Abstract
Multiple factors influence estrogen receptor α (ERα) transcriptional activity. Current models suggest that the silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor functions within a histone deactylase-containing protein complex that binds to antiestrogen-bound ERα and contributes to negative regulation of gene expression. In this report, we demonstrate that SMRT is required for full agonist-dependent ERα activation. Chromatin immunoprecipitation assays demonstrate that SMRT, like ERα and the SRC-3 coactivator, is recruited to an estrogen-responsive promoter in estrogen-treated MCF-7 cells. Depletion of SMRT, but not histone deacetylases 1 or 3, negatively impacts estradiol-stimulated ERα transcriptional activity, while exogenous expression of SMRT's receptor interaction domains blocks ERα activity, indicating a functional interaction between this corepressor and agonist-bound ERα. Stimulation of estradiol-induced ERα activity by SMRT overexpression occurred in HeLa and MCF-7 cells, but not HepG2 cells, indicating that these positive effects are cell type specific. Similarly, the ability of SMRT depletion to promote the agonist activity of tamoxifen was observed for HeLa but not MCF-7 cells. Furthermore, impairment of agonist-stimulated activity by SMRT depletion is specific to ERα and not observed for receptors for vitamin D, androgen, or thyroid hormone. Nuclear receptor corepressor (N-CoR) depletion increased the transcriptional activity of all four tested receptors. SMRT is required for full expression of the ERα target genes cyclin D1, BCL-2, and progesterone receptor but not pS2, and its depletion significantly attenuated estrogen-dependent proliferation of MCF-7 cells. Taken together, these data indicate that SMRT, in conjunction with gene-specific and cell-dependent factors, is required for positively regulating agonist-dependent ERα transcriptional activity.
Estrogens are potent mitogens in a number of target tissues, including the mammary gland, where they play a pivotal role in the development and progression of breast tumorigenesis. The effects of estrogen are mediated via estrogen receptor α (ERα) and ERβ, which are nuclear receptors that belong to a superfamily of ligand-regulated transcription factors (61). Subsequent to estradiol (E2) binding to ERα, the receptor undergoes a conformational change, dimerizes, and either binds directly to DNA via estrogen response elements (EREs) or indirectly binds DNA via interactions with other DNA-bound transcription factors, such as Sp1 or AP-1 (41, 61, 68). Although ERα binds within the promoter regions of some estrogen-sensitive target genes, it has been estimated that only ∼4% of ERα binding sites lie within 1 kb of proximal promoter regions, and the good correlation of ERα binding sites within 50 kb of the transcriptional start sites of estrogen-induced genes suggests that ERα can regulate the expression of these genes from a substantial distance (7-9, 42). The central involvement of estrogens in the genesis and progression of breast cancer has led to the identification and characterization of a number of ERα target genes, including c-myc, pS2, cathepsin D, and cyclin D1 (2, 10, 20, 66). Ultimately, multiple factors influence the ability of ERα to activate transcription of target genes, including the expression and recruitment of coregulatory molecules, ligand binding, nongenomic signaling pathways, the cellular environment, and the anatomy of the ERE present within target genes (44, 50, 61, 72).
Two distinct regions within the ERα contribute to transcriptional activity: the constitutively active activation function 1 (AF1), located in the N terminus (A/B domain), and ligand-regulatable AF2 in the C terminus (E domain). The ERα binds a variety of ligands, including agonists such as E2, selective estrogen receptor modulators (SERMs) such as 4-hydroxytamoxifen (4HT) and raloxifene, and pure antiestrogens, such as ICI 182,780 (ICI). In each case, the receptor undergoes distinct structural changes that influence the extent to which ERα interacts with coactivators or corepressors, resulting in profound effects on whether gene expression is stimulated or repressed (60). Agonist-bound ERα allows for correct exposure of AF2 and subsequent interactions with coactivator proteins to form a multiprotein complex that contacts the general transcriptional machinery and increases expression of target genes via processes involving chromatin remodeling, formation of preinitiation complexes, and an enhanced rate of RNA polymerase II reinitiation (23, 49). The ability of a number of coactivators to exert transcriptional control is influenced by intracellular signaling mediated via alterations in their posttranslational modifications, including acetylation, sumoylation, ubiquitination, phosphorylation, and methylation, that influence coactivator subcellular localization, protein-protein interactions, and protein stability (49, 67, 72).
In contrast, ERα antiestrogens with their bulky side chains render AF2 inaccessible to coactivators and disrupt transcriptional activation (60). The prevailing model suggests that the SERM-bound ERα complexes recruit corepressors and their associated histone deacetylases (HDACs) to target genes, which results in the removal of acetyl groups from histones, a more compact chromatin structure and, consequently, inhibition of gene expression (65). Although the corepressors N-CoR and SMRT were originally characterized by their ability to bind and repress unliganded RAR and TR (14, 29), the biological importance of these corepressors for unliganded ERα is less certain, and this has led to the widely accepted view that corepressors regulate ERα when it is bound to pharmacological antiestrogens. Indeed, several studies have demonstrated that N-CoR and/or SMRT interacts with ERα in the presence of 4HT and represses the weak agonistic activity of this antiestrogen (33, 43, 71, 83), and both 4HT and raloxifene have been shown to recruit N-CoR and SMRT to ERα target genes (48, 69, 70). More recently, unliganded ERα has been shown to bind to these corepressors (54, 76), and mapping experiments reveal that removal of the receptor's A domain or helix 12 improves apo-ERα binding to corepressors and suggests that the deleted regions normally compete with corepressors for binding to the same region of the ligand binding domain (36, 55). Cumulatively, these data are consistent with a role for N-CoR and/or SMRT in regulating the activities of antiestrogen bound and possibly unliganded receptor.
The molecular mechanisms by which SMRT/N-CoR repress gene expression appear quite complex. Early studies suggested that SMRT and/or N-CoR interacted with the mSin3A-HDAC1/2 complex, while subsequent data contradicted these findings and revealed that these corepressors associate with HDAC3 as well as GPS2 (25, 27, 31, 45, 58, 82). SMRT and/or N-CoR complexes were also found to contain transducin-β-like 1 (TBL1) and TBL1-related (TBLR1) proteins, and several studies indicate that these proteins contribute to inhibition of gene expression (26, 45, 77, 78, 82). However, these molecules also have been implicated in stimulating gene expression via their ability to promote exchange of cofactors on the promoters of target genes (63), and this suggests that they may play diverse and possibly context-dependent biological roles. In addition to the role of SMRT and N-CoR in inhibiting the activity of antagonist-bound steroid receptors, they also can modulate the ability of agonists to stimulate receptor transcriptional activity. For instance, N-CoR and SMRT suppress agonist-dependent activation of androgen receptor (AR) target genes (1, 6, 15, 28, 47, 79), and small interfering RNA (siRNA)-mediated inhibition of N-CoR expression enhances E2 induction of ERα transcriptional activity (35). Overlapping surfaces within NR ligand binding domains are utilized for association with SMRT/N-CoR and LXXLL-containing coactivators, and corepressor inhibition of agonist-stimulated receptor transcriptional activity likely reflects corepressor competition with coactivators for binding to this overlapping binding surface (30, 57). There are, however, limited data suggesting that coactivators and corepressors can simultaneously interact with nuclear receptors (19, 46). For instance, N-CoR binds directly to the p160 coactivator ACTR (AIB1/SRC-3/RAC3) and appears to facilitate recruitment of coactivator to unliganded thyroid hormone receptor β (TRβ) (46). Using chromatin immunoprecipitation (ChIP) assays, another study demonstrated concurrent binding of N-CoR, the coactivators SRC-1 and PCAF, and RNA polymerase II to the promoter of the androgen-regulated prostate-specific antigen gene in antiandrogen-resistant LNCaP prostate cancer cells treated with the AR antagonist bicalutamide (12). These intriguing, and incompletely understood, interactions suggest that coactivator function may not be independent of corepressors and vice versa.
In this study, we investigated the ability of SMRT to modulate ERα transcriptional activity in the presence of estrogen and antiestrogen and determined the regulatory contribution of SMRT to the expression of specific ER-responsive genes associated with cellular proliferation in breast cancer. Taken together, our data demonstrate a cell-type-specific role for SMRT in positively regulating agonist-ERα transcriptional activity via interaction with the receptor's AF2 domain. In contrast, N-CoR suppresses estrogen-stimulated ERα activity, and this highlights functional differences between these corepressor paralogs. The positive role of SMRT in regulating gene expression was specific to ERα, since the agonist-dependent activities of several other members of the nuclear receptor superfamily were negatively regulated by this corepressor. Finally, we demonstrate that the requirement of SMRT for maximal expression of ERα target genes is gene selective and that SMRT positively contributes to proliferation of ERα-positive breast cancer cells.
MATERIALS AND METHODS
Chemicals.
17β-Estradiol and the partial antiestrogen 4-hydroxytamoxifen were obtained from Sigma Chemical Company (St. Louis, MO). The pure antiestrogen ICI 182,780 was obtained from Tocris (Ellisville, MO).
Plasmids and siRNAs.
The mammalian expression construct for full-length human ERα (pCR3.1-hERα) and ERα lacking the A/B domain [pCR3.1-hERα(179C)] have been described previously (16, 59). The pBIND-ABα expression vector encodes the GAL4 DNA binding domain (DBD; amino acids 1 to 147) linked to the A/B domain of hERα (21). The ERE-E1b-Luc and pC3-Luc reporter constructs have been described previously (59, 74), while pG5-Luc is commercially available (Promega, Madison, WI). The expression vectors for mouse SMRTα (pCMX-SMRTα) and SMRTβ (pCMX-SMRTβ) have been described previously (38, 62), as has the expression vector for the first form of cloned SMRT, which encodes amino acids 1032 to 2517 (14). All constructs were sequenced and found to possess the τ deletion (24), and pCMX-SMRTα is therefore referred to as pCMX-SMRTτ in this report. Expression vectors for VDR (pAd-hVDR [53]) and AR (pCR3.1-hAR [1]), as well as the reporter genes VDRE-tk-LUC (84), ARE-E1b-Luc (59), and 28TRE-tk-Luc (77), have been described previously. The pCR3.1-TRβ expression plasmid was generated by inserting the TRβ cDNA isolated from pRST7-hTRβ (4) into the EcoRI site of pCR3.1 (Invitrogen). To generate the pBIND-ID1+2 mammalian expression vector for the GAL4 DBD fused to the region of SMRTα (amino acids 2059 to 2352) that encompasses its receptor interaction domains (IDs), pCMX-SMRTα was used as template for PCR amplification with the 5′ and 3′ primers 5′-TCTCCCACATCTGCGGCCA-3′ and 5′-GAGGTGAGTGCGTGGTCAC-3′, respectively, and the resulting PCR product was subcloned into a pCR2.1 vector using a TA cloning kit (Invitrogen, Carlsbad, CA) and sequenced. From this plasmid, an EcoRV-KpnI restriction enzyme fragment was purified and subsequently subcloned into the corresponding sites of pBIND (Promega), such that the IDs were in frame with and downstream of the GAL4 DBD.
All siRNAs were chemically synthesized by Ambion (Austin, TX) as oligonucleotide duplexes. siRNA target sequences for SMRT were directed at regions common to both SMRTα and SMRTβ (panSMRT, 5′-GGGTATCATCACCGCTGTG-3′ [77]; Sαβ2, 5′-CAGCCUUUCCUACCCAGUG-3′), or only SMRTα (5′-GGAGGAGCTGATCCAGAAC-3′; Ambion predesigned siRNA ID 17668). The N1 target sequence for N-CoR (5′-TGCTACTTCTCGAGGAAACA-3′) was published previously (77), as were sequences for HDAC1 (GCAGATGCAGAGATTCAAC [31]) and HDAC3 (TATCCCTCTACTCGTGCTGA [77]). The N2 target sequence for N-CoR (5′-GGCCTCAAGAAAGGAGAAC-3′) was a predesigned siRNA (ID 17761; Ambion). As nonspecific siRNA controls, an siRNA sequence targeting luciferase (5′-CGTACGCGGAATACTTCGA-3′) or Ambion's Silencer 2 negative control were used. Experiments measuring luciferase as an end point always employed the Silencer control siRNA.
