Abstract
We identified a mutation in the 60S ribosomal protein L33A (rpl33a-G76R) that elicits derepression of GCN4 translation (Gcd− phenotype) by allowing scanning preinitiation complexes to bypass inhibitory upstream open reading frame 4 (uORF4) independently of prior uORF1 translation and reinitiation. At 37°C, rpl33a-G76R confers defects in 60S biogenesis comparable to those produced by the deletion of RPL33A (ΔA). At 28°C, however, the 60S biogenesis defect is less severe in rpl33a-G76R than in ΔA cells, yet rpl33a-G76R confers greater derepression of GCN4 and a larger reduction in general translation. Hence, it appears that rpl33a-G76R has a stronger effect on ribosomal-subunit joining than does a comparable reduction of wild-type 60S levels conferred by ΔA. We suggest that rpl33a-G76R alters the 60S subunit in a way that impedes ribosomal-subunit joining and thereby allows 48S rRNA complexes to abort initiation at uORF4, resume scanning, and initiate downstream at GCN4. Because overexpressing tRNAiMet suppresses the Gcd− phenotype of rpl33a-G76R cells, dissociation of tRNAiMet from the 40S subunit may be responsible for abortive initiation at uORF4 in this mutant. We further demonstrate that rpl33a-G76R impairs the efficient processing of 35S and 27S pre-rRNAs and reduces the accumulation of all four mature rRNAs, indicating an important role for L33 in the biogenesis of both ribosomal subunits.
Cell growth and division are highly interconnected processes that require protein synthesis, which in turns requires the biogenesis of ribosomes, soluble translation factors, and charged tRNA species. In Saccharomyces cerevisiae, ribosome biogenesis consumes a great amount of energy and is tightly regulated according to nutrient availability and to signals depending on other macromolecular pathways (77). The production of 60S and 40S ribosomal subunits is a highly dynamic process that begins with the transcription of rRNA by RNA polymerase I (35S rRNA precursor to 25S, 18S, and 5.8S rRNAs) and RNA polymerase III (5S rRNA) in the nucleolus and ends with export of preribosomal 60S and 40S subunits to the cytoplasm, where final steps of maturation occur. The maturation pathway of the 35S pre-rRNA to yield 25S, 18S, and 5.8S rRNAs (see Fig. 4A) is closely coordinated with the assembly of 79 ribosomal proteins (r-proteins) and more than 150 trans-acting factors involved in the synthesis, maturation, and transport of the ribosomal subunits (reviewed in references 11, 41, 53, and 73). The association of 35S pre-rRNA with the U3-snoRNP complex, transiently associated trans-acting factors, and several r-proteins, mostly belonging to the 40S subunit, leads to the formation of the 90S nucleolar complexes, where cleavage at the A0, A1, and A2 processing sites occurs (16, 28; reviewed in reference 29). The pre-40S particle, which contains some newly associated maturation factors and some already present in the 90S particle, is released after A0-A1-A2 cleavage and transported to the cytoplasm, where cleavage of 20S pre-rRNA to mature 18S rRNA occurs (59). Independently, several pre-60S particles are assembled that differ in the compositions of their rRNAs (distinct 27S precursors) and trans-acting factors, which are mostly are not found in the 90S particle (4, 20, 33, 51, 58).
FIG. 4.
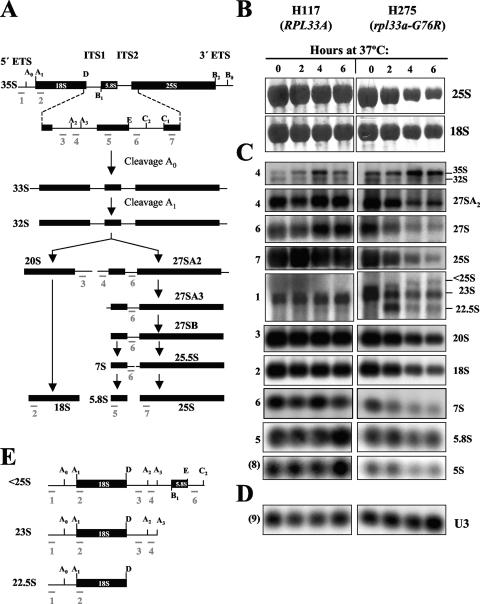
Deficient and aberrant processing of pre-rRNAs in rpl33a-G76R cells shown by Northern analysis. (A) Scheme of the pre-rRNA processing pathway. The 35S pre-rRNA contains the sequences for mature rRNAs (18S, 5.8S, and 25S) separated by two internal transcribed spaces (ITS1 and ITS2) and flanked by two external transcribed spaces (5′ETS and 3′ETS). The rRNAs are shown as filled bars and the transcribed spaces represented as lines. The processing sites are indicated above the diagram by the uppercase letters A to E. The annealing positions of oligonucleotides 1 to 7 used as hybridization probes are indicated beneath all of the rRNA species that they detect. (B to D) Strains H117 (RPL33A) and mutant H275 (rpl33a-G76R) were cultured in liquid YPD medium at 28°C to mid-logarithmic phase (OD600, ∼0.8) and shifted to 37°C for 2, 4, or 6 h. Total RNA was extracted, and samples containing 10 μg were resolved on a 1.2% agarose-4% formaldehyde gel and subjected to Northern analysis. (B) The blot was first dyed with methylene blue to test the integrity of the 25S and the 18S rRNAs. (C) The blot was consecutively hybridized with probes whose positions are depicted in panel A. The RNA species, visualized with the probes indicated at the left, are labeled on the right. The probe specific for 5S rRNA is indicated as (8) (Materials and Methods). (D) Hybridization with a probe specific for U3 (9) (Materials and Methods), which was used as internal control for loading. (E) Schematic diagram showing the potential origin of aberrant and less common pre-rRNA species indicated in panel C.
In S. cerevisiae, genetic analysis and recent biochemical approaches have shed light on the sequence of incorporation of assembly factors into preribosomal 60S and 40S particles during assembly and export (reviewed in references 47 and 67). In contrast, much less is known about the contribution of r-proteins to ribosome biogenesis, because they were found as contaminants in most protein complexes identified in comprehensive analyses of protein-protein interactions in yeast (25). Thus, it was impossible to faithfully determine their precise sequential assembly with the early 90S particle, or with 40S and 60S preribosomes (16, 28, 29, 51). However, it is well documented that insufficiency of a single r-protein often leads to aberrant processing of pre-rRNAs and to insufficient production of mature ribosomal subunits. A recent systematic study in yeast revealed that most of the 32 r-proteins of the small 40S subunit (rpS proteins) play roles in ribosome biogenesis and are essential for growth (22). Distinct rpS proteins are required for early and/or late maturation steps of the 20S pre-rRNA to 18S rRNA or for intracellular transport of the small subunit, suggesting that their correct assembly would guarantee the production of 40S particles fully functional in translation. Although a similar systematic analysis of the 46 r-proteins of the yeast 60S subunit (rpL proteins) has not yet been reported, experimental evidence that rpL proteins are also required for pre-rRNA processing exists. Thus, the absence or functional inactivation of rpL5, rpL11, rpL25, rpL28, or rpL30 affects pre-rRNA processing, resulting in similar phenotypes of accumulation of 35S and 27S pre-rRNA species suggestive of delayed processing at A0, A1, and A2 and of the late cleavage step at C2 (12, 48, 70, 72, 74).
Several r-proteins have been shown to mediate interactions with specific trans-acting factors required for the maturation of pre-rRNAs and ribosomal assembly, while others interact with components of the translation machinery (translation factors or tRNAs), and some are required for the efficient execution of the early or late steps of the translation process. For instance, (i) the physical interaction of rpS14 with the FAP7 assembly factor is required for the cleavage of the 20S pre-rRNA in pre-40S particles (30); (ii) the interaction of rpS0 with the TIF32 subunit of the translation initiation factor eIF3 may mediate eIF3 recruitment to mature 40S subunits (71); (iii) rpL5 helps to anchor the peptidyl-tRNA to the “P” site of the ribosome (45); (iv) the lack of any of the nonessential r-proteins rpL41, rpL24, and rpL39 or mutant forms of essential rpL3 affect the peptidyl-transferase activity of the resulting ribosomes (17, 18, 19, 46); and (v) the nascent polypeptide-associated factor gains access to nascent polypeptides via its interaction with rpL25 at the exit site of the ribosome (27).
