3) was a gift from Ed Leof (Mayo Clinic). Anti-ERK1/2 antibody (Transduction Laboratories), anti-phospho-p44/42 mitogen-activated protein kinase antibody (E10; Cell Signaling Technology), and anti-Erbin antibody were described previously (19).
Cell lines and transfection.
Human HeLa, HaCaT, and HEK293T cells were grown as described previously (14). HaCaT cells stably expressing enhanced green fluorescent protein (EGFP)-Smad2 were kindly provided by Caroline Hill (34). C2C12 cells were maintained as undifferentiated myoblasts in Dulbecco's modified Eagle's medium with high glucose and was supplemented with 10% fetal bovine serum. HaCaT cells were transfected with SuperFect (QIAGEN), HEK293T cells were transfected with Lipofectin (Invitrogen), HeLa cells were transfected with Transfectin (Bio-Rad), and C2C12 cells were transfected with LipofectAmine (Invitrogen).
Transcription reporter assays.
Plasmids SBE-Luc, p21-Luc (both gifts from Bert Vogelstein), and ARE-Luc (a gift from Malcolm Whitman) were used to measure TGFβ-induced transcription. Plasmid Id1-Luc (a gift from Peter ten Dijke) was used to measure BMP-induced transcription. pSVβgal (Promega), which expresses β-galactosidase under the control of the simian virus 40 early promoter, was cotransfected to allow the normalization of transfection efficiency. The amount of individual plasmid DNAs used for transfection was 0.25 μg per well (in a 12-well tray) unless specifically indicated otherwise. The total amounts of transfected DNA were adjusted to make them equal by addition of vector DNA when necessary. Reporter assays were carried out as described previously (14). Briefly, 20 to 24 h after transfection, cells were treated with TGFβ1 (2 ng/μl) or BMP2 (50 ng/μl) for 24 h. Cells then were harvested for measurement of luciferase and β-galactosidase activities. All assays were done in duplicate, and all values were normalized for transfection efficiency against β-galactosidase activities.
RNA interference.
The target sequences of short hairpin RNA 609 (shRNA-609) (GCAACTAAGTGGATTGAAA) and shRNA-795 (GCAGCTTCCTGAGACTATT) correspond to nucleotides 609 to 617 and 795 to 813, respectively, of the human Erbin coding region. Annealed sense and antisense DNA oligonucleotides corresponding to the shRNA Erbin target sequence were cloned into the pSRG vector (30). Stable HaCaT cells expressing shRNA-795 were generated and selected with 2 μg/μl of puromycin. For established shRNA Erbin stable cells, two independent lines, KD-1 and KD-2, were chosen for subsequent functional assays.
GST fusion protein, in vitro protein binding, and pull-down assays.
Glutathione S-transferase (GST) fusion proteins were prepared by using a commercial kit (Amersham Pharmacia Biotech). In vitro-translated (TNT kit; Promega) 35S-labeled protein was precleared with 5 μg of GST protein first (1 h) and then was incubated with 5 μg of different GST fusion proteins (2 h), as indicated below, in the in vitro binding buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 2 mM EDTA, 0.1% NP-40). Proteins bound to GST fusion proteins were retrieved by binding them to glutathione-Sepharose beads (Amersham Pharmacia Biotech), separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and visualized by autoradiography.
Immunoprecipitation and Western blot analysis.
Cells were harvested in 20% glycerol lysis buffer (10 mM HEPES [pH 7.9], 300 mM NaCl, 0.01 mM EGTA, 0.2% NP-40) 48 h after transfection. HA- or Myc-tagged proteins were immunoprecipitated from the cell lysate by an anti-HA or anti-Myc affinity gel, and after being extensively washed, they were eluted in SDS sample loading buffer (Bio-Rad). Eluted proteins were separated by SDS-PAGE, transferred to nitrocellulose, and detected in an immunoblot assay with appropriate antibodies as indicated below. Antibody-bound proteins were visualized by horseradish peroxidase-conjugated secondary antibody followed by chemiluminescence (Pierce).
Thymidine incorporation assays.
For thymidine incorporation assays, cells were grown in 12-well plates (20,000 cells per well) in culture medium supplemented with 0.2% fetal bovine serum for 12 h before TGFβ treatment for 48 h. [3H]thymidine was added to cells (0.025 mCi/well) 4 h before harvest. Cells were washed twice in phosphate-buffered saline, fixed in 5% trichloroacetic acid, and rinsed with water, and then DNA was extracted in 0.5 M NaOH. Radioactivity was measured by scintillation counting. The data were plotted as average values with standard errors of duplicate repeats for each condition for each independent experiment. Each independent experiment was repeated twice.
Xenopus gene expression analyses.
RNAs used for microinjection into Xenopus embryos were synthesized with SP6 RNA polymerase using an mMessage mMachine kit (Ambion). The following DNA templates were used: AscI-linearized pCS105-Erbin, pCS105-Smad1, and pCS105-Smad2; EcoRI-linearized pSP64T-activin; and AscI-linearized pCS105-BMP4(flag). RNAs of the above genes were injected alone or in combination into the animal poles of two-cell-stage embryos. The doses of RNAs used were the following: Erbin, 1 ng; Smad1, 1 ng; Smad2, 0.2 ng; activin, 1 pg; and BMP4, 20 pg. Ectodermal explants (animal caps) from injected embryos were obtained at the blastula stages (stage 9) and incubated to the gastrula stages (stage 11) or tadpole stage (stage 28), at which time total RNA was extracted from these caps, and then the gene expression pattern was analyzed by reverse transcription-PCR (RT-PCR). The primers used in RT-PCR were described previously (6).
Immunofluorescence.
Cells grown on coverslips were fixed with 4% formaldehyde for 30 min at 4°C, followed by 0.5% Triton X-100 treatment for 30 min, and then they were blocked with 5% milk in PEM buffer [400 mM potassium piperazine-N,N′-bis(2-ethanesulfonic acid) (pH 6.8), 0.8 mM EGTA, 5 mM MgCl2]. Cells then were probed with the indicated primary antibody, followed by being probed with Alexa Fluor 546 (Molecular Probes) anti-mouse antibody, and then they were examined with a Zeiss Axioplan II microscope.
RESULTS
Erbin physically interacts with TGFβ-specific Smad2 and Smad3.
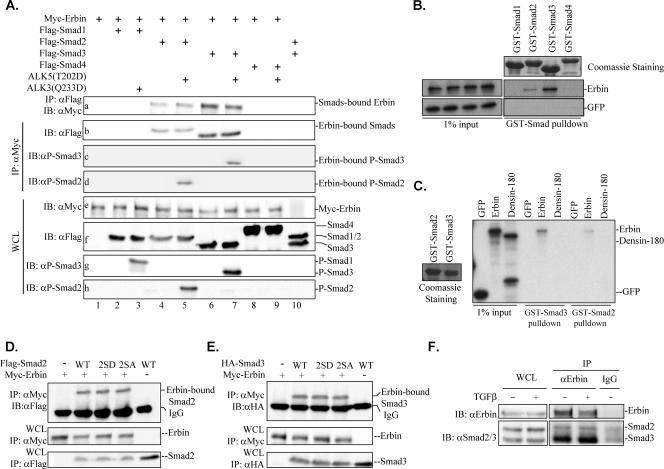
In a search for cellular proteins that interact with Erbin in a yeast two-hybrid screen, we identified Smad proteins. This finding supports a previous report on Smad3-Erbin interaction (40). To further investigate the specificity of Erbin-Smad interaction, we carried out coimmunoprecipitation experiments to examine the association between Erbin and Smad1 to Smad4. We found that Erbin was detected in the immunoprecipitates of Smad2 and Smad3 but not in those of Smad1 and Smad4 (Fig. 1A, blot a); similarly, in a reciprocal experiment, only Smad2 or Smad3 was detected in the immunocomplexes of Erbin (Fig. 1A, blot b). In addition, P-Smad2 or P-Smad3 was detected in the immunoprecipitates of Erbin (Fig. 1A, blots c and d), indicating that the active forms of Smad2/Smad3 retain their binding capacity to Erbin.