Cell culture.
MCF-7 human breast cancer cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The MCF7-XVA2-Luc cell line (a kind gift of Steffi Oesterrich) is derived from MCF-7 cells stably cotransfected with pSV-neomycin and an ERE-MAR-Luc reporter gene and was grown in DMEM with 10% FBS. ERE-MAR-Luc consists of a region of the Xenopus laevis vitellogenin A2 gene containing an ERE and scaffold/matrix attachment regions (MAR) and was inserted into the BamHI and NcoI sites of pGL3 basic (35). HeLa human cervical carcinoma cells were maintained in DMEM supplemented with 10% FBS. HepG2 human hepatoma cells were maintained in minimum essential medium supplemented with 10% FBS.
Growth assays.
MCF-7 cells were plated in six-well culture dishes at 2 × 105 cells/well 24 h prior to transfection, in phenol red-free DMEM containing either 10% FBS or charcoal-stripped FBS (sFBS). HeLa cells were plated in six-well culture dishes in phenol red-free DMEM containing 5% sFBS. On day 1, cells were transfected with siRNA targeting luciferase, SMRTα and SMRTβ (panS), or only SMRTα (Sα) using 3 μl Oligofectamine reagent per well (Invitrogen) according to the manufacturer's recommendations. After 4 hours, cells were treated with vehicle (0.1% ethanol), 1 nM E2, 100 nM 4HT, or 100 nM ICI. On day 3, fresh hormone and medium were added. On day 6, cells from duplicate wells were harvested with 0.25% trypsin-EDTA (Invitrogen), and cell number was determined with a Beckman Z1 dual Coulter Counter (Beckman Coulter, Fullerton, CA). A third well was harvested for protein to verify depletion of corepressor levels by Western blot analysis.
trans-activation assays for ERα activity.
For assays employing siRNA technologies, cells were plated in six-well culture dishes 24 h prior to transfection. At that time they were transfected with 30 pmol/well of the indicated siRNA by using Oligofectamine with Ambion's Silencer 2 as a negative control. Twenty-four hours thereafter, cells were transfected with the indicated type and amount of plasmid DNAs using TransIT-LT1 per the manufacturer's instructions (Mirus, Madison, WI). Thereafter, cells were treated with vehicle (0.1% ethanol) or the indicated ligands for 24 h. Duplicate wells were harvested, and cell extracts were prepared for luciferase activity determinations using a luciferase assay system kit (Promega) and a Luminoskan Ascent Thermo Labsystems apparatus (Thermo Electron Corporation, Milford, MA). Relative luciferase units were normalized to total cellular protein as measured in a Bio-Rad protein assay (Bio-Rad, Hercules, CA). A third well was harvested to verify corepressor downregulation in cell lysates by Western blot analysis. Duplicate samples were measured in each assay, and data are presented as the mean ± standard error of the mean (SEM) of at least three experiments.
For SMRT overexpression experiments, cells were plated in phenol red-free medium containing 10% sFBS and transfected with the indicated DNAs using Lipofectamine (Invitrogen) for HeLa cells, Lipofectamine 2000 (Invitrogen) for HepG2 cells, or TransIT-LT1 for MCF-7 cells. The total amount of DNA in each transfection experiment was kept constant by balancing DNA amounts with the empty pCMX vector. Thereafter, cells were treated with vehicle or the indicated ligands. Twenty-four hours later cells were harvested and assayed for luciferase activity. Duplicate samples were measured in each assay, and data are presented as the mean ± SEM of at least three experiments.
Western blot analyses.
Cells were harvested and then incubated for 20 min at 4°C in lysis buffer (50 mM Tris [pH 7.4] containing 150 mM NaCl, 5 mM EDTA, and 0.5% Nonidet P-40 supplemented with Complete Mini-Tablets protease inhibitors [Roche Applied Sciences, Indianapolis, IN]). The protein content of the cell lysate was determined using protein assay reagent (Bio-Rad, Hercules, CA), and equal levels of protein were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on precast NuPAGE Novex 3 to 8% or 7% Tris-acetate gels (Invitrogen) and transferred to nitrocellulose membranes (Osmonics Inc., Gloucester, MA) by electrotransfer. Nonspecific sites were saturated by incubating the blots in blocking buffer (phosphate-buffered saline containing 0.5% Tween 20 [PBS-T] and 5% dried nonfat milk powder) for 1 h at room temperature. Blots were probed with primary antibodies (listed below) in PBS-T containing 5% nonfat dry milk overnight at 4°C. After washing, blots were incubated with the appropriate horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibody (Amersham Biosciences, Piscataway, NJ) for 1 h at room temperature. The primary antibodies were as follows: anti-SMRT antibody (611386; BD Biosciences, San Jose, CA), anti-N-CoR antibody (06-892; Upstate Biotechnology, Lake Placid, NY), anti-ERα (Cell Signaling, Beverly, MA), anti-cyclin D1 (554180; BD Biosciences), anti-Bcl-2 (610538; BD Biosciences), anti-Flag M2 antibody (Sigma), or antiactin antibody (MAB1501R; Chemicon, Temicula, CA). Detection of specifically bound proteins was carried out by using ECL Plus according to the manufacturer's protocol (Amersham Pharmacia Biotech, Arlington Heights, IL) using XL-1 Blue film (Kodak, Branchburg, NJ).
[3H]estradiol binding assays.
The binding capacity of ERα expressed in MCF-7 cells was determined in vivo as previously described (75). Briefly, cells were plated in DMEM containing 10% sFBS and transfected as described above with Silencer 2 negative control or Sαβ siRNAs. Twenty-four hours thereafter, medium was aspirated from wells and replaced with phenol red-free DMEM containing 5% sFBS, ∼1.5 pmol [3H]estradiol (250 mCi; Perkin-Elmer Life Sciences, Wellesley, MA), with or without 10−3 M unlabeled E2. After incubation for 2 h at 37°C medium was aspirated, and cells were washed three times with ice-cold PBS and incubated in 100% ethanol for 10 min at room temperature to extract bound steroid. The amount of ER-bound [3H]E2 in the ethanol extract was quantified with a Beckman LS 6500 scintillation counter I (Beckman Instruments, Fullerton, CA) and biodegradable counting scintillant (Amersham Biosciences). The binding capacity in untransfected cells (100%) was compared to values obtained for cells transfected with siRNA to either luciferase or SMRTα and SMRTβ (Sαβ).
ChIP assay.
Chromatin immunoprecipitation was performed as described earlier with minor modifications (32). MCF-XVA2-Luc cells (9 × 106 cells/150-mm plate) were maintained in phenol red-free DMEM supplemented with sFBS for 48 h prior to hormone treatment. Thereafter, cells were treated with 2.5 μM α-amanitin for 90 min, washed twice with PBS, and treated with vehicle or estradiol (10 nM) for 45 min, followed by cross-linking with 1% formaldehyde for 10 min at room temperature. Cross-linking was terminated with 0.125 M glycine for 5 min at room temperature, and cells were harvested by scraping in PBS containing protease inhibitor. Nuclei were isolated using nuclei preparation buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 8.0), 85 mM KCl, 0.5% NP-40, and protease inhibitor] and then resuspended in TE buffer (10 mM EDTA, 50 mM Tris-HCl [pH 8.1], and protease inhibitor) containing 1% SDS and sonicated (10-s pulses repeated eight times). Chromatin thus prepared was diluted 10 times with TSE1 (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.1]) containing protease inhibitor, precleared by normal rabbit immunoglobulin G (IgG) and protein A/G agarose beads containing salmon sperm DNA (Upstate) and immunoprecipitated overnight at 4°C with specific antibodies (ERα with HC20 and H184, SRC-3 with C-20, and SMRT with H-300; Santa Cruz Biotechnology, Santa Cruz, CA) or rabbit IgG control. Immune complexes were then precipitated with protein A/G agarose beads, serially washed once with TSE1 containing 0.1% SDS, twice with TSE2 (1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl [pH 8.1], 0.1% SDS), once with buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and thrice with TE and then eluted with freshly prepared elution buffer (1% SDS, 100 mM NaHCO3). Formaldehyde cross-links were then reversed by incubating samples for ∼16 h at 65°C, and DNA was then purified by using a QIAGEN column. Purified DNA was quantitated by quantitative PCR (qPCR), using SYBR green chemistry and normalized against input chromatin. Primer sequences were as follows: Fwd, 5′-TTTGTTCATAAAATAGTTTTCTGCATAGC-3′; Rev, 5′-AGGTTGAGGTTACATTAACTTTGATCAG.
Endogenous gene expression.
For monitoring endogenous ER-responsive genes, cells were grown in phenol red-free DMEM supplemented with 2.5% sFBS for 24 h. Cells were then transfected as described above with the specified siRNAs and 48 h later were treated with appropriate hormones for 1.5 to 24 h. The effects of SMRT depletion on endogenous gene expression were assessed by reverse transcription and real-time quantitative PCR (RT-qPCR) or Western blot analysis (see above). For mRNA measurements total RNA was isolated by TRIzol (Invitrogen) according to the manufacturer's instructions and analyzed by real-time RT-qPCR using the ABI Prism 7700 sequence analyzer (PE Applied Biosystems, Foster City, CA) in conjunction with TaqMan or SYBR green chemistry. Primer and probe sequences for measurement of pS2 mRNA levels were described previously (11). For BCL-2, 20× primer and probe mix were purchased from Applied Biosystems (Assay on Demand; assay Hs00608023_m1). Primer and probe sequences for PR were designed using Primer Express software (PE Applied Biosystems) (forward, 5′-AGAAATGACTGCATCGTTGATAAAATC-3′; reverse, 5′-GGACCATGCCAGCCTGAC-3′; probe, 5′-6-carboxyfluorescein-CTGCCCAGCATGTCGCCTTAGAAAGTG-3′-6-carboxytetramethylrhodamine). Cyclin D1 was quantitated by SYBR green chemistry using the following primer sequences: forward, 5′-TGGAGGTCTGCGAGGAACAGAA-3′; reverse, 5′-TGCAGGCGGCTCTTTTTCA-3′. Target transcript quantities were normalized against 18S rRNA using a primer/probe set purchased from PE Applied Biosystems. Assays were performed as 25-μl reaction mixtures using TaqMan One-Step RT-PCR Master Mix reagents in MicroAmp 96-well plates (PE Applied Biosystems). Five microliters of MCF-7 total RNA (∼100 ng) was analyzed for pS2 and PR mRNA transcripts. For normalization against the 18S transcript, each sample was diluted 100-fold so that approximately 1 ng of total RNA was analyzed in a separate well. The RT reaction mixture was incubated at 48°C for 30 min to allow cDNA synthesis and terminated by heating for 10 min at 95°C. The reaction was then PCR amplified for 40 cycles consisting of 25 s at 95°C and 1 min at 60°C. Cycle threshold values for each reaction were determined using TaqMan SDS analysis software and standardized against a common total RNA sample obtained from MCF-7 cells grown in the presence of 10% sFBS. For two-step assays, RNA (∼5 μg) was reverse transcribed with SuperScript RNase H− reverse transcriptase (Invitrogen) followed by real-time quantitative PCR with SYBR green as the fluorescent dye. Cycling conditions were 50°C for 2 min and 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min.
RESULTS
Inhibition of SMRT and N-CoR expression by siRNA technology.