Mutations that disrupt the translational regulation of the distinctive GCN4 mRNA in S. cerevisiae have identified essential components of the translational apparatus, including initiation factors that control initiator tRNAiMet binding to the 40S ribosome and r-proteins (37, 38). GCN4 is one of the best-characterized transcriptional activators, first identified by its role in a major physiological response known as the general amino acid control (GAAC) response. The sophisticated mechanism that couples GCN4 expression to amino acid availability has been explored in great detail (see reference 38 for a review) and depends on four short, upstream open reading frames (uORFs) located in the GCN4 mRNA leader and on multiple trans-acting factors encoded by the GCN and GCD genes. Under conditions of amino acid sufficiency, the four uORFs prevent ribosomes from initiating translation at the GCN4 start codon, and very little GCN4 protein is produced. However, whereas solitary uORF1 reduces GCN4 expression by ∼50%, uORF3 and -4 are the critical negative elements in the leader, and ribosomes that translate these sequences cannot reinitiate at the AUG codon of GCN4. Under amino acid starvation conditions, uORF1 has a stimulatory role, allowing ribosomes to traverse inhibitory uORF3 and -4 without initiating translation and reinitiate at GCN4 instead. This up-regulation occurs because uncharged tRNA species that accumulate in amino acid-starved cells activate the protein kinase GCN2, which phosphorylates the α-subunit of translation initiation factor 2 (eIF2). This modification reduces the rates of general protein synthesis, because it inhibits the delivery of charged methionyl initiator tRNA to the 40S ribosomal subunit by the eIF2·GTP·tRNAiMet ternary complex (TC). In contrast, the delayed recruitment of the TC enables 40S subunits that have translated uORF1 and resumed scanning to bypass inhibitory uORF3 and -4 and eventually initiate at the GCN4 start codon downstream. Thus, contrary to most mRNAs, GCN4 translation is induced by a decreased availability of the TC in cells (15). However, induction of GCN4 occurs at a lower level of eIF2α phosphorylation than is required for general inhibition of protein synthesis, making it a sensitive reporter of decreased TC formation and of defects in TC recruitment to the 40S ribosome (14).
The GCD factors are required for the repression of GCN4 mRNA translation under conditions of amino acid sufficiency. The GCD genes were identified by recessive mutations that constitutively derepress GCN4 translation in the absence, or reduced function, of the eIF2α kinase GCN2 (10, 32). All known GCD proteins have functions in the initiation of protein synthesis or regulate the activities of initiation factors and are essential for growth. Consistently, all Gcd− mutants analyzed so far display slow-growth phenotypes at 28°C (Slg−) that are more noticeable at the higher temperatures of 34°C and 37°C. Here we have identified a new complementation group of Gcd− mutants defined by the gcd17-1 mutation. The GCD17 gene was cloned and shown to be identical to RPL33A, one of the two genes encoding the essential r-protein L33 in yeast (65), which belongs to a conserved family of proteins that bind tRNA. The gcd17-1 mutation, which replaces Gly-76 with Arg in rpL33A, impairs efficient early processing of 35S and 27S pre-rRNAs and greatly impedes the accumulation of 60S ribosomal subunits at the restrictive growth temperature of 37°C. This leads to the formation of half-mer polysomes, indicating a decreased rate of 60S subunit-to-40S subunit joining at the final step of translation initiation and a strong reduction in the rate of general protein synthesis. Interestingly, at 28°C, rpl33a-G76R constitutively derepresses GCN4 translation, producing a Gcd− phenotype that is considerably stronger than that produced by the null, deletion allele of RPL33A (strain Hm506 [ΔA mutation]). This is remarkable because rpl33a-G76R is less severe than ΔA in reducing 60S subunit levels at this growth temperature. Analysis of GCN4-lacZ reporters suggests that both rpl33a-G76R and ΔA mutations derepress GCN4 translation in the presence of high TC levels (i.e., in the gcn2 background) by impairing 60S-40S subunit joining at uORF4 of the GCN4 mRNA leader. Thus, rpl33a-G76R might impede the joining reaction and efficient 80S initiation complex formation. We propose that inefficient subunit joining at uORF4 allows 40S subunits to abort initiation at the uORF4 start site, resume scanning, and reinitiate downstream at GCN4. Our finding that the Gcd− phenotype of the rpl33a-G76R mutation is suppressed by overexpressing tRNAiMet suggests that the dissociation of tRNAiMet from the 40S subunit is responsible for the postulated abortive initiation events at uORF4. Thus, our data indicate that rpL33 has a critical function in the ribosome biogenesis pathway required for the efficient production of both ribosomal subunits and a second role in translation initiation at the stage of 40S subunit-60S subunit joining. Both activities contribute to the repression of GCN4 translation under nonstarvation conditions and, hence, the proper functioning of the GAAC response.
MATERIALS AND METHODS
Plasmids. (i) Cloning of GCD17 and gcd17-1 alleles.
Plasmid pPM1, bearing an ∼11-kb DNA fragment of yeast chromosome XVI, was isolated from a yeast genomic library in YCp50 (55). Subclones of the genomic insert in pPM1 were constructed in the shuttle vector pRS316 in order to define the boundaries of GCD17. Plasmid pPM2 bears a 1.9-kb SalI-NdeI genomic fragment from pPM1 cloned in pRS316, containing YPL143W (RPL33A) and the dubious ORF YPL142C encoded in the Crick strand, in the opposite orientation relative to YPL143W. Plasmid pPM26 bears an 882-bp HindIII-NdeI fragment with YPL142C cloned in pRS316 and does not complement the phenotypes of gcd17-1. Plasmid pPM13 was constructed by cloning a 1.9-kb SalI-XbaI fragment containing RPL33A in pRS426 (hcRPL33A). The gcd17-1/rpl33a-G76R mutation was identified by independent PCR amplification of the corresponding mutant allele from genomic DNAs of strains H275 and Hm337 (Table 1), with oligonucleotides F4 (5′-CGCATAACTCTTCGATAATACAG-3′) and R5 (5′-CCAAAGTCCAGAACATTCAACC-3′) used as primers, followed by automatic sequencing of the amplification products on the two DNA strands.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference or source |
|---|---|---|
| H96 | MATα gcn2-101 gcn3-101 his1-29 ura3-52 (HIS4::lacZ ura3-52) | 32 |
| H117 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4::lacZ URA3) | 32 |
| H275 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4::lacZ URA3) rpl33a-G76R | 10 |
| Hm337 | MATα gcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4::lacZ ura3-52) rpl33a-G76R | H275 × H96 |
| Hm506 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4::lacZ ura3) rpl33a::URA3 | This study |
| Hm502 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4::lacZ URA3) rpl33b::kanMX4 | This study |
| Hm505 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4::lacZ URA3) rpl33a-G76R rpl33b::kanMX4 | This study |
| Hm525 | MATagcn2-101 gcn3-101 his1-29 ino1 ura3-52 (HIS4::lacZ URA3) rpl33a::kanMX4 | This study |
| H466 | MATα gcn2-101 gcn3-101 ura3-52 | 32 |
| Hm526 | MATα gcn2-101 gcn3-101 ura3-52 rpl33a::kanMX4 | This study |
| Hm527 | MATα gcn2-101 gcn3-101 ura3-52 (rpl33a-G76R kanMX4::rpl33a) | This study |
| S288C | MATα SUC2 gal2 mal mel flo1 flo8-1 hap1 | ATCC 204508 |
| F35 | MATaura3-52 ino1 can1 (HIS4::lacZ URA3) | 43 |
| Hm531 | MATaura3-52 ino1 can1 (HIS4::lacZ URA3) rpl33a::kanMX4 | This study |
| Hm532 | MATaura3-52 ino1 can1 (HIS4::lacZ URA3) (rpl33a-G76R-kanMX4::rpl33a) | This study |
| JWY2165 | MATaura 3-52 his3-11,15 leu2-3,112 drs1-1 | 54 |
| JWY6882 | MATα his3Δ-200 leu2-Δ1 lys2-801 trp1-Δ101 ura3-52 nop7::HIS3 plus pNOP7A (CEN LEU2 nop7-A) | J. L. Woolford |
(ii) Construction of rpl33a and rpl33b null alleles.
Plasmid pPM18 was constructed as follows: the 1.9-kb XbaI-SalI fragment of chromosome XVI containing YPL143W (RPL33A), including 610 bp upstream of the ATG start codon and 493 bp downstream of the stop codon, was cloned in pRS315. A 519-bp HindIII fragment internal to the RPL33A ORF was replaced with a 1.1-kb fragment containing the URA3 gene. The 2.5-kb null allele rpl33a::URA3 is excised from pPM18 by digestion with SalI and XbaI.
The null allele rpl33a::kanMX4 was constructed by PCR-long flanking homology analysis (75). In a first PCR, plasmid pPM2 (see above) was used as the template to amplify a 164-bp sequence of the RPL33A coding region, using primers L1A and L2A, and a 601-bp sequence corresponding to the 3′ region of RPL33A, using primers L3A and L4A. To construct the rpl33b::kanMX4 null allele of RPL33B, plasmid pPM10 bearing a 3.1-kb SalI-SalI fragment of chromosome XV containing RPL33B, was used as the template to amplify by PCR (i) a 429-bp sequence of the RPL33B 5′ region, using primers L3B and L4B, and (ii) a 614-bp sequence corresponding to the 3′ region downstream of the RPL33B stop codon, using primers L1B and L2B. L1A is 5′CTTCTACTCTTCACCGGCATG3′), L2A is 5′GGGGATCCGTCGACCTGCAGCGTACCATTTTTCAATTTATTTGATTGTTGG3′, L3A is 5′AACGAGCTCGAATTCATCGATGATATGACGCATAACTCTTCGATAATACAG-3′, L4A is 5′-GCTTACAGCGAGATTCGCAG-3′, L1B is 5′CTCAGCAGCAACCACGTAAC-3′, L2B is 5′GGGGATCCGTCGACCTGCAGCGTACCATGACTTTCGGTGCCTCTG TCAG3′, L3B is 5′AACGAGCTCGAATTCATCGATGATATGAATACGTCTATGGGATTCAGC3′, and L4B is 5′GAAACGTACGGTATATTGTGAG-3′. Primers L2 and L3 carry 28-nt 5′ extensions (underlined) complementary to the kanMX4 module cloned into the pFA6a-KanMX4 plasmid (76), which was used as the template to amplify the kanMX4 sequences in a second PCR, using as primers the 164-bp and 601-bp fragments (RPL33A) or 614-bp and 429-bp fragments (RPL33B) amplified before. These reactions produce a 2.3-kb fragment containing an rpl33a::kanMX4 null allele and a 2.5-kb fragment containing an rpl33b::kanMX4 null allele, respectively.