FIG. 1.
Erbin associates with Smad2 and Smad3. (A) Erbin interacts with Smad2/Smad3 but not Smad1/Smad4 in vivo. Flag-Smads and Myc-Erbin were cotransfected into HEK293T cells with ALK5(T202D) (lanes 5, 7, and 9) or ALK3(Q233D) (lane 3). The cell lysates were split and immunoprecipitated (IP) with either anti-Myc (αMyc) or anti-FLAG (αFLAG) antibody. Erbin or Smads were detected from immunoprecipitates by Western blotting with appropriate antibodies as indicated. (B) Erbin interacts with Smad2/Smad3 but not Smad1/Smad4 in vitro. In vitro-synthesized Erbin proteins were allowed to interact with GST-Smad proteins on glutathione beads. The GST-Smad-bound Erbin was resolved by SDS-8% PAGE and detected by autoradiography. As inputs, 35S-labeled in vitro-translated Erbin proteins and control GFP (left gels) represent 1% of the total amount of protein used in each interaction assay (right gels). Equal amounts of GST-Smad1 to GST-Smad4 are indicated by Coomassie brilliant blue staining (top right). (C) Smad3 interacts with Erbin but not Densin-180. In vitro-translated Erbin or Densin-180 proteins were allowed to interact with GST-Smad2/GST-Smad3 proteins on glutathione beads. GST-Smad2- and GST-Smad3-bound proteins were resolved by 8% SDS-PAGE and detected by autoradiography. Equal amounts of GST-Smad2 and GST-Smad3 are indicated by Coomassie brilliant blue staining (left). (D and E) Erbin interacts with Smad2/Smad3 mutants. HEK293T cells were transfected with Myc-Erbin and Smad2/Smad3 (wild type or deletion mutants) as indicated, and Erbin-bound Smad2/Smad3 were immunoprecipitated with anti-Myc antibody and analyzed by Western blotting with anti-Flag or anti-HA antibody. (F) Erbin interacts with endogenous Smad2/Smad3. Cell lysates were prepared from 90% confluent HaCaT cells after 1 h of TGFβ treatment. Endogenous Smad2/Smad3 bound to Erbin were immunoprecipitated with anti-Erbin antibody (αErbin) and detected by Western blotting with anti-Smad2 (αSmad2) and anti-Smad3 (αSmad3) antibodies. WCL, whole-cell lysates; ALK5(T202D), constitutively active type I TGFβ receptor mutant; ALK3(Q233D), constitutively active type I BMP receptor mutant; WT, wild type; 2SA, inactive Smad2/Smad3 mutants with C-terminal Ser-to-Ala mutation; 2SD, active Smad2/Smad3 mutants harboring C-terminal phosphorylation-mimetic Ser-to-Asp mutation; IB, immunoblot; IgG, immunoglobulin G.
The specific interaction between Erbin and Smad2/Smad3 also was observed in GST in vitro pull-down assays in which 35S-labeled in vitro-translated Erbin could be pulled down only by GST-Smad2 or GST-Smad3 (Fig. 1B). With a similar amount of GST-Smad proteins, we noticed that more radioactive Erbin was retrieved by GST-Smad3 than GST-Smad2 (Fig. 1B), suggesting that Erbin binds to Smad3 more strongly than to Smad2.
We next examined if Smad2/Smad3 could bind to another member of the LAP protein family. Densin-180 shares the highest homology with Erbin, with 72.7% identity in the LRRs, 70.8% in the PDZ domain, and 39% in the intermediate region (1). We found that GST-Smad2/GST-Smad3 interacted with Erbin but not with Densin-180 (Fig. 1C). Taken together, our data suggest that the interaction between Smad2/Smad3 and Erbin is specific.
R-Smads are phosphorylated by TβRI upon ligand stimulation. To investigate if TGFβ or BMP regulates Erbin-Smad interaction, we utilized type I receptor mutants that constitutively phosphorylate R-Smads. After confirming constitutive phosphorylation of Smad2 and Smad3 by ALK5(T202D) (Fig. 1A, blots g and h), the effect of ALK5(T202D) on the Erbin-Smad2 and Erbin-Smad3 interactions was examined. As shown in Fig. 1A (blots a and b, lanes 4 to 7), activation of TGFβ signaling had no effect on the Erbin-Smad2 or Erbin-Smad3 association. Furthermore, constitutive activation of BMP signaling by ALK3(Q233D) did not result in detectable Erbin-Smad1 interaction (Fig. 1A, blot g, lane 3), supporting the notion that Erbin specifically interacts with Smads in the TGFβ, but not the BMP, pathway. To examine further whether Erbin binds equally to active and inactive Smad2/Smad3, we utilized Smad2/Smad3 mutants that either lack C-terminal serine phosphorylation (inactive; 2SA) or harbor phosphorylation-mimetic residues (active; 2SD). In the coimmunoprecipitation experiment, both mutants bound Erbin to an extent similar to that of wild-type Smad2/Smad3 (Fig. 1D and E).
Erbin interacts with Smad2/Smad3 under physiological conditions.
Having determined that Erbin specifically interacts with Smad2 and Smad3, we sought to investigate whether the interaction between Erbin and Smad2/Smad3 occurs at their endogenous levels and whether this interaction is regulated by TGFβ. We used a TGFβ-responsive cell line, HaCaT, in our assays. As shown in Fig. 1G, immunoprecipitation of endogenous Erbin could retrieve endogenous Smad2 and Smad3. Consistent with the stronger Erbin-Smad3 interaction observed in transfection experiments (Fig. 1A) and in vitro GST pull-down assays (Fig. 1B and C) compared to the Erbin-Smad2 interaction, endogenous Erbin also exhibited a much stronger binding to Smad3 than to Smad2 (Fig. 1F). TGFβ treatment did not influence Erbin-Smad2 and Erbin-Smad3 interaction (Fig. 1F). Our data indicate that Erbin specifically interacts with Smad3 and, to a lesser extent, with Smad2 at endogenous levels in a TGFβ-independent manner.
A novel domain in Erbin mediates its interaction with Smad2/Smad3.
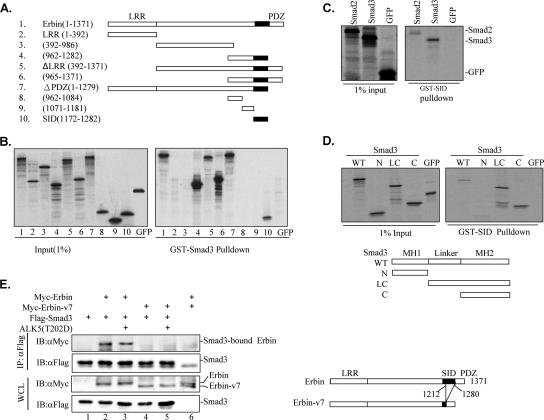
There are two well-characterized domains in Erbin, i.e., the LRR and PDZ domains, that mediate many protein-protein interactions and regulate distinct signaling pathways (26). To determine the domains required for Smad binding, a series of Erbin deletion mutants were examined (Fig. 2A). We found that a minimal region of 111 aa residues retained strong binding to GST-Smad3 (Fig. 2B, lane 10). A sequence comparison did not reveal similarity of this region to Smad-binding domains in SARA (37, 41) or Smad-interacting motifs in FoxH1 (15, 37). This newly identified domain, located in a functionally unassigned region (aa 1172 to 1282) of Erbin and immediately upstream of its PDZ domain (aa 1280 to 1371), was named SID. While all SID-containing mutants, including the N-terminal LRR domain deletion fragment (ΔLRR) (Fig. 2B, lane 5) and the C-terminal PDZ domain deletion fragment (ΔPDZ) (Fig. 2B, lane 7), interacted with Smad3 as strongly as full-length Erbin, deletion mutants lacking SID, such as the LRR domain alone (aa 1 to 392) (Fig. 2B, lane 2) and the intermediate region (aa 392 to 986) (Fig. 2B, lane 3), lost the ability to bind to Smad3. Consistent with these data, a GST-SID fusion protein sufficed to bind to 35S-labeled Smad2/Smad3 in an in vitro binding assay (Fig. 2C). Again, we observed that GST-SID bound to Smad3 more strongly than to Smad2. Our results suggest that SID is the domain in Erbin mediating Erbin-Smad interaction.