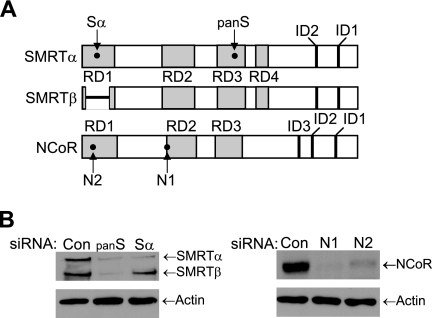
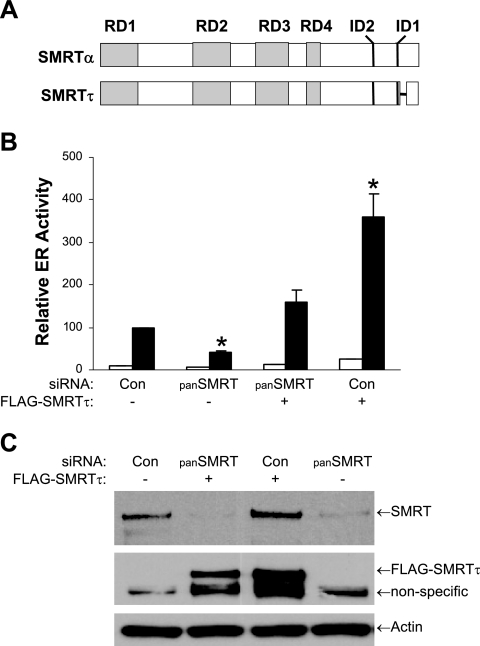
The SMRT and N-CoR corepressors, which possess N-terminal repression domains (RDs) and C-terminal nuclear receptor IDs, are represented schematically in Fig. 1A. Previously, two distinct cDNA clones were identified while screening for the N-terminal region of mouse SMRT that represented products generated via an alternative splicing event (62). Full-length SMRT is encoded by an ∼10-kb mRNA isoform and is denoted SMRTα, whereas the translation of the shorter 8.5-kb message yields SMRTβ. As indicated in the schematic, the smaller SMRTβ harbors a deletion of amino acids 34 to 254 (62), and based on sequence similarity to N-CoR, this deletion removes a majority of RD1, including TBL1/TBLR1 and GPS2 binding regions. Expression of SMRTα and SMRTβ has been demonstrated for several nonhematopoietic cell lines, including HeLa, Cos-1, Jurkat, and T47D cells (17), and Western blot analysis of whole-cell lysates of MCF-7 breast cancer cells revealed relatively equal levels of both SMRT isoforms (Fig. 1B). Western blots indicated the presence of a single N-CoR species. siRNAs are widely used to inhibit the expression of specific mRNAs, and the locations of sequences targeted by the siRNAs utilized in our studies are indicated in Fig. 1A. The panSMRT siRNA inhibited the expression of both SMRTα and SMRTβ, while the Sα siRNA, which recognizes a sequence spliced out of the SMRTβ mRNA, inhibits the expression of only SMRTα (Fig. 1B). Both siRNAs targeting N-CoR (N1 and N2) effectively inhibited the expression of this corepressor. These results demonstrate the ability of these siRNAs to effectively reduce expression of their respective corepressors.
FIG. 1.
Inhibition of SMRT and N-CoR corepressor expression by siRNA. (A) Representation of functional domains of SMRT and N-CoR and the location of target sequences for siRNA specific for SMRTα (Sα), both forms of SMRT (panSMRT or panS), and N-CoR (N1 and N2). The region within RD1 deleted in SMRTβ is represented as a short horizontal line. (B) Western blot analysis of MCF-7 lysates for SMRT (left), N-CoR (right), and actin loading controls demonstrate the ability of the siRNAs (30 pmol/well for SMRT and 40 pmol/well for N-CoR) to reduce expression of their respective corepressors in comparison to the Silencer negative control (Con) siRNA. Note the Sα siRNA does not reduce SMRTβ expression.
Inhibition of SMRT expression decreases ERα-mediated transcriptional activity.
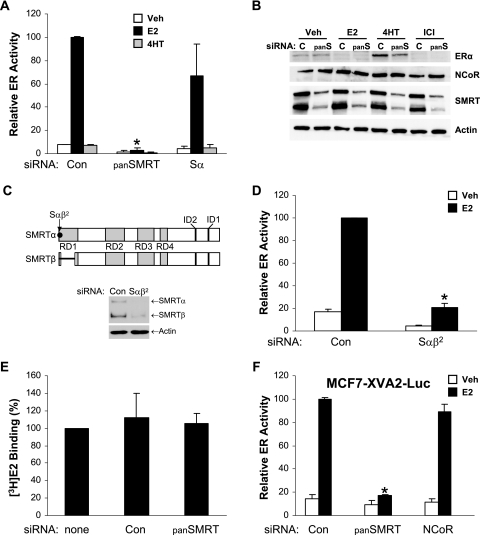
Previous data indicate that the SMRT corepressor interacts with ERα in the presence of tamoxifen as well as in the presence of estrogen or the absence of ligand (56, 71, 76). To assess the contribution of SMRT to ERα transcriptional activity under various hormonal conditions, MCF-7 cells were transfected with siRNAs for SMRTα and SMRTβ (panSMRT), only SMRTα (Sα), or a nonspecific control followed by the ERE-E1b-Luc reporter construct and treatment with vehicle, E2, or 4HT for 24 h. Depletion of both SMRTα and SMRTβ significantly attenuated the ability of E2 to stimulate ERα transcriptional activity, while inhibition of only SMRTα tended to diminish ERα-dependent gene expression (Fig. 2A). The inability of SMRTα depletion to significantly decrease ERα activity suggests that a reduction in total cellular levels of SMRT may be required to alter ERα-dependent gene expression. A change in the relative agonistic properties of 4HT was not observed. Immunoblot analysis of whole-cell lysates prepared from MCF-7 cells transfected in parallel demonstrated that the panSMRT siRNA did not alter basal expression levels of ERα or the ability of ligands to affect ERα protein levels (Fig. 2B). The expected downregulation of ERα was consistently observed in the presence of E2 or ICI, and receptor expression was stabilized in the presence of 4HT regardless of decreased SMRT protein levels. It was also noted that decreasing SMRT expression did not affect N-CoR expression. To rule out the possibility that the impact of the panSMRT siRNA on ERα transcriptional activity was due to an off-target effect, we designed a second siRNA, Sαβ2, which recognizes a sequence in the first coding exon of SMRTα and SMRTβ. This siRNA effectively reduces the expression of both SMRTα and SMRTβ in MCF-7 cells (Fig. 2C) and also was able to effectively inhibit ERα activity measured with an ERE-E1b-Luc reporter gene, confirming that depletion of SMRTα and SMRTβ significantly inhibits ERα function in MCF-7 cells (Fig. 2D).
FIG. 2.
Effect of SMRT depletion on ERα transcriptional activity in MCF-7 cells. (A) MCF-7 cells were transfected with Silencer negative control siRNA (Con) or siRNAs for SMRTα and SMRTβ (panSMRT or panS) or only SMRTα (Sα). Subsequently, cells were transfected with 1 μg ERE-E1b-Luc reporter gene and treated with 0.1% ethanol (Veh), 1 nM E2, or 100 nM 4HT for 24 h. (B) Corresponding Western blot analyses of lysates prepared from MCF-7 cells transfected with siRNAs and treated with ligands as for panel A. (C) Schematic representation of SMRT functional domains and the location of the target sequence for a second siRNA specific for SMRTα and SMRTβ (Sαβ2). Corresponding Western blot analyses are of lysates prepared from MCF-7 cells transfected with siRNAs as for panel D. (D) MCF-7 cells were transfected with Silencer negative control siRNA (Con) or Sαβ2 siRNA followed 24 h later by transfection with 1 μg ERE-E1b-Luc and treatment with 0.1% ethanol (Veh) or 1 nM E2 for 24 h. (E) [3H]estradiol binding capacity assay of MCF-7 cells transfected with no siRNA (none) or Silencer negative control or panSMRT siRNAs. (F) MCF7-XVA2-Luc cells were transfected with Silencer negative control, panSMRT, or the N1 N-CoR siRNAs and 24 h thereafter treated with either vehicle or 1 nM E2 for 24 h. Values for panels A, D, E, and F represent the average ± SEM of three experiments. *, P < 0.05 in comparison to control E2 values.
To determine whether altered SMRT expression could influence the ability of ERα to bind to its cognate ligand, MCF-7 cells were transfected with no, control, or panSMRT siRNAs and 24 h thereafter incubated with [3H]estradiol in the presence and absence of cold competitor to measure ligand binding capacity. The level of specific E2 binding within the cells was not significantly altered under any of the tested conditions, and taking this into account with the comparable expression of ERα demonstrated above, it was concluded that inhibition of SMRT does not impair the E2 binding capacity of ERα (Fig. 2E).
Finally, to assess whether these results reflected an activity of SMRT dependent on a transiently transfected (i.e., nonchromatin) target gene, similar studies were conducted in a cell line derived from MCF-7 cells, MCF7-XVA2-Luc, in which the ERE-MAR-Luc estrogen-responsive reporter gene has been stably integrated (35). The ability of E2 to stimulate expression of the luciferase gene was significantly inhibited in cells transfected with the panSMRT siRNA (Fig. 2F). In contrast, siRNA for N-CoR had no effect on ERα transcriptional activity. A previous report demonstrated that siRNA-mediated depletion of NCoR modestly increased E2-induced ERα activity as measured with transiently transfected reporter genes (35). The lack of a positive response in our experiments may reflect differences between the transient templates utilized in previous experiments (35) and the integrated reporter gene employed in this study. Taken together, these data indicate that the negative effect of SMRT depletion on ERα transcriptional activity is specific for SMRT and that reduced ERα activity is not due either to the disruption of ERα or N-CoR expression or to the ability of the receptor to bind to its ligand.
Estrogen-dependent recruitment of SMRT to an ER target gene promoter.
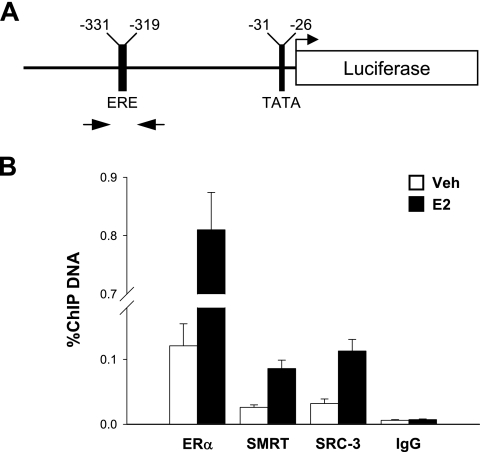
To gain insight into the role of SMRT in ERα transcriptional activity, the interaction of SMRT with an ER-responsive promoter was investigated using the MCF7-XVA2-Luc cells. This model system was used since the results above indicated that estrogen-induced luciferase expression was sensitive to SMRT depletion. Chromatin prepared from vehicle- or E2-treated cells was subjected to immunoprecipitation using specific antibodies directed against ERα, SMRT, and SRC-3 as well as normal rabbit IgG as a negative control. The positions of the primers used to quantitate chromatin are indicated in the schematic of the reporter's promoter region (Fig. 3A). As expected, ERα interaction with the promoter was strongly induced in E2-treated cells (Fig. 3B). Basal and estrogen-induced interaction of SMRT with the promoter region was also observed, and the extent of this interaction was similar to that observed for the SRC-3 coactivator. Moreover, levels of all three targets were clearly distinguishable from the minimal levels of chromatin immunoprecipitated with the normal IgG which were not affected by E2 treatment. The estrogen-dependent recruitment of SMRT to an estrogen-responsive promoter suggests that this coregulator contributes directly to stimulation of gene expression. The presence of SMRT in vehicle-treated cells indicates that this coregulator also may influence basal gene expression. This is consistent with the ability of SMRT depletion to reduce reporter gene expression under basal conditions (e.g., Fig. 2A), as well as our previous demonstration that depleting the expression of a positive-acting factor, the TIF2/SRC-2 coactivator, reduces basal expression of the pS2 gene (21).
FIG. 3.
Estrogen-induced recruitment of ERα, SMRT, and SRC3 to an ER target gene. (A) Schematic of the promoter region of the ERE-MAR-Luc reporter gene integrated in the MCF7-XVA2-Luc cell line. The positions of the primers used in the qPCRs are indicated by arrows. (B) MCF7-XVA2-Luc cells were treated with either vehicle (Veh; 0.1% ethanol) or 10 nM E2 for 45 min and subjected to ChIP assay using antibodies for ERα, SMRT, or SRC3, or rabbit IgG as a negative control. Immunoprecipitated DNA was quantitated by real-time qPCR and is expressed as a percentage of total input DNA. Values represent the average ± SEM of three to four independent experiments.