Plasmids pJW6039 (hcNOP7) and pJW3015 (hcDRS1) were kindly provided by J. W. Woolford, Jr. (Carnegie Mellon University, Pittsburgh, PA). Plasmid p2635 (hcIMT4) was previously described (2). Plasmid p1730-IMT4 (p3000) contains the SUI2, SUI3, and GCD11 genes, encoding, respectively, the α, β, and γ subunits of yeast eIF2, and the IMT4 gene, all cloned in YEp24 (3). Plasmid pAS425 contains the PAB1 gene cloned in a 2μm URA3 vector and was kindly provided by A. Sachs (University of California at Berkeley).
Yeast strain construction.
The Saccharomyces cerevisiae strains used in this study are listed in Table 1. RPL33A and RPL33B were individually replaced with the null allele rpl33a::URA3, rpl33a::kanMX4, or rpl33b::kanMX4 in the wild-type (WT) H117 strain. By contrast with the rpl33a::LEU2 null allele (65), the rpl33a::URA3 and rpl33a::kanMX4 alleles were constructed to preserve the integrity of YPL142C, a hypothetical ORF located on the strand opposite to RPL33A, which was annotated as essential (SGD). To produce strain Hm506 (rpl33a::URA3 [ΔA]), a Ura− spontaneous derivative of H117 (URA3 HIS4::lacZ) selected in 5-fluoroorotic acid was transformed with the SalI-XbaI fragment from pPM18 containing rpl33a::URA3. Correct replacement of RPL33A by rpl33a::URA3 was verified by Southern blotting, with digestion of total DNA of the corresponding transformants with PstI and SalI and the use of sequences complementary to URA3 and RPL33A as probes.
To produce strain Hm525, strain H117 (URA3 HIS4::lacZ) was transformed with a 2.3-kb fragment containing the rpl33a::kanMX4 null allele described above. To produce strains Hm502 (rpl33b::kanMX4 [ΔB mutation]) and Hm505 (rpl33a-G76RΔB), the 2.5-kb null allele rpl33B::kanMX4 was used to transform strains H117 (RPL33A) and H275 (rpl33a-G76R), respectively. Geneticin-resistant transformants were selected on yeast extract-peptone-dextrose (YPD) plates with 100 μg/ml of Geneticin (G-418), and in every case, correct replacements by the null alleles were verified by PCR using the appropriate oligonucleotides. To create ΔA and rpl33a-G76R mutations in a gcn2-101 gcn3-101 background devoid of the integrated HIS4-lacZ fusion, the RPL33A gene was replaced in strain H466 with the rpl33a::kanMX4 null allele (described above) or with a marked allele, rpl33a-G76R-kanMX4, generating the isogenic mutants Hm526 (gcn2-101 gcn3-101 rpl33a::kanMX4) and Hm527 (gcn2-101 gcn3-101 rpl33a-G76R-kanMX4), and correct replacements were verified by PCR. The rpl33a-G76R-kanMX4 marked allele was constructed by inserting a fragment of 1.4 kb from plasmid pFA6a-KanMX4 (76) containing the kanMX4 marker into the NdeI site located at the 3′ end of the functional rpl33a-G76R sequence, which was cloned as a SalI-XbaI fragment of 1.9 kb in plasmid pRS315 (pPM28). The marked allele rpl33a-G76R-kanMX4 is excised from pPM28 as a 3.1-kb EcoRI-XbaI fragment. It contains the rpl33a-G76R ORF flanked by genomic sequences upstream (276 bp) and downstream (449 bp), followed by kanMX4.
Media.
Yeast strains were grown in rich YPD medium or in standard dextrose (SD) medium supplemented as required. Starvation for histidine with 3-aminotriazol (3AT) or for tryptophan with 5-methyltryptophan (5MT) was tested as described previously (32, 39).
Biochemical techniques. (i) Assay of HIS4-lacZ and of GCN4-lacZ fusions.
β-Galactosidase assays were conducted as previously described (7) with cells grown in SD medium containing only the required supplements to an optical density at 600 nm (OD600) of ∼1. For repressing conditions, cultures were harvested in mid-logarithmic phase after 8 h of growth. For derepressing conditions, cells were grown for 3 h under repressing conditions and then for 6 h after 3AT was added to 10 mM. The values shown in Table 3 and Fig. 5D are the averages of results from three independent determinations. β-Galactosidase activities are expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside cleaved per minute per ∼1 × 107 cells.
TABLE 3.
Derepression associated with rpl33 mutations of histidine-biosynthetic enzymes
| Strainc | Relevant genotype | Growth on 3ATa |
HIS4-lacZ activity (U)b
|
|
|---|---|---|---|---|
| R conditions | DR conditions | |||
| H117 | gcn2 gcn3 | − | 150 | 170 |
| H275 | gcn2 gcn3 rpl33a-G76R | ++ | 670 | 530 |
| Hm525 | gcn2 gcn3 ΔA | +/− | 270 | 400 |
| Hm502 | gcn2 gcn3 ΔB | −/+ | 210 | 240 |
| Hm505 | gcn2 gcn3 rpl33a-G76R ΔB | ++ | 620 | 540 |
| F3 | GCN2 GCN3 | ++ | 100 | 800 |
| Hm532 | GCN2 GCN3 rpl33a-G76R | ++ | 880 | 860 |
| Hm531 | GCN2 GCN3 ΔA | + | 240 | 300 |
Strains were replica printed to 3AT (10 mM) medium and scored for growth at 28°C after 3 days. ++, confluent growth; +/− and −/+, weak and very weak growth, respectively; −, no visible growth.
β-Galactosidase activity was assayed under repressing (R; nonstarvation) and derepressing (DR; histidine starvation) conditions. Values are averages of results obtained from three independent experiments with two or three independent transformants.
All strains were isogenic.
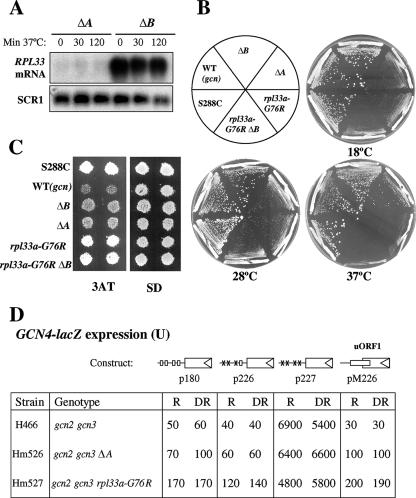
FIG. 5.
Comparison of the Gcd− and Slg− phenotypes of several rpl33 mutants. (A) Steady-state amounts of RPL33 mRNAs. Cells of Hm506 (ΔA) and Hm502 (ΔB) were grown in YPD to mid-logarithmic phase at 28°C (OD600, ∼0.8) and transferred for 30 min or 2 h at 37°C. Total RNA was extracted and 10 μg analyzed by Northern blotting using a probe that hybridizes to both RPL33A and RPL33B mRNAs and a probe for SCR1, used as the loading control (10) (Materials and Methods). (B) The Slg− phenotypes of Hm502 (ΔB), Hm506 (ΔA), and Hm505 (rpl33a-G76R ΔB) mutants are shown relative to that of the original mutant H275 (rpl33a-G76R) and to the WT phenotype (Slg+) of the parental strain H117 (gcn2 gcn3 RPL33A RPL33B). The Slg+ phenotype of the WT strain S288C (GCN2 GCN3 RPL33A RPL33B) is shown as a reference. The growth phenotypes were analyzed by streaking single colonies on YPD medium and incubating the plates for 8 days at 18°C and for 3 days at 28°C or at 37°C. (C) The Gcd− phenotype (3ATR) of the same mutants as those shown in panel B is shown relative to that of the original mutant H275 and to the 3ATS (Gcn−) phenotype of the parental strain H117. The 3ATR phenotype of the WT strain S288C is shown as a reference. Isolated colonies of each strain were replica printed to 10 mM 3AT and to SD plates and incubated for 3 days at 28°C. (D) The rpl33a-G76R and ΔA mutations lead to constitutive derepression of GCN4-lacZ independently of the positive GCN regulators. GCN4-lacZ fusions were introduced into yeast strains on low-copy-number plasmids p180, p226, p227, and pM226 (labeled boxes with a triangle at the right end). The relevant genotypes of the strains are indicated on the left below these diagrams. The gcn2-101 gcn3-101 strains H466 (RPL33A), Hm526 (ΔA), and Hm527 (rpl33a-G76R) are isogenic. The four uORFs in the leader sequence of p180 are shown as open boxes, and point mutations that remove the AUG codons of uORF1 to -3 (p226) or uORF1 to -4 (p227) are shown as ×s. uORF1 in pM226, located at the position of uORF4 and elongated to overlap the beginning of GCN4, is indicated by a rectangle. β-Galactosidase activity was measured in cells grown to mid-logarithmic phase under nonstarvation, repressing (R), or derepressing (DR) conditions of histidine starvation induced by 3AT. Values are averages of results obtained in three independent experiments with two independent transformants. Units of β-galactosidase activity were calculated as indicated in Materials and Methods.