FIG. 2.
Novel region in Erbin mediates its interaction with Smad2/Smad3. (A) Diagram of various Erbin deletion mutants used for in vitro GST pull-down assays depicted in panel B. The start and end amino acid residues for each fragment are indicated. SID is indicated by a filled box. (B) SID mediates Erbin's interaction with Smad3. The left panel shows an autoradiogram of 1% of 35S-labeled in vitro-translated Erbin deletion proteins and GFPs (as inputs). The right panel shows different in vitro-translated Erbin deletion proteins that interacted with GST-Smad3 proteins on glutathione beads. GST-Smad3-bound proteins were resolved by 10% SDS-PAGE followed by autoradiography. (C) SID directly interacts with Smad2/Smad3. In vitro-synthesized Smad2/Smad3 and GFP interacted with GST-SID proteins on glutathione beads. GST-SID-bound proteins were gel resolved and detected by autoradiography. (D) Erbin directly interacts with the MH2 domain of Smad3. The in vitro-synthesized full-length Smad3 domain (wild type [WT]), the N-terminal MH1 domain (N), the C-terminal plus linker region domain (LC), and the C-terminal MH2 domain (C) of Smad3 as well as GFP were incubated with GST-SID proteins on glutathione beads. Deletion constructs of Smad3 are shown with the MH1 and MH2 domains and the linker region as indicated. GST-SID-bound proteins were detected by SDS-PAGE and autoradiography. (E) Smad3 interacts with Erbin but not Erbin-v7. Myc-Erbin or Myc-Erbin-v7 was cotransfected into HEK293T cells with Flag-Smad3, with or without ALK5(T202D). Smad3-bound Erbin or Erbin-v7 was immunoprecipitated by anti-Flag antibody (αFLAG) and detected by Western blotting with anti-Myc (αMyc) antibody. The N-terminal LRR domain, the C-terminal PDZ domain, and SID (black box) are indicated. ΔLRR, Erbin mutant lacking the LRR domain; ΔPDZ, Erbin mutant lacking the PDZ domain; WCL, whole-cell lysate; IP, immunoprecipitate; IB, immunoblot.
Smad proteins consist of two highly conserved domains: the N-terminal MH1 domain and the C-terminal MH2 domain, linked by a more divergent linker region. We sought to determine the Erbin-interacting domain in Smad2/Smad3. In accordance with a previous report (40), our in vitro binding assays demonstrated that GST-SID specifically bound to Smad3C (MH2 domain alone) and Smad3-LC (linker region plus MH2) but not to Smad3N (MH1 domain alone) (Fig. 2D).
In most human tissues, there are several differentially spliced transcripts of Erbin, most of which differ in the coding region upstream of or within the PDZ domain (11). We noticed that a large chunk (aa 1212 to 1280) of SID is lost in Erbin transcript variant 7 (its product was designated Erbin-v7). If SID solely mediates Erbin's interaction with Smad2/Smad3, Erbin-v7 is expected to lose its interaction with Smad2/Smad3. Indeed, Erbin-v7 failed to interact with Smad3 in coimmunoprecipitation experiments (Fig. 2E).
Taken together, our data identified this domain as a novel SID that binds to the MH2 domain of Smad2/Smad3.
Erbin inhibits Smad2/Smad3-mediated transcriptional responses in mammalian cells.
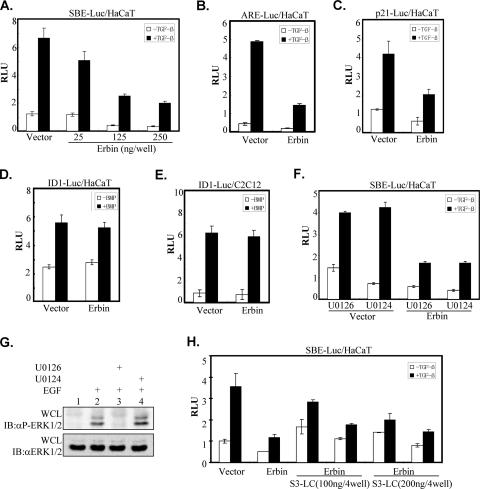
To assess the physiological consequence of Erbin-Smad interaction, we next examined the effects of Erbin on Smad2/Smad3-mediated transcriptional activation of TGFβ target genes. Direct Smad3/Smad4-dependent transcription was assessed using the SBE-Luc reporter, which contains four copies of a Smad3/Smad4 binding element, SBE (43). Increasing concentrations of Erbin caused a gradual decrease in TGFβ-dependent transcription from the SBE-Luc reporter (Fig. 3A). Overexpression of Erbin also suppressed TGFβ-induced ARE-Luc reporter activity (Fig. 3B), which depends on the transcriptional cooperation between the transcription factor FoxH1/Fast-1 and the activated Smad2 and Smad4 (27). In addition, Erbin inhibited TGFβ-induced activity of the natural p21CIP1 promoter (Fig. 3C). In sharp contrast, Erbin did not interfere with the BMP-induced Id1-Luc reporter gene in HaCaT cells (Fig. 3D) or in C2C12 cells (Fig. 3E). These data were in accordance with our demonstration that Erbin interacted with Smad2/Smad3 but not with Smad1 (Fig. 1A).
FIG. 3.
Erbin inhibits Smad-mediated transcriptional activation. (A to E) Erbin inhibits TGFβ signaling but not BMP signaling. HaCaT cells were cotransfected with Myc-Erbin with either the SBE-Luc reporter (A), ARE-Luc reporter (together with Fast-1) (B), or p21-Luc reporter (C and D). Similarly, C2C12 cells were transfected with Myc-Erbin and the Id1-Luc reporter (E). Luciferase activity was measured 20 h after stimulation with TGFβ (A, B, and C) or BMP (D and E). (F) Erbin's inhibition of TGFβ-induced transcriptional activity is independent of MEK inhibition. HaCaT cells were transfected either with SBE-Luc alone or together with Myc-Erbin. Luciferase activity was measured 20 h after TGFβ stimulation in the presence of MEK inhibitor U0126 or the control compound U0124. (G) An ERK inhibitor blocks ERK activation. HEK293T cells were treated with inhibitor U0126 or the control compound U0124 (from the same stock used for panel F) for 2 h. Cells then were stimulated with epidermal growth factor (EGF) (10 ng/ml) for 30 min. Phosphorylation of ERK was examined by Western blotting with phosphospecific ERK antibody (αP-Erk1/2). (H) Erbin inhibits Smad3-LC-mediated transcription activity. HaCaT cells were cotransfected with the SBE-Luc reporter and increasing amounts of HA-Smad3-LC (S3-LC) with or without Myc-Erbin. Luciferase activity was measured 20 h after TGFβ stimulation. RLU, relative luciferase units; WCL, whole-cell lysate; IB, immunoblot.
Previous studies had shown that Erbin inhibits ERK activation by disrupting the Sur-8-Ras-Raf interaction (8, 20), and ERK function is required for maximal activation of some target genes by TGFβ (17, 25). To exclude the possibility that Erbin inhibits TGFβ signaling through its role in ERK inactivation, we examined whether Erbin inhibits TGFβ-induced SBE-Luc expression in the presence of the MEK inhibitor U0126. U0126, but not U0124, effectively inhibited epidermal growth factor-induced ERK phosphorylation (Fig. 3G). As shown in Fig. 3F, U0126 did not inhibit TGFβ-induced activation of the SBE-Luc reporter, and importantly, it did not alter Erbin-dependent inhibition of SBE-Luc activity, suggesting that Erbin's inhibitory effect on TGFβ responses was not likely to be via the Ras-Raf-ERK pathway. We further showed that Erbin inhibits the constitutive activity of Smad3-LC (S3LC with the linker region plus the MH2 domain of Smad3). As shown in Fig. 3H, Erbin attenuated both high basal activity and TGFβ-induced activity of S3LC, suggesting that Erbin inhibits Smad3 signaling through the MH2 domain.