SMRT influences estrogen and antiestrogen regulation of ERα activity in HeLa cells.
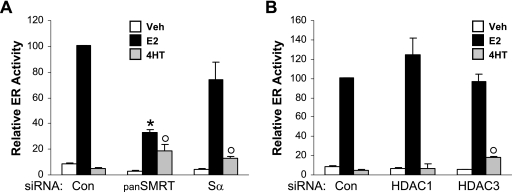
To determine whether SMRT promotion of ERα transcriptional activity was restricted to MCF-7 cells, similar ERα trans-activation assays were performed in an unrelated cell type (Fig. 4A). HeLa cells were first transfected with control, panSMRT, or Sα siRNAs followed by cotransfection with an expression vector for human ERα and the ERE-E1b-Luc reporter gene. Use of the panSMRT siRNA significantly decreased E2-induced ERα transcriptional activity, while SMRT inhibition achieved with panSMRT or Sα siRNAs promoted the agonistic activity of 4HT in HeLa cells, resulting in SERM stimulation of ERα transcriptional activity rather than repression of luciferase gene expression.
FIG. 4.
Effects of SMRT and HDAC depletion on ERα transcriptional activity in HeLa cells. HeLa cells were transfected with Silencer negative control (Con), panSMRT, or Sα siRNAs (A) or siRNA to Silencer negative control (Con), HDAC1, or HDAC3 (B). Subsequently, 10 ng of ERα expression vector and 1 μg of ERE-E1b-Luc reporter were transfected into cells, which were then treated with vehicle (Veh; 0.1% ethanol), 1 nM E2, or 100 nM 4HT for 24 h. Values represent the average ± SEM of three experiments. *, P < 0.05 versus control E2; °, P < 0.05 versus control vehicle.
Previous studies indicated that SMRT/N-CoR can be detected within at least three different transcriptional complexes (27, 45, 58, 82). One complex contains HDAC3; a second, the NURD complex, contains HDAC1; and a third, the Sin3A complex, contains both HDAC1 and Sin3A. Taking advantage of previously published siRNA target sequences specific for HDAC1 (31) and HDAC3 (77), we depleted the expression of these deacetylases to determine their contribution to the regulation of ERα activity. Depletion of neither HDAC1 nor HDAC3 significantly altered the ability of E2 to stimulate ERα transcriptional activity, indicating that the ability of SMRT to stimulate ERα-dependent gene expression is independent of the tested HDACs (Fig. 4B). In contrast, inhibition of HDAC3 expression resulted in a significant increase in the ability of 4HT to stimulate the expression of the reporter gene, suggesting that both SMRT and HDAC3, but not HDAC1, restrain the agonistic potential of 4HT in HeLa cells.
The data in Fig. 2 and 4 reveal that depletion of SMRTα and SMRTβ significantly decreases ERα transcriptional activity in both MCF-7 cells and HeLa cells. To further test whether the effect of the SMRT siRNA on ERα activity was due to SMRT depletion, a rescue experiment was performed in which endogenous SMRT was depleted by the panSMRT siRNA followed by transfection of an siRNA-insensitive SMRT expression vector. The full-length mouse SMRT cDNA (38) was selected for this purpose because it has poor homology with human SMRT in the region targeted by the siRNA and was therefore expected to be insensitive to the panSMRT siRNA. When the sequence of the full-length SMRT cDNA was compared to genomic sequence, it was found to possess a deletion 3′ to the exon encoding ID1. This deletion arises from an alternative splicing event that removes the 3′ portion of exon 44 (exon 44b), which encodes 47 amino acids (24); this form of the corepressor has been termed SMRTτ. Since the cDNA employed in our studies lacks exon 44b, we refer to the form of SMRT utilized in our overexpression experiments as SMRTτ (Fig. 5A).
FIG. 5.
Rescue of endogenous SMRT depletion. (A) Schematic representation of SMRTα and SMRTτ and their functional domains. The 47-amino-acid splice deletion in the C-terminal region of SMRTτ is represented as a short horizontal line. (B) HeLa cells were transfected with Silencer negative control siRNA (Con) or panSMRT siRNA, and 24 h later cells were transfected with 1 μg ERE-E1b-Luc reporter, 2.5 ng pCR3.1-ERα, and either 500 ng of mouse Flag-SMRTτ (+) or its corresponding parent (-) expression vector. Cells were subsequently treated with 0.1% ethanol (Veh) or 1 nM E2 for 24 h. (C) Corresponding Western blot analyses of lysates for endogenous SMRT (top), Flag epitope-tagged SMRTτ (middle), or actin (bottom) prepared from HeLa cells transfected with siRNAs as for panel B. For the Flag antibody blot, a nonspecific band is present in all lanes. Values for panel B represent the average ± SEM of three experiments. *, P < 0.05 in comparison to control E2 values.
HeLa cells were first transfected with control or panSMRT siRNAs followed by cotransfection with expression vectors for FLAG-tagged SMRTτ and ERα and the ERE-E1b-Luc reporter gene. As shown in Fig. 5B, reduction of endogenous SMRT expression reduced E2-induced luciferase gene expression by more than 50%. When exogenous SMRTτ was added back to the panSMRT siRNA-treated cells, ERα-mediated gene expression was restored. Expression of SMRTτ in control siRNA-treated cells resulted in a strong stimulation of luciferase activity, indicating that exogenous expression of SMRTτ could further stimulate ERα activity in cells maintaining endogenous SMRT expression. Immunoblot analysis of whole-cell lysates of parallel cultures with a human-specific SMRT antibody demonstrated effective depletion of endogenous SMRT levels, while Flag M2 antibody revealed the expected overexpression of FLAG epitope-tagged SMRTτ in these cells (Fig. 5C).
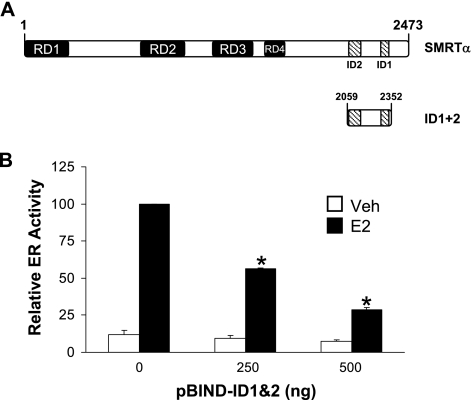
To confirm that SMRT can interact with ERα and contribute to the receptor's transcriptional activity, a plasmid encoding the SMRT receptor interaction domains 1 and 2 (ID1+2) (Fig. 6A) was cotransfected into HeLa cells along with the ERE-E1b-Luc reporter and a human ERα expression vector. As the level of ID1+2 expressed in the cells was increased, the ability of E2 to stimulate reporter gene expression was reduced in a dose-dependent manner (Fig. 6B), indicating that this fragment inhibits E2-stimulated ERα transcriptional activity, possibly by blocking the interaction of endogenous SMRT and other factors, such as coactivators, with ERα. Indeed, a recent report demonstrating the binding of a CoRNR box motif with the coactivator binding groove within the ERα ligand binding domain (73) suggests that the ID1+2 fragment may block coactivator interaction with ERα. This experiment also indicates that a region(s) N-terminal to the ID2 site is required for SMRT to stimulate E2-dependent ERα activity.
FIG. 6.
Effect of expression of the receptor interaction domain of SMRT (ID1+2) on ER transcriptional activity in HeLa cells. (A) Schematic representation of full-length SMRTα and the location of its two nuclear receptor interaction domains (ID1+2). (B) HeLa cells were transfected with increasing amounts of expression vector for the SMRT receptor interaction domain (pBIND-ID1+2) along with 1 μg ERE-E1b-Luc reporter and 10 ng human ERα expression plasmid. The total amount of DNA was kept constant with pBIND parent vector. Twenty-four h after transfection cells were treated with 0.1% ethanol (Veh) or 1 nM E2 for 24 h and subsequently assayed for luciferase activity. Values are normalized to that obtained for the empty vector pBIND in the presence of E2, which was defined as 100. Bars represent the average ± SEM of three experiments. *, P < 0.05 versus control E2.
SMRTτ overexpression increases ERα transcriptional activity in a cell-specific manner.
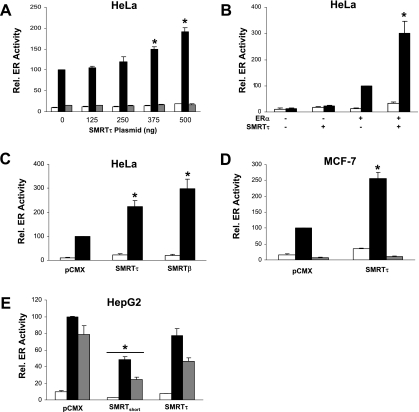
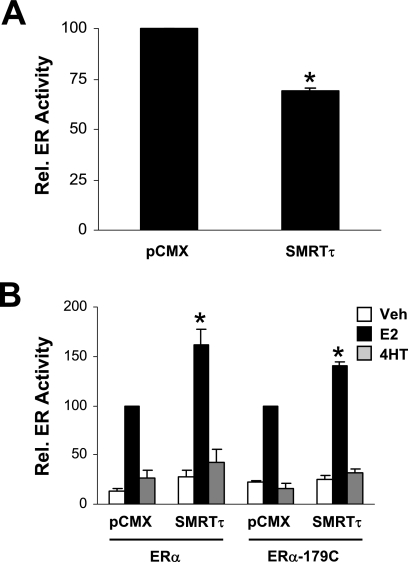
To substantiate the above data, overexpression experiments were conducted in MCF-7, HeLa, and HepG2 cells using the expression construct pCMX-mSMRTτ in the presence of vehicle, E2, or 4HT. HeLa cells were transiently cotransfected with the ERE-E1b-Luc reporter and expression vectors for ERα and increasing amounts of SMRTτ. Figure 7A demonstrates that SMRTτ overexpression significantly increased E2-stimulated reporter gene activity in a dose-dependent manner, while basal and 4HT-stimulated transcriptional activities were unaffected. The effect of SMRTτ overexpression on reporter gene activity was dependent on ERα expression, as no increase in luciferase activity was detected for cells transfected with the control vector lacking the ERα cDNA (Fig. 7B). To test whether the absence of the RD1 region affected SMRT stimulation of ERα transcriptional activity, experiments were conducted with overexpressed SMRTτ or SMRTβ in HeLa cells. Both forms of SMRT stimulated ERα-dependent gene expression (Fig. 7C). Similar stimulation of reporter gene expression was obtained for ERα-positive MCF-7 cells transiently transfected with the ERE-E1b-Luc reporter and SMRTτ expression vector, demonstrating that SMRTτ stimulation of E2-dependent ERα transcriptional activity can be observed in cells with endogenous ERα expression (Fig. 7D). Overexpression of SMRTτ did not alter the ability of 4HT to block ERα-dependent gene expression.
FIG. 7.
Effect of SMRT overexpression on ERα transcriptional activity is cell type dependent. (A) HeLa cells were transfected with expression vectors for ERα (10 ng), SMRTτ (0 to 500 ng), and 1 μg ERE-E1b-Luc. (B) HeLa cells were transfected with expression vectors for ERα and/or SMRTτ (+) or their respective empty control vectors (−) in addition to 1 μg ERE-E1b-Luc. (C) HeLa cells were transfected with 500 ng expression vector for SMRTτ, SMRTβ, or the pCMX control vector, as well as constructs for ERα and the ERE-E1b-Luc reporter gene. (D) MCF-7 cells were transfected with an expression vector for SMRTτ (500 ng) and 1 μg ERE-E1b-Luc. (E) HepG2 cells were transfected with expression vectors for ERα (50 ng) or SMRTshort or SMRTτ (500 ng) and 1 μg of the pC3-Luc reporter gene. The total amount of DNA transfected into each well was balanced with pCMX empty vector. Cells were treated with vehicle (0.1% ethanol, white boxes), 1 nM (HeLa and MCF-7) or 10 nM (HepG2) E2 (black boxes), or 100 nM 4HT (gray boxes) and harvested 24 h thereafter for luciferase measurements. Data represent the average ± SEM of three to four experiments. *, P < 0.05 compared to the corresponding pCMX-alone groups.