(ii) Northern analysis.
To measure the steady-state levels of pre-rRNA and rRNA in WT H117 and in the rpl33a-G76R mutant H275, Northern analysis was carried as follows. In all experiments, cells were grown in liquid YPD medium at 28°C to and OD600 of ∼1 (time zero) and then transferred to 37°C for several hours. Total RNA was extracted from equivalent numbers of cells (5 × 108) of these strains by the phenol-acid method (60). Samples containing 10 μg of total RNA were electrophoresed in 1.2% agarose-4% formaldehyde gels for analysis of rRNAs. The RNAs were blotted to positively charged nylon membranes (Roche) and immobilized by UV cross-linking with a UV Stratalinker 2400 (Stratagene). The blots were hybridized sequentially with ∼20-nucleotide-long oligonucleotides labeled at their 5′ ends with [γ-32P]ATP (6,000 Ci/mmol), and direct quantification of the corresponding hybridization signals was performed by PhosphorImaging analysis using MacBas-v2.5 and a BAS-1500 PhosphorImager. The oligonucleotides used as probes are listed below, and those designated 1 to 7 are complementary to rRNA sequences indicated in Fig. 4A. Oligonucleotide 1 is 5′TCAGGTCTCTCTGCTGC3′, 2 is 5′AGCCATTCGCAGTTTCACTG3′, 3 is 5′TTAAGCGCAGGCCCGGCTGG3′, 4 is 5′TGTTACCTCTGGGCCC3′, 5 is 5′TGCGTTCTTCATCGATGCGAGAACC3′, 6 is 5′GGCCAGCAATTTCAAGTTA3′, 7 is 5′TACTAAGGCAATCCGGTTGG3′, 8 (5S) is 5′CAGTTGATCGGACGGGAAAC3′, 9 (U3) is 5′GGATTGCGGACCAAGCTAA3′, and 10 (SCR1) is 5′GAGGGAACGGCCACAATGTG3′. When mRNAs were analyzed, total yeast RNA was prepared and RNA blot hybridization was carried out as described previously (35). A 3.1-kb PCR product containing the entire HIS4 gene sequence was used as the radiolabeled probe for HIS4 mRNA, and a 6.7-kb HindIII fragment containing the entire pyruvate kinase-coding sequence (PYK1) was used as the probe for PYK1 mRNA. A 450-bp KpnI-MluI fragment was used as the probe for GCN4 mRNA, and a 601-bp PCR fragment corresponding to the 3′ end of the RPL33A gene was used as the probe for RPL33A and RPL33B mRNAs.
(iii) Polysome analysis.
Polysome analysis by sucrose gradient centrifugation was basically done as previously described (23). Cells were grown in YPD at 28°C to mid-logarithmic phase, and then cultures were incubated at 37°C and an OD600 of ∼1. When the cells were harvested, cycloheximide was added at a final concentration of 100 μg/ml. Whole-cell extracts (WCE) were extracted as described in reference 23 and loaded onto 7% to 50% gradients, which were scanned at 254 nm. For ribosomal-subunit quantification, low-Mg2+ sucrose gradients and WCE were prepared in the absence of cycloheximide.
(iv) Measurement of radioactive-methionine incorporation into protein.
Radioactive-methionine incorporation was measured as described in reference 5, with some modifications. Cells were grown in 200 ml of SD medium containing the necessary supplements and lacking methionine at 28°C to an OD600 of ∼0.6 and then transferred to 37°C for 3 or 6 h. Duplicate 1-ml aliquots were removed at the times indicated and incubated with 5 μCi of l-[35S]methionine (>1,000 Ci/mmol; Amersham) and unlabeled methionine to a final concentration of 0.25 mM for 10 min at 28°C. Incorporation of the radiolabeled methionine was monitored by trichloroacetic acid (TCA) precipitation. TCA (3 ml of a 5% solution) was added to each aliquot, followed by heating at 90°C for 15 min and incubation on ice. The precipitates were collected on GF/C filters, which were washed three times with 10 ml of 5% TCA and 15 ml of 95% ethanol, and dried under a heat lamp, and cells were counted by scintillation spectrometry.
RESULTS
Phenotype of gcd17 mutants.
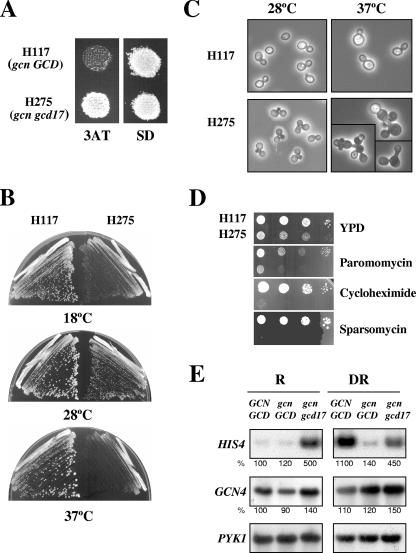
The Gcd− mutant H275 was isolated as a spontaneous suppressor of the Gcn− phenotype of the gcn2-101 gcn3-101 strain H117 (10, 32). Further genetic analysis showed that H275 carries a monogenic gcd mutation, which confers a semidominant Gcd− phenotype in heterozygous gcd/GCD diploids (10). In this study, we found that H275 defines a new complementation group of Gcd− mutants, designated gcd17 mutants. All gcn2 and gcn3 mutations confer sensitivity to 3AT, because they impair derepression of GCN4 and of histidine biosynthetic genes regulated by GCN4 in response to histidine starvation imposed by 3AT (39). The gcd17-1 mutation confers 3AT resistance in a gcn2-101 gcn3-101 background (Fig. 1A), suggesting that it derepresses GCN4 translation. In addition, H275 exhibits a severe Slg− phenotype on minimal media at 28°C, which is exacerbated at both higher (32°C to 37°C) and lower (13°C to 18°C) temperatures, such that very tiny colonies arise on plates of minimal medium only after incubation for 4 days (37°C) or 10 to 12 days (18°C) (Fig. 1B). Aberrant cell morphologies were also observed for H275 at restrictive and permissive temperatures (Fig. 1C). The Gcd− phenotype, slow growth, and cell morphology defects of the gcd17 mutant suggest that, in addition to having a role in general amino acid control, the GCD17 product has an essential function that indirectly affects cell cycle or morphogenetic events. In support of a possible role for GCD17 in translation, we found that H275 showed an increased sensitivity to drugs that affect protein synthesis (Fig. 1D).
FIG. 1.
Gcd− (3ATR), Slg−, cell morphology, and antibiotic-sensitivity phenotypes of the H275 mutant (gcd17-1/rpl33a-G76R). (A) 3ATR phenotype of H275 relative to the 3ATS phenotype of the isogenic WT GCD17 strain H117 (gcn2-101 gcn3-101). Isolated colonies of each strain were replica printed to plates containing 10 mM 3AT and to minimally supplemented SD medium plates and incubated for 3 days at 28°C. (B) Slg− phenotype of H275 at 18°C, 28°C, and 37°C. Cells were streaked for single colonies on SD plates that were incubated at 18°C (10 days), 28°C (3 days), or 37°C (4 days). (C) Aberrant cell morphologies of the H275 mutant. Cells of H117 and of the H275 mutant were grown for 24 h in liquid YPD at 28°C or 37°C, and pictures were taken under a phase-contrast microscope (magnification, ×40). (D) Increased sensitivity of the H275 mutant to the protein synthesis inhibitors paromomycin (500 μg/ml), cycloheximide (0.025 μg/ml), and sparsomycin (5 μg/ml) relative to that of H117. Serial dilutions of cells (104 to 10 cells) grown for 12 h in YPD were plotted on YPD plates and on YPD plates containing the indicated concentrations of antibiotics and incubated for 3 or 4 days at 28°C. (E) The gcd17-1/rpl33a-G76R mutation increases the transcription of the HIS4 gene. Cells of isogenic strains H117 (gcn GCD17) and H275 (gcn gcd17-1) and of the WT strain F35 (GCN GCD) were grown under repressing (R) (SD medium) and derepressing (DR) conditions of tryptophan starvation induced by 0.5 mM 5MT. Total RNA was extracted and 10 μg analyzed by Northern blot analysis in three independent blots, using radiolabeled probes specific to visualize HIS4, GCN4, and PYK1 mRNAs (Materials and Methods). The hybridization signals were quantified by PhosphorImaging analysis, and values normalized relative to PYK1 mRNA are given in percentages below each panel relative to the corresponding values in F35, which were set to 100%.