SID is necessary and sufficient for Erbin-mediated inhibition of TGFβ signaling.
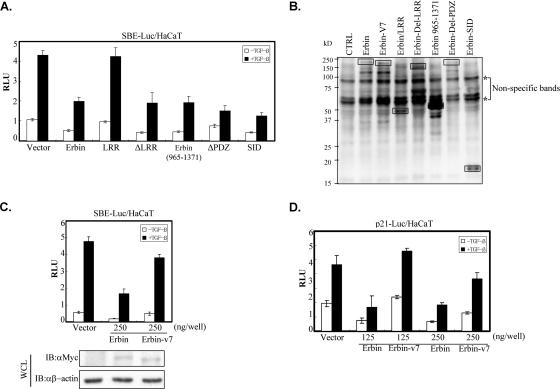
We next sought to evaluate how SID contributes to Erbin-dependent inhibition of TGFβ signaling. Various deletion mutants of Erbin were examined for their ability to inhibit TGFβ-induced transactivation of the SBE-Luc reporter. As shown in Fig. 4A, the LRR domain alone had no inhibitory effect on TGFβ-induced SBE-Luc expression, although it was expressed to a level similar to that of wild-type Erbin (Fig. 4B). Other deletion mutants containing SID exhibited inhibitory effects similar to that of wild-type Erbin. Notably, ectopic expression of SID alone could suppress TGFβ-induced SBE-Luc expression, suggesting that SID is sufficient for mediating Erbin's inhibitory effect on TGFβ signaling.
FIG. 4.
Erbin inhibits TGFβ signaling through SID. (A) SID is necessary for Erbin's inhibitory effect on SBE promoter activity. HaCaT cells were cotransfected with the SBE-Luc reporter and either wild-type Erbin or Erbin deletion mutants as indicated. Luciferase activity was measured 20 h after TGFβ stimulation. (B) Cell lysates also were subjected to Western blot analysis to examine the expression of deletion mutants. The protein band corresponding to each construct is indicated by a solid box. Asterisks indicate apparent nonspecific bands. (C and D) Erbin, but not Erbin-v7, inhibits TGFβ-induced transcriptional activation of SBE-Luc. Myc-Erbin or Myc-Erbin-v7 was cotransfected into HaCaT cells with SBE-Luc (C) or p21-Luc (D). Luciferase activity was measured 20 h after TGFβ stimulation. The expression level of Erbin or Erbin-v7 from cell lysates shown in panel C was determined with Western blot analysis. RLU, relative luciferase activity; WCL, whole-cell lysate; IB, immunoblot; CTRL, control. αMyc, anti-Myc antibody; αβ-actin, anti-β-actin antibody.
We then examined the effect of Erbin-v7, which lacks a large chunk of SID (Fig. 2E), on TGFβ transcriptional responses. Parallel comparison between Erbin and Erbin-v7 revealed that Erbin-v7 failed to inhibit TGFβ-induced transcriptional activation of SBE-Luc (Fig. 4C) or p21-Luc (Fig. 4D), suggesting that SID is essential for Erbin to inhibit TGFβ signaling.
Erbin suppresses activin-responsive but not BMP-responsive endogenous gene expression in Xenopus embryos.
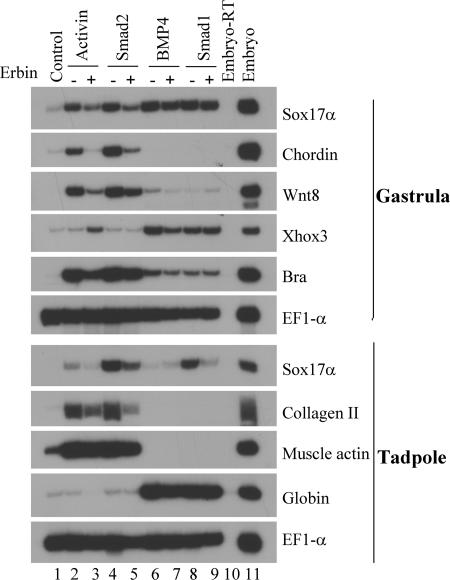
During early vertebrate development, activin/nodal and BMP signals are known to control embryonic patterning and cell fate determination (7). Activation of TGFβ signaling induces mesoderm- and endoderm-specific gene expression in a dose-dependent fashion in early Xenopus embryos. To examine whether Erbin can regulate TGFβ signaling during Xenopus embryogenesis, we determined the effects of Erbin overexpression on activin- or BMP-induced endogenous gene expression in Xenopus ectodermal explants (animal caps). As shown in Fig. 5, activin and Smad2 induced a whole range of mesendodermal markers, including the ventrolateral mesodermal marker Xwnt8, the dorsal mesodermal marker chordin, and the endodermal marker Sox17α, at gastrula stages as well as the dorsal mesodermal marker type II collagen at tadpole stages (compare lanes 2 and 4 with lane 1). Coexpression of Erbin with activin or Smad2 suppressed expression of these genes (compare lane 3 with 2 and lane 5 with 4). In contrast, when Erbin was coexpressed with BMP4 or Smad1, the genes activated by these molecules, including the ventral mesodermal marker Xhox3 at gastrula stages and the blood maker globin at tadpole stages, were not significantly affected (compare lane 9 with 8 and lane 7 with 6). The results of three independent experiments were similar. Taken together, these results are consistent with the cell culture data and indicate that Erbin preferentially blocks activin, but not BMP, signals in Xenopus animal caps.
FIG. 5.
Erbin inhibits mesodermal and endodermal marker induction by activin/Smad2, but not BMP4/Smad1, signals in Xenopus ectodermal explants. Mesendodermal gene induction by 1 pg of activin (lane 2) or 0.2 ng of Smad2 (lane 4) RNA was inhibited by the presence of 1 ng of Erbin RNA (lanes 3 and 5), whereas several BMP4 (20 pg)-induced (lane 6) or Smad1 (1 ng)-induced (lane 8) mesendodermal markers in animal caps were not significantly affected by the presence of Erbin (1 ng RNA). In all of these experiments, RNAs were injected into the animal poles of two-cell-stage frog embryos. Animal caps from the injected embryos were dissected at blastula stages and harvested at gastrula stages or tadpole stages for RT-PCR assays. The gene expression in uninjected control animal caps is shown in lane 1, and the expression pattern of whole embryos processed in the absence (Embryo-RT) or presence (Embryo) of reverse transcriptase in RT-PCRs is shown in lanes 10 and 11, respectively. The RT-PCR assay of the gene EF1-α was used as the loading control.
Knockdown of Erbin expression enhances TGFβ growth-inhibitory and transcriptional responses.
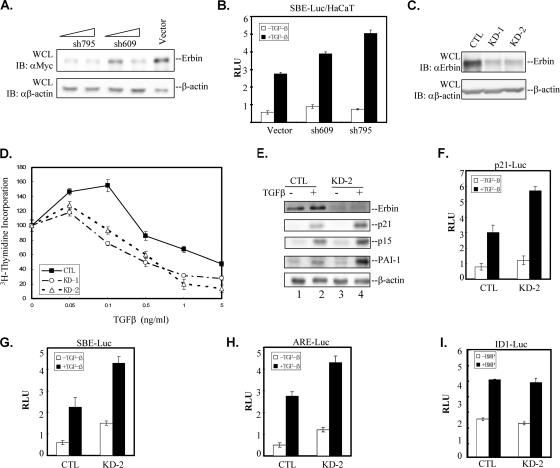
We next took a loss-of-function approach to address whether Erbin depletion enhances TGFβ-dependent transcriptional responses. We made two shRNAs against Erbin. Expression of these shRNAs reduced Erbin protein levels in HEK293T cells (Fig. 6A) and significantly increased TGFβ-induced SBE-Luc reporter activity in HaCaT cells (Fig. 6B). We then established HaCaT stable cells that express Erbin shRNA-795. A significant reduction in Erbin expression was detected in two independent stable clones (KD-1 and KD-2) compared to the level of Erbin expression in a vector control cell line (cytotoxic T lymphocytes [CTL]) (Fig. 6C). The stable control CTL cell line carrying shRNA expression vector pSRG behaved similarly to its parent HaCaT cells in several aspects, including TGFβ-induced Smad2/Smad3 phosphorylation, growth inhibition, and transcription induction (data not shown). These stable cells were used to compare their growth-inhibitory and transcriptional profiles in response to TGFβ.