To further examine whether the cell environment plays a role in SMRTτ regulation of ERα activity, SMRTτ was overexpressed in HepG2 hepatocarcinoma cells, where both E2 and 4HT function as ERα agonists and ERα transcriptional activity is predominately mediated by the ligand-independent AF1 domain, as opposed to HeLa and MCF-7 cells, which require AF2 function for robust ERα transcriptional activity (52, 74). HepG2 cells were transiently transfected with ERα expression vectors and pC3-Luc reporter along with expression vector for SMRTτ or the first isolated, but incomplete, SMRTshort cDNA which corresponds to amino acids 1032 to 2517 and therefore lacks RD1 and RD2. This form of SMRT is not known to exist in nature. Overexpression of mSMRTτ in HepG2 cells had no effect on basal or ligand-stimulated ERα activity, while overexpression of SMRTshort significantly decreased basal and ligand-stimulated ERα activity (Fig. 7E). The results of the SMRTshort overexpression are similar to a previous report (71) and confirm that this artificial form of SMRT represses ERα transcriptional activity. Taking the results from three cell lines together, our data suggest that cell-dependent factors influence the ability of SMRT to regulate ERα transcriptional activity, possibly reflecting differences between the receptor's requirements for AF1 versus AF2 activity.
SMRT enhancement of nuclear receptor transcriptional activity is ERα specific.
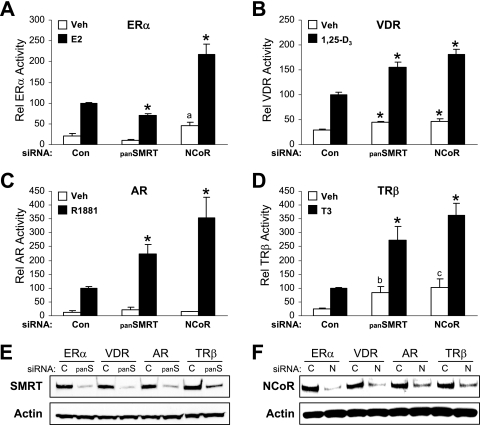
To ascertain whether the ability of SMRT to stimulate nuclear receptor transcriptional activity is specific for ERα or is common to other members of the nuclear receptor superfamily, such as the vitamin D receptor (VDR), AR, or TRβ, HeLa cells, which are deficient for many NRs, were chosen as an amenable model in which to test the specificity of corepressor depletion. Tests were also conducted to determine whether SMRT and its paralog, N-CoR, exerted similar effects on the activities of these receptors. HeLa cells were transfected with siRNA against all forms of SMRT (panSMRT) or N-CoR (N1) and subsequently cotransfected with different NR expression plasmids and their corresponding luciferase reporter genes followed by treatment with appropriate agonistic ligands. As expected, SMRT depletion in ERα/ERE-transfected cells resulted in a significant decrease in E2-induced luciferase activity (Fig. 8A). However, the opposite result was obtained for VDR-, AR-, or TR-transfected and hormone-treated cells (Fig. 8B to D). Silencing of SMRT expression increased luciferase activity, which was consistent with previous studies for AR and VDR (40, 79). This demonstrates that SMRT stimulation of receptor-dependent gene expression is specific for ERα. Furthermore, N-CoR depletion increased the transcriptional activity of all the tested NRs, including ERα, which is consistent with the results of other AR and ERα experiments employing transient reporter genes (35, 79). Similar experiments conducted with the N2 siRNA for ERα activity also increased luciferase expression (data not shown). Western blot analyses revealed that the panSMRT and N1 siRNAs effectively reduced SMRTα and N-CoR protein levels, respectively (Fig. 8E and F), and confirmed that the predominant form of SMRT expression in HeLa cells is SMRTα (17). Taken together, these results support the finding that SMRT depletion and the ensuing impairment of agonist-dependent NR transcriptional activity is ERα specific and that SMRT and N-CoR are not functional homologs in this regard.
FIG. 8.
Effect of SMRT or N-CoR depletion on ERα, VDR, AR, and TRβ transcriptional activity. HeLa cells were transfected with siRNAs for Silencer negative control (Con), SMRTα and SMRTβ (panSMRT), or N-CoR (N1). After 24 h, cells were transfected with expression vectors for ERα (A), VDR (B), AR (C), or TRβ (D) and the respective response element containing luciferase reporter genes and treated with vehicle (0.1% ethanol) or 1 nM of the corresponding hormone for 24 h. Data are the mean ± SEM of three experiments. *, P < 0.05 in comparison to the corresponding vehicle or ligand control; a, P = 0.071; b, P = 0.054; c, P = 0.066. Expression levels of SMRT (E) and N-CoR (F) were monitored by Western blotting for a parallel set of cells transfected with the indicated siRNAs and plasmids.
SMRTτ stimulates the AF2 activity of ERα.
To elucidate whether SMRTτ mediates ERα transcriptional activity via the constitutively active AF1 and/or the ligand-regulatable AF2 regions, functional interaction assays were performed in the HeLa cell line. To first examine the effect of SMRTτ expression on ERα-AF1 activity, HeLa cells were transiently cotransfected with pBIND-ABα, which encodes the GAL4 DNA binding domain fused to the A/B domains of ERα, which encompass the ligand-independent AF1 region, and the pG5-Luc reporter gene with either the SMRTτ expression plasmid or its corresponding parental control. Coexpression of SMRTτ significantly reduced the hormone-independent transcriptional activity of pBIND-ABα (Fig. 9A). Next, SMRT was tested in HeLa cells transiently transfected with an expression construct for full-length human ERα or an ERα construct encoding amino acids 179 to 595 (encompassing the DNA and ligand binding domains and the ligand-dependent AF2 region) and the ERE-E1b-Luc reporter gene. Overexpression of SMRTτ stimulated the E2-dependent activity of the full-length ERα as well as the N-terminally truncated 179C construct (Fig. 9B). Coexpression of SMRTτ had no effect on 4HT-stimulated transcriptional activity. Thus, SMRTτ positively regulates ERα transcriptional activity via the receptor's AF2 domain.
FIG. 9.
SMRTτ stimulates ERα AF2 transcriptional activity. (A) HeLa cells were transfected with 50 ng expression vector for GAL4 DNA binding domain linked to the ERα AB domain and a pG5-Luc reporter gene along with either 500 ng of pCMX empty vector or SMRTτ expression vector. Cells were harvested 48 h after transfection and luciferase activity measured. ABα transcriptional activity in the presence of pCMX was defined as 100. Data represent the averages ± SEM of three experiments. (B) HeLa cells were transfected with 10 ng expression vector for wild-type ERα or an ERα mutant consisting of amino acids 179 to 595 along with 1 μg ERE-E1b-Luc reporter gene and either 500 ng of pCMX empty vector or SMRTτ expression vector. Cells were treated with 0.1% ethanol (Veh), 1 nM E2, or 100 nM 4HT and harvested for luciferase activity measurements 24 h later. The activity for each form of ERα in the presence of pCMX was defined as 100. Data represent the average ± SEM of three experiments. *, P < 0.05 compared to the corresponding pCMX control treatment.
Effects of SMRT depletion on endogenous ERα target gene transcription are gene specific.
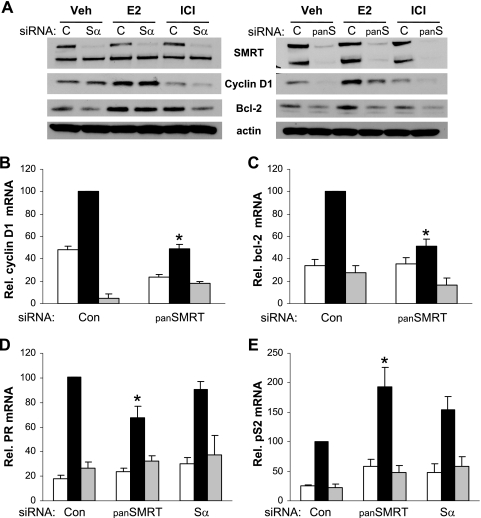
To understand the biological ramifications of the positive role that SMRT plays in promoting estrogen/ERα activities, the expression of endogenous estrogen target genes was assessed after depletion of SMRT protein levels. MCF-7 cells were transfected with Sα or panSMRT siRNAs for 48 h and subsequently treated with vehicle, E2, or ICI 182,780, and the expression levels of the estrogen-inducible genes for BCL-2, cyclin D1, PR, and pS2 were analyzed. As expected, transfection with panSMRT siRNA significantly reduced expression of both SMRT isoforms, whereas Sα only reduced SMRTα protein levels (Fig. 10A). Western blot analysis showed that reducing levels of both SMRT isoforms resulted in decreased cyclin D1 and Bcl-2 protein levels. This reduction was not seen after depletion of only SMRTα. Quantitative RT-PCR revealed that panSMRT siRNA treatment also reduced expression of mRNAs for cyclin D1 and BCL-2, indicating that loss of all SMRT expression compromised the expression of these genes (Fig. 10B and C). A similar pattern was observed for PR mRNA transcripts. Knockdown of both SMRTα and SMRTβ expression reduced PR transcript levels by ∼30%, while inhibition of only SMRTα expression had no effect (Fig. 10D). The ability of SMRT to positively affect estrogen-regulated mRNA expression is not universal, as our data demonstrated that reducing levels of SMRT increases pS2 mRNA levels by up to twofold (Fig. 10E).
FIG. 10.
SMRT regulation of ERα target genes is gene specific. (A) Western blot analyses of SMRT, cyclin D1, and Bcl-2 protein levels from MCF-7 whole-cell lysates. MCF-7 cells were transfected with siRNA against both SMRT isoforms (panS), only SMRTα (Sα), or luciferase (c). After 48 h, cells were treated with 0.1% ethanol (Veh), 1 nM E2, or 100 nM ICI for 3 h for cyclin D1 or 1.5 h for Bcl-2 measurements. Actin was used as a loading control. (B and C) MCF-7 cells were transfected with siRNAs for Silencer control (Con) or SMRTα and SMRTβ (panSMRT) for 48 h and subsequently treated with vehicle (0.1% ethanol; white bars), 1 nM E2 (black bars), or 100 nM ICI (gray bars) for 3 h (cyclin D1) or 24 h (Bcl-2). (D and E) MCF-7 cells were transfected with siRNAs for luciferase (Con), SMRTα and SMRTβ (panSMRT), or only SMRTα (Sα) for 48 h and subsequently treated with vehicle (0.1% ethanol; white bars), 1 nM E2 (black bars), or 100 nM ICI (gray bars) for 24 h (PR) or 8 h (pS2). For panels B to E, RNA was isolated and the indicated mRNAs were quantitated by RT-qPCR and normalized to signals obtained for 18S RNA. Values are the averages of up to eight replicates and are normalized to those obtained for control cells in the presence of E2. *, P < 0.05 in comparison to the corresponding group for the control siRNA.
Inhibition of SMRT expression decreases proliferation of MCF-7 breast cancer cells.