Under amino acid starvation conditions, GCN4 activates the transcription of the HIS4 gene and a large number of amino acid-biosynthetic genes (36). More recently, whole-genome studies have shown that GCN4 activates 77 amino acid-biosynthetic genes and directly or indirectly regulates a very large set of 539 genes, encompassing ∼1/10 of the yeast genome (50). Gcd− mutants exhibit high levels of HIS4 transcription under conditions of amino acid sufficiency owing to constitutive derepression of GCN4 expression. Consistent with this, under amino acid-replete conditions, levels of β-galactosidase synthesized from a HIS4-lacZ fusion construct were fivefold higher in the gcd17 mutant H275 than in the isogenic GCD17 strain H117 (10). Here, we show that the gcd17-1 mutation elicits high levels of HIS4 mRNA in the gcn2-101 gcn3-101 background both under nonstarvation conditions of growth in minimal medium (SD) and under tryptophan limitation imposed by 5MT (Fig. 1E). Relative to the levels of PYK1 mRNA used as the internal control, the levels of HIS4 mRNA were 5-fold higher in the gcd17 mutant H275 than in the GCD17 parent H117, while those of GCN4 mRNA were only ∼1.5-fold higher in the mutant. The WT GCN GCD strain F35 exhibits levels of HIS4 mRNA ∼11 times higher when it is grown in 5MT than when it is grown in SD medium (Fig. 1E). These data are consistent with the idea that gcd17-1 leads to a partial derepression of GCN4 translation, with attendant derepression of the GCN4 target gene HIS4.
Cloning of GCD17 and of the gcd17-1 mutant allele.
The WT allele of GCD17 was cloned in plasmid pPM1 from a yeast genomic library (Materials and Methods) by complementing the recessive Slg− phenotype at 34°C of the gcd17-1 ura3-52 strain Hm337 (Table 1). Subcloning of the genomic insert contained in pPM1 and complementation analysis identified YPL143W, encoding the 60S ribosomal-subunit protein rpL33A (Materials and Methods). To distinguish between complementation by GCD17 and dosage-dependent suppression of the gcd17-1 mutation, an 882-bp HindIII-NdeI fragment of the pPM1 insert corresponding to the 3′ end of the RPL33A gene (http://db.yeastgenome.org/cgi-bin/locus.pl?locus=rpl33a) was subcloned into a nonreplicating URA3 plasmid and shown to direct plasmid integration into the gcd17 locus (rpl33a) in Hm337 (data not shown). As the integration event restored WT growth to Hm337, it verified by marker rescue that RPL33A is the WT allele of GCD17 and mapped the gcd17-1 mutation to the corresponding 882-bp HindIII-NdeI fragment of the rpl33a mutant allele.
rpL33A, previously designated L37 (or Rp47) in S. cerevisiae (44), is encoded by duplicate genes, RPL33A (XVI) and RPL33B (XV), which are differentially expressed. The RPL33A gene was estimated to produce mRNA levels sixfold higher than those generated from RPL33B when fused to a LAC4 reporter gene. Accordingly, a null Δrpl33a::LEU2 mutant is viable but severely impaired in growth, and a Δrpl33b::URA3 mutant exhibits normal growth, while Δrpl33a Δrpl33b double mutants are inviable, indicating that rpL33 is an essential protein (65). The coding region of RPL33A is very similar to that of RPL33B, with only one conservative amino acid difference (Asp-40 in rpL33A and Glu-40 in rpL33B); however, overexpression of either RPL33A or RPL33B complements the phenotypes of the deletion mutants, indicating that both products are functional (65). Recently, it was shown that rpL33A is haploinsufficient in rich medium (YPD), while the loss of rpL33B only slightly reduces fitness (13).
We determined that the gcd17-1 mutation in mutants H275 and Hm337 is a single-point mutation (see Materials and Methods) consisting of a transversion from G to C at nucleotide 750 of the RPL33A ORF, replacing a glycine codon (GGT) with an arginine codon (CGT) at amino acid residue 76 of rpL33A.
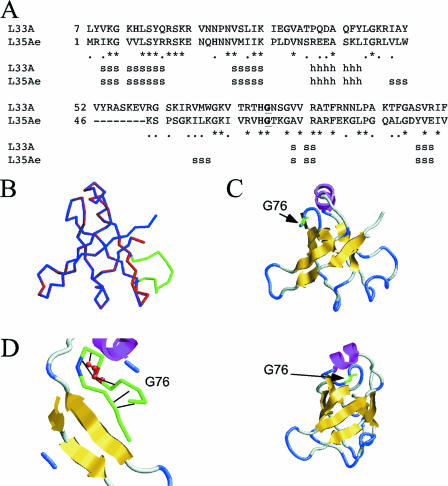
Structural features of rpL33 and prediction of a three-dimensional (3D) model.
rpL33 is a small basic protein of 106 amino acid residues (excluding the N-terminally acetylated methionine that is processed) with orthologs in archaeabacteria (like L35Ae of Pyrococcus furiosus) and eukaryotes (like L32 of Xenopus spp. and L35a of rat and human) but not in bacteria. Interestingly, the rpl33a-G76R mutation maps to a motif of 22 amino acids that belongs to a putative RNA-binding domain located in the carboxy-terminal region of rpL33A and is well conserved in the 99 members of the “ribosomal protein L35Ae family” (http://www.ebi.ac.uk/interpro/IEntry?ac=IPR001780). However, the position of rpL33 in the cryo-electron microscopy structure of the yeast 80S ribosome has not been determined (63).
The nuclear magnetic resonance structure of the orthologous 50S r-protein L35Ae from the archaebacterium Pyrococcus furiosus (36% identity) has been obtained (62). We generated a theoretical model of the 3D structure of rpL33A using the coordinates of 1SQRm1.pdb for L35Ae and the ProModII program (N. Guex and M. C. Peitsch, SWISS-MODEL server). The sequence alignment of rpL33A with L35Ae and secondary-structure predictions are shown in Fig. 2A. Superposition of the main-chain backbones of the two proteins revealed only an extra loop in rpL33A, as shown in Fig. 2B. The predicted 3D structure of rpL33A, shown by a ribbon depiction in Fig. 2C, exhibits a β-barrel, one α-helix, and several long loops and turns. The G76R mutation alters a rigid loop close to the α-helix (Fig. 2D) and is not expected to disturb the overall structure of the protein (J. de las Rivas, personal communication).
FIG. 2.
Theoretical 3D model for rpL33. (A) Alignment of rpL33A's primary sequence with that of Pyrococcus furiosus 1SQR and secondary-structure predictions for rpL33. h, predicted alpha helix, s, predicted beta sheet. (B) Overlap of the main-chain backbones of 1SQR (blue) and rpL33A (red), showing a loop of rpL33A absent in 1SQR (green). (C) Predicted β-barrel 3D structure of rpL33 (yellow), with the position of an α-helix (dark pink) and of the G76R mutation in rpl33a-G76R (green). The diagram shown in panel B and the diagram in the upper portion of panel C are in the same orientation, and that in the bottom of panel C is rotated 90° to the right. (D) Glycine 76 (red) is predicted to form hydrogen bonds inside a rigid loop; its primary sequence is part of the 22-amino-acid carboxy-terminal domain conserved in the 99 members of the r-protein L35Ae family.
The rpl33a-G76R mutation causes severe defects in polysome assembly.