FIG. 6.
Knockdown of Erbin expression enhances TGFβ signaling. (A) Different shRNAs against Erbin reduce Erbin protein levels in HEK293T cells. Increasing amounts of Erbin shRNA construct shRNA-795 or shRNA-609 or the empty vector pSRG were transiently transfected into HEK293T cells together with Myc-Erbin. Knockdown of Erbin expression was examined by Western blotting. Immunoblotting with anti-β-actin antibody (αβ-actin) served as a loading control. (B) Reducing Erbin expression enhances TGFβ-induced SBE-Luc expression. HaCaT cells were cotransfected with the SBE-Luc reporter and Erbin shRNA construct shRNA-795 (sh795) or shRNA-609 (sh609). Luciferase activity was measured 20 h after TGFβ stimulation. (C) Erbin expression is effectively reduced in Erbin shRNA stable cell lines. Whole-cell lysates (WCL) were prepared from two independent HaCaT cell lines stably expressing Erbin shRNA-795 (KD-1 and KD-2) or an empty pSRG vector (CTL). The endogenous Erbin expression level was detected by Western blotting. Immunoblotting (IB) with anti-β-actin antibody served as a loading control. (D) Reducing Erbin expression enhances TGFβ-induced growth inhibition. KD-1, KD-2, or CTL cells were stimulated with various concentrations of TGFβ as indicated. Cell proliferation was assessed by incorporation of [3H]thymidine. Data are expressed as mean percentages ± standard deviations of thymidine incorporation relative to basal counts of each cell line from duplicate experiments. (E) Knockdown of endogenous Erbin expression enhances ligand-induced expression of TGFβ target genes. Whole-cell extracts from control HaCaT cells or KD-2 cells treated with TGFβ for 24 h were immunoblotted with the indicated antibodies shown on the right. The β-actin immunoblot served as a loading control. (F to H) Reducing Erbin expression enhances Smad2/Smad3-mediated transcriptional activity. HaCaT line KD-2 or CTL cells were transfected with p21-Luc (F), SBE-Luc (G), or ARE-Luc (H). Luciferase activity was measured 20 h after TGFβ stimulation. (I) Reducing Erbin expression did not affect BMP signaling. HaCaT line KD-2 or CTL cells were transfected with Id1-Luc. Luciferase activity was measured 20 h after BMP2 stimulation. RLU, relative luciferase activity; αMyc, anti-Myc antibody; αErbin, anti-Erbin antibody.
TGFβ potently inhibits cell cycle progression at the G1 phase. Smad3 has a key function in mediating the TGFβ growth-inhibitory response (10, 14). Thus, we investigated whether knockdown of Erbin expression affected the ability of cells to undergo growth inhibition in response to TGFβ. KD-1 and KD-2 cells were cultured in the presence of various amounts of TGFβ for 48 h, and [3H]thymidine incorporation into cellular DNA was quantified. As shown in Fig. 6D, knockdown of Erbin expression rendered cells more sensitive to TGFβ-induced growth inhibition. This enhanced growth-inhibitory response to TGFβ was apparent especially at low concentrations of TGFβ (0.1 ng/ml), and the cells reached a maximal level of growth arrest between 1 and 5 ng/ml of TGFβ, whereas in control cells the peak growth inhibition occurred at approximately 0.5 ng/ml.
We also examined the effect of Erbin knockdown on expression of TGFβ target genes. As shown in Fig. 6E, cyclin-dependent kinase inhibitors p15 and p21, which are thought to mediate TGFβ cell cycle arrest, exhibited a strong induction in response to TGFβ in control HaCaT cells (Fig. 6E, lanes 1 and 2). Importantly, knockdown of Erbin profoundly enhanced TGFβ-induced p15 and p21 expression, as the protein levels of these genes were significantly higher in Erbin knockdown KD-2 cells than they were in control cells. Similarly, TGFβ-induced expression of plasminogen activator inhibitor 1 (PAI-1) also was enhanced. It has been shown that the p15, p21, and PAI-1 genes are induced by TGFβ at transcriptional levels. To further prove that Erbin knockdown specifically enhances TGFβ gene responses at the transcriptional level, we analyzed several TGFβ-responsive reporters. We found that knockdown of Erbin expression resulted in an increase in TGFβ-dependent transcriptional activation from both the natural promoter, as in the case of p21-Luc (Fig. 6F), and synthetic promoters, as in the cases of SBE-Luc (Fig. 6G) and ARE-Luc (Fig. 6H), but not in BMP-dependent transcriptional activation from Id1-Luc (Fig. 6I).
Erbin blocks downstream oligomerization and nuclear translocation of Smad2/Smad3.
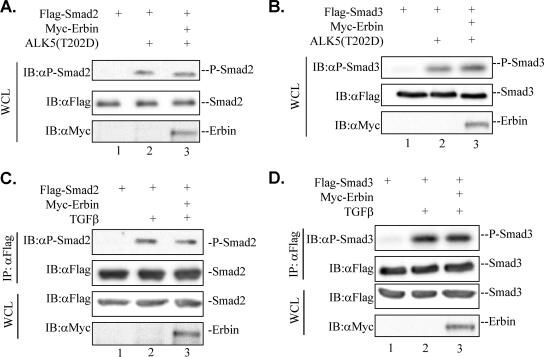
Activation of R-Smad proteins involves receptor-mediated phosphorylation and oligomerization with Smad4, followed by subsequent nuclear translocation. To further characterize the mechanism of Erbin's inhibitory effect on TGFβ signaling, we sought to determine the steps that Erbin acts on in the TGFβ signaling pathway. We first examined whether Erbin affected TGFβ-induced phosphorylation of Smad2/Smad3. The level of Smad2/Smad3 phosphorylation, mediated by TβRI/ALK5, was examined by Western blotting with an antibody specific for phospho-Smad2/Smad3. Ectopic expression of Erbin did not affect the level of P-Smad2 (Fig. 7A and C) or P-Smad3 (Fig. 7B and D) in the presence of either activated TβRI(T202D) or soluble TGFβ ligand.
FIG. 7.
Erbin does not affect phosphorylation of Smad2/Smad3. (A and B) Erbin does not affect ALK5(T202D)-mediated phosphorylation of Smad2/Smad3. Cell lysates were prepared from HEK293T cells cotransfected with Flag-Smad2, Flag-Smad3, ALK5(T202D), and Myc-Erbin as indicated. Phosphorylation of Smad2/Smad3 was detected by Western blotting with anti-P-Smad2 (αP-Smad2) and anti-P-Smad3 (αP-Smad3) antibodies. Expression levels of Myc-Erbin or Flag-Smad2 and Flag-Smad3 were detected by anti-Myc (αMyc) or anti-Flag (αFlag) antibodies. (C and D) Erbin does not affect TGFβ-induced phosphorylation of Smad2/Smad3. Cell lysates were prepared from HEK293T cells that were cotransfected with Flag-Smad2 and Flag-Smad3 with or without Myc-Erbin and treated with or without TGFβ (1 h). Flag-Smad proteins first were immunoprecipitated by anti-Flag antibody and then were subjected to Western blot analysis. WCL, whole-cell lysate; IB, immunoblot; IP, immunoprecipitate.