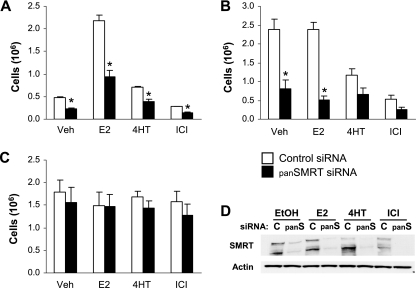
Most data imply a role for SMRT in inhibiting the activity of tamoxifen-bound ERα (33, 43, 70, 71). The growth of MCF-7 breast cancer cells is estrogen and ERα dependent, and cell growth assays were therefore utilized as an efficient method for monitoring cellular responses to SMRT depletion. MCF-7 cells transfected with control siRNA responded appropriately to hormone treatment; cells grown in stripped serum did not proliferate significantly until treated with E2 (Fig. 11A), whereas MCF-7 cells maintained in full serum grew similarly in the presence or absence of E2 (Fig. 11B). Furthermore, these MCF-7 cells were growth inhibited by the antiestrogens 4HT and ICI. To examine whether reducing SMRT expression alters biological responses, panSMRT siRNA targeting both SMRT isoforms was transfected into cells which were subsequently treated with estrogen or antiestrogen. Consistent with the results of the trans-activation assays, depletion of both SMRTα and SMRTβ expression abrogated cell growth in MCF-7 cells grown in stripped serum by 50% in the absence of ligand and by 57% in the presence of agonist. The ability of SMRT knockdown to reduce proliferation of vehicle-treated cells is consistent with SMRT siRNA-mediated reductions in basal ERα activity (Fig. 2). SMRT depletion in MCF-7 cells grown in full serum resulted in growth inhibition of 66% in the absence of ligand and 78% in the presence of E2. Western blot analysis of whole-cell extracts prepared from MCF-7 cells indicated that the siRNA reduced both SMRT isoforms (Fig. 11D). To determine whether the observed reduction in cell number resulting from SMRT silencing would be observed in an ERα-negative cell line, similar cell growth experiments were conducted in ER-negative HeLa cervical carcinoma cells (Fig. 11C). HeLa cells treated with hormone grew similarly to vehicle-treated cells, consistent with the lack of ERα expression. Additionally, transfection with panSMRT siRNA did not alter proliferation of these ERα-negative cells. Growth assays were repeated in both cell lines with siRNA targeting the SMRTα isoform, and similar results were obtained (data not shown). Taken together, these results indicate that the ability of SMRT depletion to inhibit cell proliferation is not a general response of all cells and suggest that the observed growth-inhibitory effects in MCF-7 cells are the result of reduced SMRT stimulation of ERα activity in these cells.
FIG. 11.
Effect of SMRT depletion on proliferation of MCF-7 and HeLa cells. MCF-7 (A and B) and HeLa (C) cells were transfected with siRNA for luciferase (control) or SMRTα and SMRTβ (panSMRT) and treated with 0.1% ethanol (Veh), 1 nM E2, 100 nM 4HT, or 100 nM ICI for 5 days prior to cell number determination by Coulter Counter. Cells were cultured in medium containing 10% sFBS (A), 10% FBS (B), or 5% sFBS (C). Data are expressed as the mean ± SEM for three experiments. *, P < 0.05 for SMRT siRNA-treated cells in comparison to the respective controls. (D) Representative Western blot analysis of SMRT expression for MCF-7 cells treated with control (c) or SMRTα and SMRTβ (panS) siRNAs.
DISCUSSION
SMRT and N-CoR are related corepressors that influence the transcriptional activity of many members of the nuclear receptor superfamily. Current working models suggest that in the presence of a SERM such as 4HT, SMRT and N-CoR and their associated inhibitory molecules are recruited to ERα and block its activation of receptor-dependent gene expression. However, the data presented in this report clearly indicate that SMRT plays a positive role in regulating ERα transcriptional activity. Inhibition of SMRT expression by siRNA significantly abrogated agonist-induced ERα transcriptional activity in HeLa and MCF-7 breast cancer cells. This effect is specific, as evidenced by the inability of N-CoR depletion to repress ERα-dependent gene expression. In addition, our data demonstrate that overexpression of SMRTα potentiates ERα activity in both MCF-7 and HeLa cells, but not HepG2 cells. Taken together with the ability of the receptor interaction domain of SMRT (ID1+2) to act as an inhibitor of receptor-dependent gene expression, these data indicate that SMRT is a cell-type-specific coactivator of ERα, contrary to its assumed role as an obligatory corepressor. We therefore conclude that SMRT is required for full estrogen-dependent ERα transcriptional activity in a cell-specific manner.
The first cDNA published for SMRT (14), which lacked a significant portion of the N terminus of full-length SMRT (∼1,031 amino acids [62]), has been widely used to test the effects of SMRT overexpression on the transcriptional activity of nuclear receptors. Our current results confirm the ability of the truncated SMRT (i.e., SMRTshort) to inhibit ERα transcriptional activity in HepG2 cells (71) and further demonstrate functional differences between SMRT with a complete N terminus (SMRTτ) and SMRTshort in this cell environment. More importantly, SMRTτ stimulation of ERα activity in HeLa and MCF-7 cells reveals that its activation of ERα is cell type specific. The transcriptional activity of ERα in HepG2 cells on the C3 promoter is largely AF1 dependent (74), and the ability of SMRTτ to repress the transcriptional activity of this activation domain is consistent with the lack of SMRTτ stimulation of full-length ERα in these cells. There are no known binding sites for SMRT within the A/B domain of ERα, and it is possible that this inhibition occurs via an indirect mechanism. In contrast, SMRTτ stimulated AF2 activity, and this region is important for ERα-dependent gene expression in HeLa and MCF-7 cells.
MCF-7 cells express both full-length SMRTα as well as the SMRTβ splice variant lacking RD1. In general, depletion of both SMRTα and SMRTβ with the panSMRT siRNA reduced ERα transcriptional activity to a greater extent than depletion of just SMRTα. Although our transient SMRT overexpression experiments suggest that full-length SMRT and SMRTβ do not differ in their ability to stimulate the transcriptional activity of E2-bound ERα, at present we are unable to conclusively discern whether this is the case. The inability of the SMRTα siRNA to significantly reduce ERα activity may reflect the consequences of partial SMRT depletion and suggest that a reduction in total SMRT levels is required for a significant impact on ERα transcriptional activity to be obtained. Efforts to generate a SMRTβ-specific siRNA have not been successful, and we are therefore unable to test the effects of depleting only SMRTβ on ERα-dependent gene expression. It is, however, possible that variable expression of alternatively spliced forms of SMRT may differ significantly in their biological potential, as has been observed for SMRT expression during Xenopus development (51). In a mammalian system, Cote et al. recently showed that overexpression of SMRTβ increased ligand binding to and transcriptional activity of a PML/RARα-I410T mutant while overexpression of SMRTα repressed the activity of this mutant fusion protein (17), and this suggests a functional difference between these two forms of SMRT. Consistent with the fusion protein effects, it was also demonstrated that SMRTβ overexpression increased the transcriptional activity of wild-type RARα in Jurkat cells (17). Ongoing investigations in this laboratory are examining the abilities of SMRTα versus SMRTβ to regulate ERα activity.
The ability to stimulate agonist-dependent transcriptional activity was selective for ERα and was not observed for AR, VDR, or TRβ. Indeed, the increased expression of the reporter genes for these receptors in cells depleted of N-CoR or SMRT by siRNA is consistent with the previously demonstrated abilities of both N-CoR and SMRT to repress their transcriptional activity (40, 77, 79). Depletion of N-CoR stimulated ERα transcriptional activity in HeLa cells, which also highlights the specificity of ERα-SMRT stimulation of agonist-induced gene expression. N-CoR and SMRT are large proteins (>2,400 amino acids) that only share ∼37% sequence identity (13, 62), and several biological systems reveal functional differences between these molecules. For instance, while SMRT and N-CoR bind to a common group of proteins, each corepressor also makes specific protein contacts (e.g., only SMRT interacts with Ku70, while interaction with JMJD2A is unique to N-CoR [80, 81]). Furthermore, SMRT is unable to compensate for loss of N-CoR expression in N-CoR null mice, which are embryonic lethal (34). Thus, regions of sequence divergence likely account for corepressor-specific functions and selective interactions with other cellular factors and are therefore candidates for mediating differences in the abilities of N-CoR and SMRT to regulate agonist-bound ERα activity.
Unexpectedly, depletion of both SMRT isoforms did not increase the agonist potential of 4HT on ERα activity measured in a trans-activation assay in MCF-7 breast cancer cells, and this implies that endogenous SMRT is not a significant contributor to tamoxifen's antagonist activity, as measured with this reporter, in this cell environment. However, knockdown of SMRT expression in HeLa cells enhanced the ERα agonist activity of 4HT, and this indicates that endogenous SMRT contributes to the antagonistic biocharacter of 4HT in this cell type. Increased 4HT agonist activity in cells depleted of HDAC3 implicates a SMRT-HDAC3 complex as a mediator of 4HT's antagonist activity in HeLa cells. This is consistent with the recruitment of HDAC3 to the promoters of genes inhibited by this antiestrogen (3, 48). A number of other corepressors are potential repressors of 4HT-ERα activity, including N-CoR and REA (18), and it is possible that one or more of these molecules maintains the antagonist activity of 4HT in HeLa cells. In antibody microinjection experiments performed in Rat-1 cells, inhibiting SMRT or N-CoR revealed 4HT agonist activity measured on an ERE-lacZ reporter (43). Other investigators also have reported that SMRT depletion by siRNA enhanced 4HT stimulation of the expression of the XBP-1 gene, but not three other tested genes (39). These findings, taken together with our data, lead us to conclude that the ability of endogenous SMRT to repress 4HT-bound ERα activity is cell type and gene specific.
The effect of SMRT depletion on the regulation of endogenous ERα-responsive genes was examined in MCF-7 cells. Similar to the trans-activation experiments, reduction of both SMRT isoforms had a greater impact on expression of endogenous ERα target genes than depletion of only SMRTα. Moreover, not all ERα target genes were equally affected by SMRT depletion, suggesting that SMRT differentially regulates ERα target genes. Expression of the pS2 gene was enhanced in SMRT-depleted cells, indicating that this molecule inhibits expression of this gene. Chromatin immunoprecipitation assays have indicated that low levels of SMRT are present on the promoter of the pS2 gene under basal conditions (54, 70), and it is possible that loss of SMRT binding to basal promoters increases pS2 expression following E2 stimulation. In contrast, siRNA-mediated depletion of SMRT inhibited the expression of Bcl-2, PR, and cyclin D1. We are not, however, aware of any studies that have examined the interaction of SMRT with these genes. Estrogen induction of cyclin D1 has been assessed previously in MCF-7 cells transfected with a different siRNA for SMRT, and while there was some decrease in cyclin D1 mRNA levels, this was not thought to be significant (39). In these experiments, SMRTβ levels were not tested, and our own data clearly demonstrate that changes in gene expression are more pronounced in cells depleted of SMRTα and SMRTβ. The differential effects of SMRT depletion on gene expression suggest that differences relating to the structure of the EREs as well as combinatorial transcription factor networks between ERα, SMRT, and other transcription factors and/or coregulators may affect the recruitment and/or activity of SMRT and enable it to exert gene-specific effects.
We also utilized cell growth assays to monitor the effects of SMRT on an estradiol-stimulated cellular response. Depletion of SMRT expression inhibited the estrogen-dependent growth of ERα-positive MCF-7 cells while the growth of ERα-negative HeLa cells was not altered regardless of ligand exposure, which indicates that SMRT silencing does not have a general growth-inhibitory effect. The MCF-7 results are consistent with SMRT's role in stimulating ERα activity and expression of cyclin D1, a protein required for breast cancer cell proliferation (64). In a previous MCF-7 cell study, simultaneous siRNA-mediated knockdown of both N-CoR and SMRT did not affect the ability of E2 to promote cell cycle entry (39). It is possible that those results differ from our SMRT depletion studies because the positive effects of N-CoR loss are balanced by the negative effects of SMRT depletion.
Depletion of SMRT also reduced cellular proliferation in the absence of ligand or presence of ERα antagonist, indicating that SMRT can contribute to basal cell growth and is not required for the antiproliferative activity of tamoxifen in MCF-7 cells. The former is likely related to SMRT depletion effects on basal ERα activity, as suggested by our reporter gene assay results; however, the possibility of additional SMRT depletion effects on other cell cycle regulators cannot be ruled out. Earlier studies have explored the role of N-CoR and SMRT in tamoxifen-stimulated cell cycle entry and proliferation, with conflicting results (22, 39, 56). While one study found that overexpressing a dominant negative form of N-CoR (amino acids 1586 to 2211) did not lead to tamoxifen-stimulated cell cycle entry or increased proliferation (56), another report demonstrated that expression of a different but overlapping C-terminal region of N-CoR (amino acids 1944 to 2453) blocked the inhibitory effect of 4HT on cell growth (22). The basis for this discrepancy is unknown, but differences in the region of N-CoR or culture conditions could be contributing factors. It also is possible that the two different N-CoR fragments were expressed at different levels and/or their interaction with ERα differentially affected ERα binding to other coregulators required for transcription. Another study reported that individual siRNA silencing of SMRT in MCF-7 cells did not lead to tamoxifen-stimulated cell cycle entry measured as the percent change in cells in S/G2/M, a finding consistent with our data, but demonstrated that simultaneous silencing of both SMRT and N-CoR led to a significant increase in cell cycle entry (39). This increase for cells lacking both N-CoR and SMRT is intriguing, and further studies will be required to determine whether corepressor depletion effects on tamoxifen-induced cell cycle entry reflect SMRT and N-CoR actions at common or unique gene targets that coordinately affect proliferation.