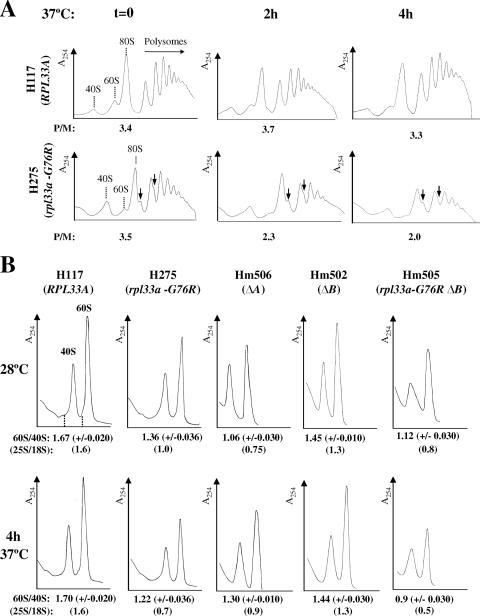
Because rpL33 is an essential constituent of the 60S ribosomal subunit, we investigated whether the rpl33a-G76R mutation causes defects in the general translation of mRNAs. To that end, we analyzed total polysome profiles from WT and mutant rpl33a-G76R strains by fractionating WCE on sucrose gradients by velocity sedimentation (see Materials and Methods). Consistent with a reduction in the rate of colony formation on solid medium at all temperatures, the rpl33a-G76R mutant grew in liquid YPD medium at 37°C with an approximately twofold decrease in the growth rate relative to that of WT H117 (RPL33A). Therefore, we analyzed polysome profiles after shifting mutant and WT cells from 28°C to 37°C for 2 and 4 h. Quantification of the profiles revealed nearly identical polysome-to-monosome (P/M) ratios in mutant (P/M ratio, 3.5) and WT (P/M ratio, 3.4) cells grown at 28°C to mid-logarithmic phase. However, the pool of free 40S ribosomal subunits was elevated and that of 60S subunits was moderately decreased in the mutant at 28°C, concomitant with the appearance of half-mers in the 80S monosome and disome (2-mer) peaks of the profiles (Fig. 3A, time zero graph). The presence of half-mers, which contain one or more 80S ribosomes plus a 43S preinitiation complex (PIC) in the mRNA leader (34), is a hallmark of a reduced level of 60S subunits (56) resulting in a delay in 60S-subunit joining at the last step of translation initiation (23). When cells were grown to mid-logarithmic phase at 28°C and then incubated for 2 or 4 h at 37°C, the WT strain showed very little difference in polysome content (Fig. 3A, top panels). In contrast, after incubation at 37°C, the rpl33a-G76R mutant showed a notable reduction in the P/M ratio and a decrease in the average size of the polysomes compared to what was seen in WT cells. Half-mers are still observed in the monosome peaks of the mutant profiles, and the reduction in the 60S subunit is more apparent after 2 and 4 h of incubation at 37°C (Fig. 3A, bottom panels).
FIG. 3.
The rpl33a-G76R mutation leads to a shortage in 60S ribosomal subunits and to the accumulation of half-mer polysomes. (A) The rpl33a-G76R strain H275 (bottom panels) and the isogenic RPL33A WT strain H117 (top panels) were grown in YPD to mid-logarithmic phase at 28°C (OD600, ∼0.8) and then shifted for 2 or 4 h at 37°C. Cycloheximide was added at 100 μg/ml before harvesting of cells, and WCE containing ribosomes and polyribosomes were prepared in the presence of 30 mM Mg2+ and separated by velocity sedimentation on 7% to 50% sucrose gradients. In the top panels, peaks representing free-ribosomal 40S and 60S subunits and 80S monosomes are indicated, and half-mer polysomes appearing in the H275 mutant profiles are marked by vertical arrows. The P/M ratios were calculated for profiles obtained from cells grown at 28°C (at time zero [graphs marked t = 0]) and after incubation at 37°C (graphs marked 2h and 4h), and the averages of values obtained in three independent experiments are indicated on the A254 tracings. (B) Strains H117 (RPL33A), H275 (rpl33a-G76R), Hm506 (ΔA), Hm502 (ΔB), and Hm505 (rpl33a-G76R ΔB) were cultured as described for panel A, but WCE were prepared in the absence of cycloheximide and Mg2+ and resolved by velocity sedimentation through 7% to 50% sucrose gradients. The mean ratios of total 60S/40S subunits determined from three replicate experiments are indicated below the A254 tracings, and the 25S/18S rRNA ratios estimated with an Agilent Technologies model 2100 bioanalyzer are indicated inside parentheses below the 60S/40S rRNA ratios.
Quantification of total ribosomal subunits in low-Mg2+ sucrose gradients, where polysomes and 80S ribosomes dissociate to free subunits, revealed a deficit in 60S relative to 40S ribosomal subunits in the rpl33a-G76R mutant (Fig. 3B). A nearly constant ratio of 60S subunits to 40S subunits of ∼1.70 was observed in the WT strain at 28°C and 37°C; however, the 60S/40S rRNA ratio was only 1.36 for the rpl33a-G76R mutant at 28°C and diminished further to 1.22 after 4 h at 37°C. Independent measurements of the 25S/18S rRNA ratios with an Agilent bioanalyzer revealed that the levels of rRNAs were nearly constant in the WT strain H117 (∼1.6) but decreased in the rpl33a-G76R strain with incubation at 37°C (from ∼1.0 at 28°C to 0.7 after 4 h at 37°C) in a manner similar to a reduction in the ratio of 60S subunits to 40S subunits (Fig. 3B). A moderate decrease in the total amount of 40S ribosomal subunits per A260 unit of extract was also observed in the rpl33a-G76R mutant under these conditions (Fig. 3B, bottom panels), as reported for other yeast mutants impaired in 60S-subunit production (73).
Together, the data in Fig. 3 suggest that the slow-growth phenotype of the rpl33a-G76R mutant results at least partially from a reduction in the level of 60S subunits, with a concomitant decrease in the rate of 60S-subunit joining that reduces the rate of general protein synthesis. Consistent with this, the rate of incorporation of radioactive methionine into acid-insoluble material was decreased by ∼45% at 28°C and by 60% after 6 h of incubation at 37°C in rpl33a-G76R cells compared to that of isogenic RPL33 cells (Table 2).
TABLE 2.
Rates of protein synthesis in rpl33 mutants
| Strain | Relevant genotype (37°C) |
l-[35S]methionine incorporation (cpm/103 cells/OD600) ata:
|
||
|---|---|---|---|---|
| 0 h | 3 h | 6 h | ||
| H117 | gcn2 gcn3 | 75.5 (±0.5) | 31 (±2.0) | 51 (±2.0) |
| H275 | gcn2 gcn3 rpl33a-G76R | 40 (±1.4) | 17 (±1.4) | 20 (±0.7) |
| Hm506 | gcn2 gcn3 ΔA | 51 (±2.0) | 25 (±1.0) | 31 (±2.4) |
| Hm502 | gcn2 gcn3 ΔB | 71 (±2.0) | 29 (±2.0) | 41 (±0.9) |
| Hm505 | gcn2 gcn3 rpl33a-G76R ΔB | 32 (±2.0) | 10 (±2.0) | 12 (±2.0) |
Values are averages of results obtained from two or three independent experiments (standard errors are shown inside parentheses).
Defects in pre-rRNA processing caused by the rpl33a-G76R mutation.
Mutations in genes for trans-acting factors involved early in the assembly of 60S subunits may cause some disassembly of early pre-60S ribosomal particles and attendant destabilization of pre-rRNA intermediates (reviewed in reference 41). Because some of those factors are believed to promote the correct folding of pre-rRNAs or the correct assembly of r-proteins, the mutations lead to defects in pre-rRNA processing and stability as secondary consequences (73). In the same manner, it is conceivable that the assembly of the mutant rpL33a-G76R protein into 60S preribosomes impairs the processing or stability of the pre-rRNAs that give rise to 25S rRNA.
To investigate defects in the pre-rRNA maturation pathway caused by the rpl33a-G76R mutation, we conducted Northern analysis of pre-rRNA species in mutant and WT cells grown to mid-logarithmic phase at 28°C and transferred to 37°C for 2, 4, or 6 h (Fig. 4). The processing pathway of pre-rRNA in S. cerevisiae and the probes for Northern analysis are shown in Fig. 4A. Staining of the blot with methylene blue revealed that the proportion of stable RNA comprised of rRNA declined in the mutant throughout the time course at 37°C, with substantial reductions in the steady-state levels of 25S rRNA and smaller reductions in those of 18S rRNA (Fig. 4B). This suggests a stronger effect of the rpl33a-G76R mutation on the production and accumulation of rRNAs in the 60S pathway than in the 40S pathway, in accordance with the results shown in Fig. 3B.
The pre-rRNAs and rRNAs were quantified relative to the level of polymerase II (Pol II)-transcribed U3 RNA (Fig. 4D). Hybridization with probes 1 and 4 revealed that cleavage at A0-A1-A2 of the 35S pre-rRNA is defective in the rpl33a-G76R mutant. The steady-state levels of 35S were higher in the mutant than in the WT at 28°C and after 2 h at 37°C but not after 4 h and 6 h at 37°C. In addition, strong reductions were observed in the levels of 27SA2 pre-rRNA (∼40%; probe 4) and 20S pre-rRNA (∼60%; probe 3) after 4 h at 37°C. Accordingly, the 35S/27SA2 and 35S/20S rRNA ratios were elevated in the mutant. The reduction in levels of 27SA2 and 20S pre-rRNA species at 37°C is consistent with a reduced rate of cleavage at sites A0, A1, and A2 (Fig. 4A).
Hybridization with probe 6 (between processing sites E and C2) (Fig. 4A) revealed that the levels of 27S pre-rRNA species were also diminished by a factor of ∼50% and that those of the 7S pre-rRNA species was reduced by ∼70% in the rpl33a-G76R mutant relative to levels in the WT after 4 h at 37°C (Fig. 4C). However, the 27SA2/25S and 27S/25S rRNA ratios were higher in the mutant than in the WT, suggesting that their processing is slower in the mutant, leading to lower rates of 25S and 5.8S rRNA production.