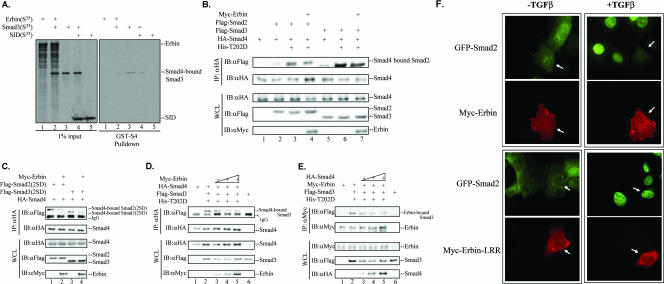
To investigate whether Erbin affects oligomerization of R-Smads with Smad4, we examined the effect of Erbin on Smad3-Smad4 association. The GST-Smad4 fusion protein could bind to 35S-labeled Smad3 in vitro (Fig. 8A, lane 3). Interestingly, addition of 35S-labeled Erbin caused a decrease in the level of GST-Smad4-bound Smad3, indicating that Erbin could disrupt the interaction between Smad3 and Smad4 (Fig. 8A, lane 2). Furthermore, in accordance with our observation that SID repressed the TGFβ-responsive reporter activation (Fig. 3F), we found that SID also could decrease the association between GST-Smad4 and GST-Smad3 to similar extents (Fig. 8A, lane 4).
FIG. 8.
Erbin physically sequesters R-Smad2/R-Smad3 from binding to Smad4. (A) Erbin disrupts Smad3-Smad4 interaction in vitro. The left blot shows an autoradiogram of 1% input of 35S-labeled in vitro-translated proteins as indicated. The right blot shows in vitro-translated Smad3 in the absence or presence of in vitro-translated Erbin; SID was allowed to interact with GST-Smad4 proteins on glutathione beads. GST-Smad4-bound Smad3 proteins were gel resolved and detected by autoradiography. (B and C) Erbin disrupts Smad3-Smad4 interaction in vivo. HEK293T cells were transfected with expression plasmids for Flag-Smad2/Smad3, HA-Smad4, ALK5(T202D), and Myc-Erbin, as indicated. Smad4-bound Smad2/Smad3 was immunoprecipitated with anti-HA (αHA) antibody and detected by anti-Flag (αFlag) Western blotting. (D) Overexpression of Erbin reduces association of Smad4 with Smad3. HEK293T cells were transfected with Flag-Smad3, HA-Smad4, ALK5(T202D), and increasing amounts of Myc-Erbin. Smad4-bound Smad3 was immunoprecipitated with anti-HA antibody and detected by anti-Flag Western blotting. (E) Overexpression of Smad4 reduces association of Erbin with Smad3. HEK293T cells were transfected with Myc-Erbin, Flag-Smad3, ALK5(T202D), and increasing amounts of HA-Smad4. Erbin-bound Smad3 was immunoprecipitated with anti-Myc antibody (αMyc) and detected by anti-Flag Western blotting. (F) Erbin blocks Smad2 nuclear translocation in response to TGFβ. HaCaT cells stably expressing GFP-Smad2 were transiently transfected with Myc-Erbin (top) or Myc-Erbin-LRR (bottom). Twenty-four hours after transfection, cells were treated with TGFβ or were left untreated for 1 h and fixed. Erbin or the Erbin LRR domain was detected by indirect immunofluorescence staining using anti-Myc antibodies. 4′,6′-Diamidino-2-phenylindole (DAPI)-stained DNA in the nucleus is shown. IP, immunoprecipitate; WCL, whole-cell lysate; IB, immunoblot; IgG, immunoglobulin G.
To examine whether Erbin could decrease the complex formation between Smad2/Smad3 and Smad4 in vivo, we carried out coimmunoprecipitation experiments. As expected, ALK5(T202D) stimulated the interaction between Smad4 and Smad2/Smad3. Coexpression of Erbin reduced the Smad2/Smad3-Smad4 association (Fig. 8B), indicating that Erbin could block oligomerization of Smad2/Smad3 with Smad4 in vivo. To confirm that Erbin reduced Smad3-Smad4 association in a phosphorylation-independent manner, we utilized the phosphorylation-mimetic mutant Smad2/Smad3(2SD), which readily associates with Smad4 in the absence of ALK5(T202D). As shown in Fig. 8C, coexpression of Erbin resulted in a reduced formation of the Smad3(2SD)-Smad4 or Smad2(2SD)-Smad4 complex. Taken together, these data indicated that Erbin could physically sequester Smad2/Smad3 from forming a complex with Smad4.
These results prompted us to carry out titration experiments to further examine if Erbin or Smad4 competes for Smad3. Indeed, an increased amount of Erbin reduced the association of Smad4 with Smad3 in coimmunoprecipitation experiments (Fig. 8D). Reciprocally, increased doses of Smad4 reduced the association of Erbin with Smad3 (Fig. 8E). These data suggest that the relative ratio of Erbin to Smad4 or their relative affinity for R-Smads influences the level of TGFβ responses in a given cell.
Erbin resides at the cell membrane and/or cytoplasm in mammalian cells. We next asked whether Erbin inhibited the nuclear accumulation of Smad2/Smad3 in response to TGFβ. We used GFP-Smad2-expressing HaCaT stable cells, which exhibit an easily detectable Smad2 nuclear translocation upon TGFβ stimulation (34). GFP-Smad2 stable cells were transfected with Erbin or Erbin-LRR and treated with TGFβ. Expression of Erbin effectively blocked TGFβ-induced GFP-Smad2 accumulation in the nucleus, as Erbin-positive cells (white arrowhead) exhibited a comparatively weak green fluorescence all over the cells (Fig. 8F), while a strong green fluorescence could be observed easily in the nuclei of Erbin-negative cells. In spite of its expression level, which was similar to that of wild-type Erbin, Erbin-LRR, which was unable to interact with Smad2/Smad3 (Fig. 2B), failed to block GFP-Smad2 translocation in response to TGFβ. These results suggest that Erbin-mediated inhibition of Smad2 nuclear translocation requires its interaction with Smad2.
DISCUSSION
One notable difference among the members of the LAP protein family is their divergent intermediate regions between the N-terminal LRRs and the C-terminal PDZ domains, which might confer simply member-specific interactions. Using a series of Erbin deletion mutants containing an LRR, a PDZ domain, and several intermediate regions, we have identified a region in Erbin, designated SID (for Smad-interacting domain or Smad-inhibitory domain), that is crucial for Erbin-R-Smad2/Erbin-R-Smad3 interaction as well as for Erbin's inhibitory role in TGFβ signaling. SID locates in a functionally unassigned region in Erbin, which is different from established protein-interacting domains such as the PDZ domain for direct binding to ErbB2 (4, 19) and the LRR domain for direct binding to Sur-8 (8). Thus, identification of this unique SID suggests that the function of Erbin in TGFβ signaling is independent of its function either as an ErbB2-interacting protein or as an ERK inhibitor. Erbin and Densin-180 share high levels of similarity overall in the LRR domain (72.7% identity in the N-terminal 400 aa) and in the PDZ domain (70.8% identity in the C-terminal 88 aa). In contrast, the amino acid sequence of SID in Erbin shares only 20.7% identity with its counterpart in Densin-180. The ability of Erbin, but not Densin-180, to bind to Smad2/Smad3 is consistent with the SID sequences being divergent. Interestingly, multiple alternatively spliced variants of Erbin exist in humans, with their sequences differing mainly in the intermediate region (11). This suggests that these isoforms of Erbin may have differential abilities to modulate TGFβ signaling. Our study also implies that spatially and/or temporally regulated expression of different Erbin variants may contribute to differential TGFβ sensitivity in different cell or tissue types.