The molecular mechanisms that enable SMRT to stimulate the transcriptional activity of ERα, and not the activity of the other nuclear receptors tested, and the basis for its gene-selective stimulatory activity are not known but could be mediated at the level of receptor, coactivators, or chromatin. Indeed, the ability of SMRT to stimulate the expression of a gene via unliganded TRα and a negative thyroid hormone response element (5) suggests that SMRT regulation of gene expression is highly context dependent. It is possible that SMRT plays a role in activation of gene expression prior to E2 stimulation, perhaps through modulation of chromatin structure (e.g., a priming effect), and there is some evidence that SMRT binds to target gene promoters under basal conditions and is released following E2 treatment (54, 69, 70). However, our ChIP data suggest an alternative possibility in which SMRT's stimulatory effects occur in concert with its recruitment to target genes, possibly as a component of a molecular exchange protein complex or through association with other factors that positively affect transcription. Perissi et al. have demonstrated that TBL1 and TBLR1 facilitate exchange of corepressor molecules for coactivators on a variety of gene promoters and contribute to activation of ERα-dependent gene expression (63). However, TBL1 and TBLR1 bind to SMRT and N-CoR (26, 45, 82), and since the latter does not stimulate ERα transcriptional activity, additional mechanisms would be required to distinguish the positive effects of SMRT from the negative role of N-CoR in ERα-dependent gene expression. Interactions between SMRT and coactivators, such as SRC-3 and NCOA6, have also been demonstrated (37, 46), and these protein complexes may work in concert to efficiently activate transcription. Our data demonstrate that estrogen-induced ERα activity does not require HDACs 1 or 3 and that SMRT stimulation of ERα is cell and gene specific and therefore suggest that a milieu of cellular factors other than these two deacetylases acts in a selective manner with SMRT to promote ERα regulation of transcription. We are currently exploring the role of SMRT in regulating receptor and coactivator association with ERα target genes. It should be noted that N-CoR also has been shown to stimulate RAR activity on a DR +1 element (34), which implies there is a larger role for corepressor activation of gene expression than previously thought.
Our results show that ERα transcriptional activity and modulation of estrogen-responsive target genes are positively influenced by the expression of SMRT and suggest that the role of SMRT in regulating ERα activity and cell proliferation in vivo should be carefully considered, for instance, in ERα-positive breast tumors. In addition, these data demonstrate that SMRT's effects on ERα do not necessarily depend on pharmacological antiestrogens, and this substantially extends the repertoire of SMRT-ERα-regulated biology. Finally, our results suggest that the role of SMRT in mediating tamoxifen antagonist activity in vivo should be considered on a tissue-by-tissue basis and that cell- and gene-selective SMRT positive or negative regulation of ERα transcriptional activity may be one of the mechanisms by which tissue-specific effects of antiestrogens are obtained.
Acknowledgments
We thank Zeming Jin for constructing the ID1+2 expression vector used in this study and David Lonard for designing the primers for PR qPCR. The assistance of Qiang He for the relative binding affinity assays and the technical support of Judy Roscoe and Cheryl Parker for cell culture are gratefully acknowledged.
T.J. was supported by NIH training grant HD07165. This work was supported by Public Health Service grant DK53002 to C.L.S.
Footnotes
Published ahead of print on 25 June 2007.
REFERENCES
- 1.Agoulnik, I. U., W. C. Krause, W. E. I. Bingman, H. T. Rahman, M. Amrikachi, G. E. Ayala, and N. L. Weigel. 2003. Repressors of androgen and progesterone receptor action. J. Biol. Chem. 278:31136-31148. [DOI] [PubMed] [Google Scholar]
- 2.Augereau, P., F. Miralles, V. Cavailles, C. Gaudelet, M. Parker, and H. Rochefort. 1994. Characterization of the proximal estrogen-responsive element of human cathepsin D gene. Mol. Endocrinol. 8:693-703. [DOI] [PubMed] [Google Scholar]
- 3.Baek, S. H., K. A. Ohgi, D. W. Rose, E. H. Koo, C. K. Glass, and M. G. Rosenfeld. 2002. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell 110:55-67. [DOI] [PubMed] [Google Scholar]
- 4.Baniahmad, A., I. Ha, D. Reinberg, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1993. Interaction of human thyroid hormone receptor beta with transcription factors TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc. Natl. Acad. Sci. USA 90:8832-8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berghagen, H., E. Ragnhildstveit, K. Krogsrud, G. Thuestad, J. Apriletti, and F. Saatcioglu. 2002. Corepressor SMRT functions as a coactivator for thyroid hormone receptor T3Rα from a negative hormone response element. J. Biol. Chem. 277:49517-49522. [DOI] [PubMed] [Google Scholar]
- 6.Berrevoets, C. A., A. Umar, J. Trapman, and A. O. Brinkmann. 2004. Differential modulation of androgen receptor transcriptional activity by the nuclear receptor co-repressor (N-CoR). Biochem. J. 379:731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourdeau, V., J. Deschênes, R. Métivier, Y. Nagai, D. Nguyen, N. Bretschneider, F. Gannon, J. H. White, and S. Mader. 2004. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol. Endocrinol. 18:1411-1427. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33-43. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, J. S., C. A. Meyer, J. Song, W. Li, T. R. Geistlinger, J. Eeckhoute, A. S. Brodsky, E. K. Keeton, K. C. Fertuck, G. F. Hall, Q. Wang, S. Bekiranov, V. Sementchenko, E. A. Fox, P. A. Silver, T. R. Gingeras, X. S. Liu, and M. Brown. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38:1289-1297. [DOI] [PubMed] [Google Scholar]
- 10.Castro-Rivera, E., I. Samudio, and S. Safe. 2001. Estrogen regulation of cyclin D1 gene expression in ZR-75 breast cancer cells involves multiple enhancer elements. J. Biol. Chem. 276:30853-30861. [DOI] [PubMed] [Google Scholar]
- 11.Cavarretta, I. T. R., R. Mukopadhyay, D. M. Lonard, L. M. Cowsert, C. F. Bennett, B. W. O'Malley, and C. L. Smith. 2002. Reduction of coactivator expression by antisense oligodeoxynucleotides inhibits ERα transcriptional activity and MCF-7 proliferation. Mol. Endocrinol. 16:253-270. [DOI] [PubMed] [Google Scholar]
- 12.Chen, C. D., D. S. Welsbie, C. Tran, S. H. Baek, R. Chen, R. Vessella, M. G. Rosenfeld, and C. L. Sawyers. 2004. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 10:33-39. [DOI] [PubMed] [Google Scholar]
- 13.Chen, D., K. Umesono, and R. M. Evans. 1996. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc. Natl. Acad. Sci. USA 93:7567-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, J. D., and R. M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454-457. [DOI] [PubMed] [Google Scholar]
- 15.Cheng, S., S. Brzostek, S. R. Lee, A. N. Hollenberg, and S. P. Balk. 2002. Inhibition of the dihydrotestosterone-activated androgen receptor by nuclear receptor corepressor. Mol. Endocrinol. 16:1492-1501. [DOI] [PubMed] [Google Scholar]
- 16.Coleman, K. M., M. Dutertre, A. El-Gharbawy, B. G. Rowan, N. L. Weigel, and C. L. Smith. 2003. Mechanistic differences in the activation of estrogen receptor-α (ERα)- and ER-β-dependent gene expression by cAMP signaling pathway(s). J. Biol. Chem. 278:12834-12845. [DOI] [PubMed] [Google Scholar]
- 17.Cote, S., S. McNamara, D. Brambilla, A. Bianchini, G. Rizzo, S. Victoria del Rincon, F. Grignani, C. Nervi, and W. H. Miller. 2004. Expression of SMRTβ promotes ligand-induced activation of mutated and wild-type retinoid receptors. Blood 104:4226-4335. [DOI] [PubMed] [Google Scholar]
- 18.Dobrzycka, K. M., S. M. Townson, S. Jiang, and S. Oesterreich. 2003. Estrogen receptor corepressors—a role in human breast cancer? Endocr. Relat. Cancer 10:517-536. [DOI] [PubMed] [Google Scholar]
- 19.Dotzlaw, H., M. Papaioannou, U. Moehren, F. Claessens, and A. Baniahmad. 2003. Agonist-antagonist coactivator and corepressor interplay on the human androgen receptor. Mol. Cell. Endocrinol. 213:79-85. [DOI] [PubMed] [Google Scholar]
- 20.Dubik, D., and R. P. Shiu. 1992. Mechanism of estrogen activation of c-myc oncogene expression. Oncogene 7:1587-1594. [PubMed] [Google Scholar]
- 21.Dutertre, M., and C. L. Smith. 2003. Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen receptor-α: regulation by phosphorylation sites in the A/B region depends on other receptor domains. Mol. Endocrinol. 17:1296-1314. [DOI] [PubMed] [Google Scholar]
- 22.Fujita, T., Y. Kobayashi, O. Wada, Y. Tateishi, L. Kitada, Y. Yamamoto, H. Takashima, A. Murayama, T. Yano, T. Baba, S. Kato, Y. Kawabe, and J. Yanagisawa. 2003. Full activation of estrogen receptor alpha activation function-1 induces proliferation of breast cancer cells. J. Biol. Chem. 278:26704-26714. [DOI] [PubMed] [Google Scholar]
- 23.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 24.Goodson, M. L., B. A. Jonas, and M. L. Privalsky. 2005. Alternative mRNA splicing of SMRT creates functional diversity by generating corepressor isoforms with different affinities for different nuclear receptors. J. Biol. Chem. 280:7493-7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guenther, M. G., O. Barak, and M. A. Lazar. 2001. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21:6091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 27.Heinzel, T., R. M. Lavinsky, T.-M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W.-M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 28.Hodgson, M. C., I. Astapova, S. Cheng, L. J. Lee, M. C. Verhoeven, E. Choi, S. P. Balk, and A. N. Hollenberg. 2005. The androgen receptor recruits nuclear receptor corepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J. Biol. Chem. 280:6511-6519. [DOI] [PubMed] [Google Scholar]
- 29.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kanei, M. Soderstrom, and C. K. Glass. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 30.Huang, H.-J., J. D. Norris, and D. P. McDonnell. 2002. Identification of a negative regulatory surface within estrogen receptor a provides evidence in support of a role for corepressors in regulating cellular responses to agonists and antagonists. Mol. Endocrinol. 16:1778-1792. [DOI] [PubMed] [Google Scholar]
- 31.Ishizuka, T., and M. A. Lazar. 2003. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol. Cell. Biol. 23:5122-5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaber, B. M., T. Gao, L. Huang, S. Karmakar, and C. L. Smith. 2006. The pure estrogen receptor antagonist ICI 182,780 promotes a novel interaction of estrogen receptor-α with the 3′,5′-cyclic adenosine monophosphate response element-binding protein-binding protein/p300 coactivators. Mol. Endocrinol. 20:2695-2710. [DOI] [PubMed] [Google Scholar]
- 33.Jackson, T. A., J. K. Richer, D. L. Bain, G. S. Takimoto, L. Tung, and K. B. Horwitz. 1997. The partial agonist activity of antagonist-occupied steroid receptors is controlled by a novel hinge domain-binding coactivator L7/SPA and the corepressors N-CoR or SMRT. Mol. Endocrinol. 11:693-705. [DOI] [PubMed] [Google Scholar]
- 34.Jepsen, K., O. Hermanson, T. M. Onami, A. S. Gleiberman, V. Lunyak, R. J. McEvilly, R. Kurokawa, V. Kumar, F. Liu, E. Seto, S. M. Hedrick, G. Mandel, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102:753-763. [DOI] [PubMed] [Google Scholar]
- 35.Jiang, S., R. Meyer, K. Kang, C. K. Osborne, J. Wong, and S. Oesterreich. 2006. Scaffold attachment factor SAFB1 suppresses estrogen receptor a-mediated transcription in part via interaction with nuclear receptor corepressor. Mol. Endocrinol. 20:311-320. [DOI] [PubMed] [Google Scholar]
- 36.Jung, D.-J., S.-K. Lee, and J. W. Lee. 2001. Agonist-dependent repression mediated by mutant estrogen receptor a that lacks the activation function 2 core domain. J. Biol. Chem. 276:37280-37283. [DOI] [PubMed] [Google Scholar]
- 37.Jung, S. Y., H. Luo, A. Malovannaya, J. Zhang, J. Qin, and M. A. Lazar. 2005. Characterization of nuclear receptor corepressor complexes. NURSA datasets. [Online.] http://www.nursa.org/10.1621/datasets.01002.