Another three pre-rRNA species were detected in samples of the rpl33a-G76R mutant, whose potential structures are depicted in Fig. 4E. First, 23S pre-rRNA was visualized with probes 1 and 4 in samples of the H275 mutant and of isogenic WT H117 (Fig. 4C). In yeast, alternative processing at A3 versus A2 seems to depend at least partly on the strain background (reference 24 and references therein). The 23S pre-rRNA accumulated ∼1.5-fold at 28°C but diminished progressively during the incubation at 37°C in rpl33a-G76R cells. Second, an anomalous rRNA species that migrates just above 23S and below 25S in WT samples was visualized with probes 1, 4, and 6, while in mutant samples it suffers a progressive change of mobility during the incubation at 37°C, which allowed its identification (Fig. 4C). A similar “<25S” species was detected in other WT yeast strains, but its identity was not investigated further (i.e., see references 22 and 40). Based on our hybridization data, we deduced that the <25S species would be produced in our gcn2-101 gcn3-101 genetic background by alternative processing at C2 when 35S pre-rRNA was not yet cleaved at A0-A1-A2 (Fig. 4A). A third anomalous species was detected with probe 1 in rpl33a-G76R at 37°C that may correspond to a 22.5S precursor produced by the cleavage of 35S or 23S pre-rRNAs at site D (9). This precursor is seen only in the mutant at 37°C, suggesting that in rpl33a-G76R cells, processing at site D occurs when early processing at A0-A1 is inhibited (22, 52).
Analysis of low-molecular-weight rRNA species of the 60S subunit in the same blot revealed that levels of 5.8S rRNA (probe 5) were diminished by ∼30% in the rpl33a-G76R mutant relative to levels in the WT after 4 h at 37°C. However, the ratio of 7S rRNA to 5.8S rRNA was slightly lower in the mutant than in the WT, indicating that this processing reaction is not affected by rpl33a-G76R. Notably, levels of Pol III-transcribed 5S rRNA (probe 8) were reduced in the same proportion as 5.8S rRNA in mutant cells (Fig. 4C). The 5S rRNA is short-lived, probably reflecting its lack of nucleotide modification, unless it is complexed with yeast rpL5 (12). Thus, perhaps a fraction of 5S rRNA is degraded because its rate of assembly into the corresponding preribosomal 60S particles is reduced in rpl33a-G76R cells.
Finally, in the 40S maturation branch, there was a greater reduction in the level of 20S pre-rRNA (∼60%; probe 3) than of 18S rRNA (∼40%; probe 2) in the mutant after 4 h at 37°C (Fig. 4C), suggesting that processing of 20S to 18S rRNA is normal in rpl33a-G76R cells.
In summary, the Northern analysis revealed multiple defects indicative of impaired processing of 35S pre-rRNA at sites A0-A1-A2 that would contribute to the reduced production of both 25S and 18S rRNAs in rpl33a-G76R cells at the restrictive temperature. It appears that the reduction is more severe for 25S rRNA because of additional defects in 27S to 25S rRNA processing and to the instability of the 27SA2 precursor. Together, these data support the notion that rpL33 is required early in the ribosome biogenesis pathway for correct and efficient processing and for the optimal stability of several pre-rRNA species and, hence, the normal accumulation of the four mature rRNAs. It is possible, however, that defective biogenesis of the 60S rRNA has an indirect effect on the early processing of 35S pre-rRNA at A0-A1-A2 (73). Substantial reductions in the steady-state levels of mature 25S, 5.8S, and 5S rRNAs are consistent with the severe deficit of 60S subunits observed in the polysome profiles of the rpl33a-G76R mutant. The strong decrease in levels of 60S subunits and the less severe reduction in 40S subunits most likely contribute to the translation defect and Slg− phenotype of the rpl33a-G76R mutant.
Phenotype of reduced dosage of WT rpL33 compared to that of the rpl33a-G76R mutant.
The Slg− phenotype of rpl33a-G76R mutants is recessive in heterozygous diploids (rpl33a-G76R/RPL33A), probably due to the loss of rpL33A function in rRNA maturation and 60S biogenesis. In contrast, the Gcd− phenotype of rpl33a-G76R shows dominance, as noted above, suggesting an alteration in rpL33A function in translation initiation. We investigated whether a reduced dosage of WT rpL33, due to chromosomal deletion of RPL33A or RPL33B, leads to a Gcd− phenotype in the gcn2-101 gcn3-101 background and whether the deletions impact 60S subunit biogenesis to the extent observed for the rpl33a-G76R mutation. First, total RNAs obtained from the single-deletion mutants Hm506 (rpl33a::URA3 RPL33B [ΔA]) and Hm502 (RPL33A rpl33b::kanMX4 [ΔB]) were subjected to Northern analysis, using a genomic probe that hybridizes to both RPL33A and RPL33B mRNAs (Materials and Methods). Quantification of the hybridization signals relative to those of the Pol III-transcribed SCR1 RNA analyzed as an internal control revealed that the steady-state levels of RPL33A mRNA in the ΔB mutant is ∼85% higher than that of RPL33B mRNA in the ΔA mutant (Fig. 5A). This result is in agreement with a previous quantification of β-galactosidase activity synthesized from RPL33A-lacZ and RPL33B-lacZ hybrid transcripts in deletion mutants constructed in another yeast genetic background (65) in which RPL33A expression was estimated to be sevenfold higher (86%) than that of RPL33B (∼14%). Thus, ΔA and ΔB should have different consequences for cellular processes in which rpL33 is required, a prediction borne out by the growth phenotypes of the corresponding deletion mutants (Fig. 5B). Replacing RPL33B with the null allele rpl33b::kanMX4 in strain H275 (rpl33a-G76R) generated the viable rpl33a-G76R rpl33b::kanMX4 double mutant Hm505 (rpl33a-G76R ΔB), which has an Slg− phenotype similar to that of the rpl33a-G76R single mutant (Fig. 5B). Because rpL33 is essential, this result implies that the mutant protein rpL33a-G76R is assembled into mutant 60S subunits that are partially functional in translation.
We next compared the phenotypes of the rpl33a-G76R mutant, ΔA mutant, ΔB mutant, and a strain with the combined rpl33a-G76R and ΔB mutations on cell growth, ribosome biogenesis, and general protein synthesis. The rpl33a-G76R and ΔA mutations produce similar growth defects at 28°C, but rpl33a-G76R confers a more severe Slg− phenotype than does the ΔA mutation at 37°C (Fig. 5B). By contrast, the ΔB mutant grows indistinguishably from the WT at all temperatures. Notably, at 28°C, the rpl33a-G76R mutant exhibits a smaller reduction in the 60S/40S ratio (declining to 1.36 from the WT value of 1.67) than does the ΔA strain (ratio of 1.06) (Fig. 3B, time zero graph), thus indicating that the complete absence of rpL33A leads to a stronger 60S subunit biogenesis defect than does rpl33a-G76R at this temperature. However, after 4 h at 37°C, the 60S/40S ratio was lower in the rpl33a-G76R mutant (1.22) than in the ΔA mutant (1.3), showing that rpl33a-G76R impairs 60S subunit biogenesis to an extent comparable to that given by ΔA at the restrictive temperature of 37°C. As expected, the ΔB mutant exhibits 60S/40S ratios lower than that of the WT but higher than that of the ΔA mutant (Fig. 3B), indicating that the absence of rpL33B causes a less severe 60S subunit biogenesis defect than does ΔA. The rpl33a-G76R ΔB double mutant has a 60S biogenesis defect comparable to that of the ΔA mutant at 28°C but much more severe than that of the ΔA mutant at 37°C (Fig. 3B).
In contrast to the effects on ribosome biogenesis, rpl33a-G76R leads to a greater reduction in [35S]methionine incorporation than does ΔA at 28°C (Table 2). This difference was also observed at 37°C, but in this case it was consistent with the relative effects of the ΔA mutation and rpl33a-G76R on 60S subunit biogenesis (Fig. 3B). The rpl33a-G76R ΔB double mutant displays a much stronger reduction in protein synthesis than does the ΔA single mutant at 28°C, even though these two strains have comparable 60S subunit biogenesis defects at this temperature. At 37°C, the rpl33a-G76R ΔB double mutant exhibits the strongest defect of all in protein synthesis, commensurate with the most severe 60S subunit biogenesis defect observed for any of the mutants at this temperature (Table 2 and Fig. 3B, respectively). The facts that at 28°C the rpl33a-G76R and rpl33a-G76R ΔB mutants display defects in 60S subunit biogenesis that are less significant than or similar to those of the ΔA mutant, but reductions in methionine incorporation were significantly larger than in the ΔA mutant at this temperature suggest that altering rpL33A with G76R impairs translation more strongly than does the elimination of rpL33A by ΔA.
Strong evidence supporting the last conclusion came from comparing the Gcd− phenotypes of the rpl33a-G76R and ΔA mutants. As shown in Fig. 5C, the ΔA mutation leads to a weak Gcd− phenotype in Hm506 (3ATR/3ATS), showing lower 3AT resistance than that conferred by rpl33a-G76R in the isogenic H275 mutant, even though the two mutants exhibit nearly identical Slg− phenotypes at 28°C on rich medium (Fig. 5B) and minimal medium (data not shown). This result implies that at 28°C, rpl33a-G76R produces an additional defect in translation initiation that elicits greater derepression of GCN4 translation than does the ΔA mutation. The ΔB mutation also confers a weak Gcd− phenotype (3ATS/3ATR), suggesting that it reduces rpL33 levels enough to weakly derepress GCN4 but not enough to significantly affect general protein synthesis.