Unlike most Smad-interacting partners, such as nuclear transcription cofactors (12), Erbin mainly is found on the plasma membrane and/or in the cytoplasm in cultured cells (4, 19, 29). Thus, Erbin's action on TGFβ signaling most likely is achieved outside of the nucleus. One mechanism by which Erbin could influence Smad2/Smad3 activity is through modulation of Smad2/Smad3 activation by TGFβ receptors on the cell membrane. However, several lines of evidence argue against this possibility. First, Erbin's inhibitory effect on TGFβ-induced gene transcription is independent of the LRR or the PDZ domain (Fig. 4). Second, we did not observe any association between Erbin and TGFβ receptors in coimmunoprecipitation experiments with HEK293T cells (data not shown). Erbin also did not interact with the membrane-associated adaptor molecule SARA, which facilitates the interaction of Smad2 and Smad3 with the activated TGFβ receptors (data not shown). Finally, Erbin has no effect on the level of phosphorylated Smad2/Smad3 directly induced by constitutively active TβRI(T202D) or ligand stimulation (Fig. 7). Therefore, Erbin seems to inhibit TGFβ signaling at a step downstream of Smad2/Smad3 phosphorylation by disrupting the Smad complex in the cytoplasm. Erbin and Smad4 compete for binding to Smad3 (Fig. 8D and E), suggesting that the relative ratio of Erbin and Smad4 or their relative affinity for R-Smads influences the strength of TGFβ responses in a given cell.
Since Erbin's basolateral localization (4, 19, 29) as well as the nucleocytoplasmic shuttling of R-Smads (21, 34, 38, 42) has been well characterized, the localization of Erbin-Smad interaction presumably is restricted by the location at which Erbin resides. Erbin-mediated inhibition of TGFβ signaling logically would occur on the basolateral side. However, it is not clear how expression or activities of TGFβ signaling components and other regulators (other than Erbin) are distributed in a polarized epithelial cell. Murphy et al. reported that Smad2/Smad3 phosphorylation primarily occurs through ligand addition to the basolateral surface (33). The endogenous TGFβ ligand secretion was from the apical surfaces, while the endogenous type I receptor and a chimeric type II receptor (the extracellular ligand-binding domains of granulocyte-macrophage colony-stimulating factor receptors fused with the cytoplasmic and transmembrane domains of the type II receptor) were localized at the basolateral surfaces of polarized MDCK cells (33). Therefore, a combined spatial and temporal localization of ligand, receptors, Smads, and other signaling modulators (e.g., Erbin) would define the overall strength of TGFβ responses in these locales in polarized cells.
As a negative modulator of Smad2/Smad3 function, proper expression of Erbin and its alternative transcripts may play an important role in modulating TGFβ-induced cellular response, e.g., cell growth inhibition and cell fate determination. Several studies revealed that Erbin expression could be regulated at the transcriptional level by different stimuli. Erbin was induced by proinflammatory stimuli such as tumor necrosis factor alpha (32) or by BRCA1 overexpression (2). It also is conceivable that inhibition of TGFβ antiproliferative responses, rather than ErbB2/Her2 proproliferative actions, by increased expression of Erbin likely contributes to tumorigenesis. Indeed, DNA copy numbers of Erbin in cervical tumor samples were found to be significantly different from those of normal controls (18), and increased expression of Erbin genes has been observed in in vitro colon carcinoma and glioblastoma models (9).
TGFβ and the related activin and BMPs are well-known regulators of embryonic patterning and cell fate specification (16). Our studies have revealed that a novel and unique function of Erbin is restricted to the regulation of Smad2/Smad3-dependent activin/TGFβ signaling. Consistent with its inhibitory role in TGFβ growth-inhibitory and transcriptional responses in cultured human cells, Erbin also profoundly inhibits activin/Smad2-mediated, but not BMP/Smad1-mediated, induction of endogenous genes required for cell fate specification in Xenopus embryos (Fig. 5). Despite the finding that Erbin inhibits the TGFβ signaling pathway, further detailed studies are needed to dissect the physiological roles of Erbin during development and/or in human diseases.
Acknowledgments
We thank Edward Leof, Peter ten Dijke, Bert Vogelstein, Malcolm Whitman, and Caroline S. Hill for various reagents.
This research was supported by NIH grants (R01GM63773 and R01CA108454 to X.-H.F., R01DK073932 and R21CA11293 to X.L., R01HD43345 to C.C., and R01NS44521 to L.M.). X.L. was supported by a Baylor Breast Center SPORE career development award. X.-H.F. is a Leukemia and Lymphoma Society Scholar.
Footnotes
Published ahead of print on 25 June 2007.
REFERENCES
- 1.Apperson, M. L., I. S. Moon, and M. B. Kennedy. 1996. Characterization of densin-180, a new brain-specific synaptic protein of the O-sialoglycoprotein family. J. Neurosci. 16:6839-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atalay, A., T. Crook, M. Ozturk, and I. G. Yulug. 2002. Identification of genes induced by BRCA1 in breast cancer cells. Biochem. Biophys. Res. Commun. 299:839-846. [DOI] [PubMed] [Google Scholar]
- 3.Bilder, D., D. Birnbaum, J. P. Borg, P. Bryant, J. Huigbretse, E. Jansen, M. B. Kennedy, M. Labouesse, R. Legouis, B. Mechler, N. Perrimon, M. Petit, and P. Sinha. 2000. Collective nomenclature for LAP proteins. Nat. Cell Biol. 2:E114. [DOI] [PubMed] [Google Scholar]
- 4.Borg, J. P., S. Marchetto, B. A. Le, V. Ollendorff, F. Jaulin-Bastard, H. Saito, E. Fournier, J. Adelaide, B. Margolis, and D. Birnbaum. 2000. ERBIN: a basolateral PDZ protein that interacts with the mammalian ERBB2/HER2 receptor. Nat. Cell Biol. 2:407-414. [DOI] [PubMed] [Google Scholar]
- 5.Bryant, P. J., and A. Huwe. 2000. LAP proteins: what's up with epithelia? Nat. Cell Biol. 2:E141-E143. [DOI] [PubMed] [Google Scholar]
- 6.Chang, C., P. A. Wilson, L. S. Mathews, and A. Hemmati-Brivanlou. 1997. A Xenopus type I activin receptor mediates mesodermal but not neural specification during embryogenesis. Development 124:827-837. [DOI] [PubMed] [Google Scholar]
- 7.Chang, H., C. W. Brown, and M. M. Matzuk. 2002. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr. Rev. 23:787-823. [DOI] [PubMed] [Google Scholar]
- 8.Dai, P., W. C. Xiong, and L. Mei. 2006. Erbin inhibits RAF activation by disrupting the sur-8-Ras-Raf complex. J. Biol. Chem. 281:927-933. [DOI] [PubMed] [Google Scholar]
- 9.Dardousis, K., C. Voolstra, M. Roengvoraphoj, A. Sekandarzad, S. Mesghenna, J. Winkler, Y. Ko, J. Hescheler, and A. Sachinidis. 2007. Identification of differentially expressed genes involved in the formation of multicellular tumor spheroids by HT-29 colon carcinoma cells. Mol. Ther. 15:94-102. [DOI] [PubMed] [Google Scholar]
- 10.Datto, M. B., J. P. Frederick, L. H. Pan, A. J. Borton, Y. Zhuang, and X. F. Wang. 1999. Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol. Cell. Biol. 19:2495-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favre, B., L. Fontao, J. Koster, R. Shafaatian, F. Jaunin, J. H. Saurat, A. Sonnenberg, and L. Borradori. 2001. The hemidesmosomal protein bullous pemphigoid antigen 1 and the integrin beta 4 subunit bind to ERBIN. Molecular cloning of multiple alternative splice variants of ERBIN and analysis of their tissue expression. J. Biol. Chem. 276:32427-32436. [DOI] [PubMed] [Google Scholar]