- 38.Kao, H. Y., M. Downes, P. Ordentlich, and R. M. Evans. 2000. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 14:55-66. [PMC free article] [PubMed] [Google Scholar]
- 39.Keeton, E. K., and M. Brown. 2005. Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-α and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT. Mol. Endocrinol. 19:1543-1554. [DOI] [PubMed] [Google Scholar]
- 40.Khanim, F. L., L. M. Gommersall, V. H. J. Wood, K. L. Smith, L. Montalvo, L. P. O'Neill, Y. Xu, D. M. Peehl, P. M. Stewart, B. M. Turner, and M. J. Campbell. 2004. Altered SMRT levels disrupt vitamin D3 receptor signalling in prostate cancer cells. Oncogene 23:6712-6725. [DOI] [PubMed] [Google Scholar]
- 41.Kushner, P. J., D. A. Agard, G. L. Greene, T. S. Scanlan, A. K. Shiau, R. M. Uht, and P. Webb. 2000. Estrogen receptor pathways to AP-1. J Steroid Biochem. Mol. Biol. 74:311-317. [DOI] [PubMed] [Google Scholar]
- 42.Laganière, J., G. Deblois, C. Lefebvre, A. R. Bataille, F. Robert, and V. Giguère. 2005. Location analysis of estrogen receptor a target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. USA 102:11651-11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavinsky, R. M., K. Jepsen, T. Heinzel, J. Torchia, T.-M. Mullen, R. Schiff, A. L. Del-Rio, M. Ricote, S. Ngo, J. Gemsch, S. G. Hilsenbeck, C. K. Osborne, C. K. Glass, M. G. Rosenfeld, and D. W. Rose. 1998. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc. Natl. Acad. Sci. USA 95:2920-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levin, E. R. 2005. Integration of the extranuclear and nuclear actions of estrogen. Mol. Endocrinol. 19:1951-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, J., J. Wang, J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, X., E. A. Kimbrel, D. J. Kenan, and D. P. McDonnell. 2002. Direct interactions between corepressors and coactivators permit the integration of nuclear receptor-mediated repression and activation. Mol. Endocrinol. 16:1482-1491. [DOI] [PubMed] [Google Scholar]
- 47.Liao, G., L.-Y. Chen, A. Zhang, A. Godavarthy, F. Xia, J. C. Ghosh, H. Li, and J. D. Chen. 2003. Regulation of androgen receptor activity by the nuclear receptor corepressor SMRT. J. Biol. Chem. 278:5052-5061. [DOI] [PubMed] [Google Scholar]
- 48.Liu, X. F., and M. K. Bagchi. 2004. Recruitment of distinct chromatin-modifying complexes by tamoxifen-complexed estrogen receptor at natural target gene promoters in vivo. J. Biol. Chem. 279:15050-15058. [DOI] [PubMed] [Google Scholar]
- 49.Lonard, D. M., and B. W. O'Malley. 2006. The expanding cosmos of nuclear receptor coactivators. Cell 125:411-414. [DOI] [PubMed] [Google Scholar]
- 50.Loven, M. A., V. S. Likhite, I. Choi, and A. M. Nardulli. 2001. Estrogen response elements alter coactivator recruitment through allosteric modulation of estrogen receptor β conformation. J. Biol. Chem. 276:45282-45288. [DOI] [PubMed] [Google Scholar]
- 51.Malartre, M., S. Short, and C. Sharpe. 2006. Xenopus embryos lacking specific isoforms of the corepressor SMRT develop abnormal heads. Dev. Biol. 292:333-343. [DOI] [PubMed] [Google Scholar]
- 52.McDonnell, D. P., D. L. Clemm, T. Hermann, M. E. Goldman, and J. W. Pike. 1995. Analysis of estrogen receptor function in vitro reveals three distinct classes of antiestrogens. Mol. Endocrinol. 9:659-669. [DOI] [PubMed] [Google Scholar]
- 53.McDonnell, D. P., R. A. Scott, S. A. Kerner, B. W. O'Malley, and J. W. Pike. 1989. Functional domains of the human vitamin D3 receptor regulate osteocalcin gene expression. Mol. Endocrinol. 3:635-644. [DOI] [PubMed] [Google Scholar]
- 54.Métivier, R., G. Penot, R. P. Carmouche, M. R. Hübner, G. Reid, S. Denger, D. Manu, H. Brand, M. Kos, V. Benes, and F. Gannon. 2004. Transcriptional complexes engaged by apo-estrogen receptor-α isoforms have divergent outcomes. EMBO J. 23:3653-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Métivier, R., A. Stark, G. Flouriot, M. R. Hübner, H. Brand, G. Penot, D. Manu, S. Denger, G. Reid, M. Kos, R. B. Russell, O. Kah, F. Pakdel, and F. Gannon. 2002. A dynamic structural model for estrogen receptor-α activation by ligands, emphasizing the role of interactions between distant A and E domains. Mol. Cell 10:1019-1032. [DOI] [PubMed] [Google Scholar]
- 56.Morrison, A. J., R. E. Herrick, E. C. Heinsohn, R. Schiff, and C. K. Osborne. 2003. Dominant-negative nuclear receptor corepressor relieves transcriptional inhibition of retinoic acid receptor but does not alter the agonist/antagonist activities of the tamoxifen-bound estrogen receptor. Mol. Endocrinol. 17:1543-1554. [DOI] [PubMed] [Google Scholar]
- 57.Nagy, L., H.-Y. Kao, J. D. Love, C. Li, E. Banayo, J. T. Gooch, V. Krishna, K. Chatterjee, R. M. Evans, and J. W. R. Schwabe. 1999. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 13:3209-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 59.Nawaz, Z., D. M. Lonard, C. L. Smith, E. Lev-Lehman, S. Y. Tsai, M.-J. Tsai, and B. W. O'Malley. 1999. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol. Cell. Biol. 19:1182-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nettles, K. W., and G. L. Greene. 2005. Ligand control of coregulator recruitment to nuclear receptors. Annu. Rev. Physiol. 67:309-333. [DOI] [PubMed] [Google Scholar]
- 61.Nilsson, S., S. Mäkelä, E. Treuter, M. Tujague, J. Thomsen, G. Andersson, E. Enmark, K. Pettersson, M. Warner, and J.-Å. Gustafsson. 2001. Mechanisms of estrogen action. Physiol. Rev. 81:1535-1565. [DOI] [PubMed] [Google Scholar]
- 62.Ordentlich, P., M. Downes, W. Xie, A. Genin, N. B. Spinner, and R. M. Evans. 1999. Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc. Natl. Acad. Sci. USA 96:2639-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perissi, V., A. Aggarwal, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2004. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116:511-526. [DOI] [PubMed] [Google Scholar]
- 64.Prall, O. W., E. M. Rogan, E. A. Musgrove, C. K. Watts, and R. L. Sutherland. 1998. c-Myc or cyclin D1 mimics estrogen effects on cyclin E-Cdk2 activation and cell cycle reentry. Mol. Cell. Biol. 18:4499-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Privalsky, M. L. 2004. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu. Rev. Physiol. 66:315-360. [DOI] [PubMed] [Google Scholar]
- 66.Rio, M. C., and P. Chambon. 1990. The pS2 gene, mRNA, and protein: a potential marker for human breast cancer. Cancer Cells 2:269-274. [PubMed] [Google Scholar]
- 67.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 20:1405-1428. [DOI] [PubMed] [Google Scholar]
- 68.Safe, S. 2001. Transcriptional activation of genes by 17β-estradiol through estrogen receptor-Sp1 interactions. Vit. Horm. 62:231-252. [DOI] [PubMed] [Google Scholar]
- 69.Shang, Y., and M. Brown. 2002. Molecular determinants for the tissue specificity of SERMs. Science 295:2465-2468. [DOI] [PubMed] [Google Scholar]
- 70.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 71.Smith, C. L., Z. Nawaz, and B. W. O'Malley. 1997. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol. Endocrinol. 11:657-666. [DOI] [PubMed] [Google Scholar]
- 72.Smith, C. L., and B. W. O'Malley. 2004. Coregulator funtion: a key to understanding tissue specificity of selective receptor modulators. Endocr. Rev. 25:45-71. [DOI] [PubMed] [Google Scholar]
- 73.Soller, M. J. 2006. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer 45:717-719. [DOI] [PubMed] [Google Scholar]
- 74.Tzukerman, M. T., A. Esty, D. Santiso-Mere, P. Danielian, M. G. Parker, R. B. Stein, J. W. Pike, and D. P. McDonnell. 1994. Human estrogen receptor transactivational capacity is determined by both cellular and promoter context and mediated by two functionally distinct intramolecular regions. Mol. Endocrinol. 8:21-30. [DOI] [PubMed] [Google Scholar]
- 75.Walters, M. R., M. Dutertre, and C. L. Smith. 2002. SKF-82958 is a subtype-selective estrogen receptor-α (ERa) agonist that induces functional interactions between ERα and AP-1. J. Biol. Chem. 277:1669-1679. [DOI] [PubMed] [Google Scholar]
- 76.Webb, P., P. Nguyen, and P. J. Kushner. 2003. Differential SERM effects on corepressor binding dictate ERα activity in vivo. J. Biol. Chem. 278:6912-6920. [DOI] [PubMed] [Google Scholar]
- 77.Yoon, H.-G., D. W. Chan, Z. Q. Huang, J. Li, J. D. Fondell, J. Qin, and J. Wong. 2003. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 22:1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoon, H.-G., Y. Choi, P. A. Cole, and J. Wong. 2005. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 25:324-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoon, H.-G., and J. Wong. 2006. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol. Endocrinol. 20:1048-1060. [DOI] [PubMed] [Google Scholar]
- 80.Yu, J., C. Palmer, T. Alenghat, Y. Li, G. Kao, and M. A. Lazar. 2006. The corepressor silencing mediator for retinoid and thyroid hormone receptor facilitates cellular recovery from DNA double-strand breaks. Cancer Res. 66:9316-9322. [DOI] [PubMed] [Google Scholar]
- 81.Zhang, D., H.-G. Yoon, and J. Wong. 2005. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2). Mol. Cell. Biol. 25:6404-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, J., M. Kalkum, B. T. Chait, and R. G. Roeder. 2002. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell 9:611-623. [DOI] [PubMed] [Google Scholar]
- 83.Zhang, X., M. Jeyakumar, S. Petukhov, and M. K. Bagchi. 1998. A nuclear receptor corepressor modulates transcriptional activity of antagonist-occupied steroid hormone receptor. Mol. Endocrinol. 12:513-524. [DOI] [PubMed] [Google Scholar]
- 84.Zou, A., M. G. Elgort, and E. A. Allegretto. 1997. Retinoid X receptor (RXR) ligands activate the human 25-hydroxyvitamin D3-24-hydroxylase promoter via RXR heterodimer binding to two vitamin D-responsive elements and elicit additive effects with 1,25-dihydroxyvitamin D3. J. Biol. Chem. 272:19027-19034. [DOI] [PubMed] [Google Scholar]