- 12.Feng, X. H., and R. Derynck. 2005. Specificity and versatility in TGF signaling through Smads. Annu. Rev. Cell Dev. Biol. 21:659-693. [DOI] [PubMed] [Google Scholar]
- 13.Feng, X. H., Y. Y. Liang, M. Liang, W. Zhai, and X. Lin. 2002. Direct interaction of c-Myc with Smad2 and Smad3 to inhibit TGFβ-mediated induction of the CDK inhibitor p15Ink4B. Mol. Cell 9:133-143. [DOI] [PubMed] [Google Scholar]
- 14.Feng, X. H., X. Lin, and R. Derynck. 2000. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J. 19:5178-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Germain, S., M. Howell, G. M. Esslemont, and C. S. Hill. 2000. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 14:435-451. [PMC free article] [PubMed] [Google Scholar]
- 16.Harland, R., and J. Gerhart. 1997. Formation and function of Spemann's organizer. Annu. Rev. Cell Dev. Biol. 13:611-667. [DOI] [PubMed] [Google Scholar]
- 17.Hayashida, T., M. Decaestecker, and H. W. Schnaper. 2003. Cross-talk between ERK MAP kinase and Smad signaling pathways enhances TGF-beta-dependent responses in human mesangial cells. FASEB J. 17:1576-1578. [DOI] [PubMed] [Google Scholar]
- 18.Huang, F. Y., P. M. Chiu, K. F. Tam, Y. K. Kwok, E. T. Lau, M. H. Tang, T. Y. Ng, V. W. Liu, A. N. Cheung, and H. Y. Ngan. 2006. Semi-quantitative fluorescent PCR analysis identifies PRKAA1 on chromosome 5 as a potential candidate cancer gene of cervical cancer. Gynecol. Oncol. 103:219-225. [DOI] [PubMed] [Google Scholar]
- 19.Huang, Y. Z., Q. Wang, W. C. Xiong, and L. Mei. 2001. Erbin is a protein concentrated at postsynaptic membranes that interacts with PSD-95. J. Biol. Chem. 276:19318-19326. [DOI] [PubMed] [Google Scholar]
- 20.Huang, Y. Z., M. Zang, W. C. Xiong, Z. Luo, and L. Mei. 2003. Erbin suppresses the MAP kinase pathway. J. Biol. Chem. 278:1108-1114. [DOI] [PubMed] [Google Scholar]
- 21.Inman, G. J., F. J. Nicolas, and C. S. Hill. 2002. Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol. Cell 10:283-294. [DOI] [PubMed] [Google Scholar]
- 22.Izawa, I., M. Nishizawa, Y. Tomono, K. Ohtakara, T. Takahashi, and M. Inagaki. 2002. ERBIN associates with p0071, an armadillo protein, at cell-cell junctions of epithelial cells. Genes Cells 7:475-485. [DOI] [PubMed] [Google Scholar]
- 23.Jaulin-Bastard, F., J. P. Arsanto, B. A. Le, C. Navarro, F. Vely, H. Saito, S. Marchetto, M. Hatzfeld, M. J. Santoni, D. Birnbaum, and J. P. Borg. 2002. Interaction between Erbin and a catenin-related protein in epithelial cells. J. Biol. Chem. 277:2869-2875. [DOI] [PubMed] [Google Scholar]
- 24.Jaulin-Bastard, F., H. Saito, B. A. Le, V. Ollendorff, S. Marchetto, D. Birnbaum, and J. P. Borg. 2001. The ERBB2/HER2 receptor differentially interacts with ERBIN and PICK1 PSD-95/DLG/ZO-1 domain proteins. J. Biol. Chem. 276:15256-15263. [DOI] [PubMed] [Google Scholar]
- 25.Kim, Y. K., G. U. Bae, J. K. Kang, J. W. Park, E. K. Lee, H. Y. Lee, W. S. Choi, H. W. Lee, and J. W. Han. 2006. Cooperation of H2O2-mediated ERK activation with Smad pathway in TGF-beta1 induction of p21WAF1/Cip1. Cell. Signal. 18:236-243. [DOI] [PubMed] [Google Scholar]
- 26.Kolch, W. 2003. Erbin: sorting out ErbB2 receptors or giving Ras a break? Sci. STKE 2003:e37. [DOI] [PubMed] [Google Scholar]
- 27.Labbé, E., C. Silvestri, P. A. Hoodless, J. L. Wrana, and L. Attisano. 1998. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell 2:109-120. [DOI] [PubMed] [Google Scholar]
- 28.Laura, R. P., A. S. Witt, H. A. Held, R. Gerstner, K. Deshayes, M. F. Koehler, K. S. Kosik, S. S. Sidhu, and L. A. Lasky. 2002. The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J. Biol. Chem. 277:12906-12914. [DOI] [PubMed] [Google Scholar]
- 29.Legouis, R., F. Jaulin-Bastard, S. Schott, C. Navarro, J. P. Borg, and M. Labouesse. 2003. Basolateral targeting by leucine-rich repeat domains in epithelial cells. EMBO Rep. 4:1096-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, X., X. Duan, Y. Y. Liang, Y. Su, K. H. Wrighton, J. Long, M. Hu, C. M. Davis, J. Wang, F. C. Brunicardi, Y. Shi, Y. G. Chen, A. Meng, and X. H. Feng. 2006. PPM1A functions as a Smad phosphatase to terminate TGFβ signaling. Cell 125:915-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massagué, J., J. Seoane, and D. Wotton. 2005. Smad transcription factors. Genes Dev. 19:2783-2810. [DOI] [PubMed] [Google Scholar]
- 32.McDonald, C., F. F. Chen, V. Ollendorff, Y. Ogura, S. Marchetto, P. Lecine, J. P. Borg, and G. Nunez. 2005. A role for Erbin in the regulation of Nod2-dependent NF-κB signaling. J. Biol. Chem. 280:40301-40309. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, S. J., J. J. Dore, M. Edens, R. J. Coffey, J. A. Barnard, H. Mitchell, M. Wilkes, and E. B. Leof. 2004. Differential trafficking of transforming growth factor-beta receptors and ligand in polarized epithelial cells. Mol. Biol. Cell 15:2853-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicolás, F. J., K. De Bosscher, B. Schmierer, and S. C. Hill. 2004. Analysis of Smad nucleocytoplasmic shuttling in living cells. J. Cell Sci. 117:4113-4125. [DOI] [PubMed] [Google Scholar]
- 35.Ohno, H., S. Hirabayashi, T. Iizuka, H. Ohnishi, T. Fujita, and Y. Hata. 2002. Localization of p0071-interacting proteins, plakophilin-related armadillo-repeat protein-interacting protein (PAPIN) and ERBIN, in epithelial cells. Oncogene 21:7042-7049. [DOI] [PubMed] [Google Scholar]
- 36.Pawson, T., and J. D. Scott. 1997. Signaling through scaffold, anchoring, and adaptor proteins. Science 278:2075-2080. [DOI] [PubMed] [Google Scholar]
- 37.Randall, R. A., S. Germain, G. J. Inman, P. A. Bates, and C. S. Hill. 2002. Different Smad2 partners bind a common hydrophobic pocket in Smad2 via a defined proline-rich motif. EMBO J. 21:145-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmierer, B., and C. S. Hill. 2005. Kinetic analysis of Smad nucleocytoplasmic shuttling reveals a mechanism for transforming growth factor beta-dependent nuclear accumulation of Smads. Mol. Cell. Biol. 25:9845-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ten Dijke, P., and C.-H. Heldin. 2006. Smad signal transduction: Smads in proliferation, differentiation and disease. Springer, Berlin, Germany.
- 40.Warner, D. R., M. M. Pisano, E. A. Roberts, and R. M. Greene. 2003. Identification of three novel Smad binding proteins involved in cell polarity. FEBS Lett. 539:167-173. [DOI] [PubMed] [Google Scholar]
- 41.Wu, G., Y. G. Chen, B. Ozdamar, C. A. Gyuricza, P. A. Chong, J. L. Wrana, J. Massague, and Y. Shi. 2000. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science 287:92-97. [DOI] [PubMed] [Google Scholar]
- 42.Xu, L., Y. Kang, S. Col, and J. Massague. 2002. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol. Cell 10:271-282. [DOI] [PubMed] [Google Scholar]
- 43.Zawel, L., J. L. Dai, P. Buckhaults, S. Zhou, K. W. Kinzler, B. Vogelstein, and S. E. Kern. 1998. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell 1:611-617. [DOI] [PubMed] [Google Scholar]