To quantify the strength of the Gcd− phenotype conferred by each mutation in gcn2-101 gcn3-101 cells at 28°C, we first measured the β-galactosidase activity expressed from a HIS4-lacZ fusion integrated at the URA3 locus of the mutant and WT strains (Table 3). In the absence of amino acid starvation, the β-galactosidase activity was ∼4.5-fold higher in rpl33a-G76R mutant than in WT cells but only ∼2-fold higher in ΔA cells, indicating that the rpL33a-G76R mutant protein has a stronger effect on HIS4-lacZ expression than does the complete absence of rpL33A. As expected, the absence of rpL33B (ΔB) increased HIS4 expression by the least amount, only ∼1.5-fold relative to the level expressed in H117, but did not exacerbate the derepression produced by rpl33a-G76R in the double mutant. Under amino acid starvation conditions (Table 3), the values of β-galactosidase were similar to those measured in the absence of starvation, indicating that the rpl33a-G76R, ΔA, and ΔB mutations constitutively derepress HIS4-lacZ expression in gcn2 gcn3 cells (Gcd− phenotype).
We also measured the β-galactosidase activity synthesized from a HIS4-lacZ fusion integrated at the URA3 locus of the GCN2 GCN3 WT strain F35 (RPL33A) and of isogenic Hm531 (ΔA) and Hm532 (rpl33a-G76R) mutants generated in the same GCN background (Table 3). As expected, the values of β-galactosidase were approximately eightfold higher under starvation conditions than under nonstarvation conditions in WT strain F35. Nearly identical high levels of β-galactosidase activity were observed in the rpl33a-G76R mutant under both repressing and derepressing conditions, indicating that this mutation constitutively increases HIS4-lacZ expression in GCN cells to an extent similar to that given by starvation in isogenic WT cells. In contrast, ΔA increased HIS4-lacZ expression by a much smaller amount under repressing and derepressing conditions. These data confirm that rpl33a-G76R elicits much stronger derepression of a GCN4 target gene than does ΔA at 28°C.
To obtain direct evidence that rpl33a-G76R and the ΔA mutations derepress GCN4 expression at the translational level, we measured β-galactosidase expression from a plasmid-borne GCN4-lacZ fusion in the isogenic gcn2-101 gcn3-101 strains H466 (RPL33A), Hm526 (ΔA), and Hm527 (rpl33a-G76R) grown at 28°C. The fusion construct in p180 contains the WT GCN4 mRNA leader with the four uORFs and, thus, exhibits WT translational regulation of GCN4 expression (49). As shown in Fig. 5D, low-level expression of GCN4-lacZ on p180 was observed in the WT strain H466 under nonstarvation and histidine starvation conditions, because the gcn2-101 gcn3-101 alleles present in this strain impair derepression of GCN4 translation (36). The rpl33a-G76R mutation led to GCN4-lacZ expression in Hm527 at levels ∼3-fold greater than those observed in H466, whereas ΔA in Hm526 produced only ∼1.5-fold greater expression. These results, together with those in Tables 2 and 3 described above, indicate that the rpl33a-G76R mutation impairs 60S subunit function in translation initiation in a manner distinguishable from a simple reduction in the abundance of 60S subunits. Given that 60S subunits participate only at the last step of 80S initiation complex assembly, it is likely that rpl33a-G76R impairs 60S-subunit joining to the 48S PIC.
The derepression of GCN4 translation under amino acid starvation requires uORF1, so that GCN4-lacZ expression is low and unregulated from the construct in p226 containing uORF4 alone in WT cells (Fig. 5D) (49). Interestingly, rpl33a-G76R increased the expression of the p226 construct approximately threefold compared to that seen in WT strain H466 under repressing and derepressing conditions, whereas ΔA led to a smaller increase of less than twofold. These findings suggest that the derepression of GCN4-lacZ expression conferred by rpl33a-G76R and ΔA results from leaky scanning of uORF4 by fully assembled PICs (i.e., containing the TC) because of a defect in subunit joining. (The term leaky scanning signifies the bypass of an AUG codon by a scanning PIC and does not refer to the nature of the scanning process at sequences preceding the start codon.) However, this effect could also arise from increased reinitiation following uORF4 termination. To distinguish between these two possibilities, we measured GCN4-lacZ expression from the pM226 construct in which uORF1 is the sole uORF, located in the position of uORF4 and elongated to overlap the beginning of GCN4 (31). Mutations that cause leaky scanning would increase GCN4-lacZ expression from pM226, whereas a mutation that allowed increased reinitiation after terminating at uORF4 would not affect the expression of this construct. As shown in Fig. 5D, pM226 gives very low expression in WT cells, because the ribosomes cannot reinitiate after terminating at the elongated version of uORF1 far downstream from the GCN4 AUG codon. In contrast, rpl33a-G76R increased the expression of the pM226 construct approximately sevenfold compared to that seen in WT strain H466, under repressing and derepressing conditions, and ΔA led to a smaller increase of less than fourfold, consistent with increased leaky scanning of uORF1 in the two mutants.
Finally, if rpL33A represses the translation of GCN4 via the uORFs, eliminating all of them should abolish the derepressing effect of the rpl33a-G76R and ΔA mutations on GCN4-lacZ expression. Consistent with this prediction, rpl33a-G76R and ΔA had little effect on the expression of the GCN4-lacZ construct lacking all four uORFs (p227), thus indicating that rpL33 regulates GCN4 expression at the translational level via the uORFs. Together, these data show that rpL33A is required for the repression of GCN4 translation under conditions of amino acid sufficiency by preventing leaky scanning of the uORFs, most likely by ensuring efficient 60S-subunit joining.
Genetic partners of rpL33.
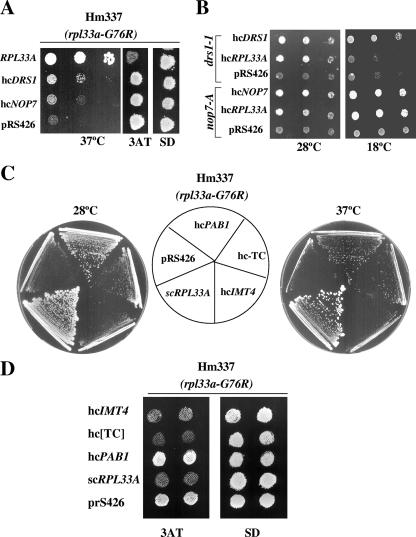
To discover possible interactions of rpL33 with other components of the pre-60S particles, we tested for functional suppression of the rpl33a-G76R mutation by overexpressing NOP7, which encodes a nucleolar protein essential for the biogenesis of the 60S subunit (1). The nop7-1 mutation or depletion of NOP7 leads to pre-rRNA processing defects similar to those described above for rpl33a-G76R mutants (1), and rpL33 copurifies with TAP-NOP7 (33). The nop7-1 mutation was identified in a genetic screen for mutations synthetically lethal with drs1 mutations (1). DRS1 encodes a DEAD box RNA helicase required for 60S subunit biogenesis (54), and most of the drs1 mutants are defective in the processing of 27S pre-rRNA to 25S rRNA (1, 6, 54). Overexpression of NOP7 or DRS1 (Materials and Methods) weakly suppressed the Slg− phenotype at 37°C but not the 3ATR phenotype of the rpl33a-G76R mutant Hm337 (Fig. 6A). Conversely, the hcRPL33A plasmid suppressed the Slg− phenotype produced by nop7-A at 28°C and 18°C (Fig. 6B) and that produced by nop7-C and nop7-F but not that produced by 10 other nop7 mutations (data not shown). The same test was conducted with the hcRPL33A plasmid on several drs1 mutants (drs1-1, -3, -5, and -6 mutants) (1). Only the Slg− phenotypes at 28°C of the drs1-1 (Fig. 6B) and drs1-6 (not shown) mutants were suppressed by overexpressing rpL33A. The allele-specific suppression of the nop7 and drs1 mutations by rpL33A overexpression observed here may indicate functional or physical interactions of rpL33A with two nucleolar proteins required for essential steps leading to the synthesis of 60S ribosomal subunits. rpL33A might facilitate the function of NOP7 in the exonucleolytic processing of the 27SA3 pre-rRNA to later processing intermediates and the mature 5.8Ss rRNA or stimulate the function of DRS1 in the production of 25S rRNA from 27S pre-rRNAs.
FIG. 6.
Genetic interactions of rpl33a-G76R. (A) Weak suppression of rpl33a-G76R by hcNOP7 and hcDRS1 plasmids. Transformants of the Hm337 mutant (rpl33a-G76R) carrying an empty vector (pRS426) or high-copy-number plasmids bearing RPL33A (pPM13), NOP7 (pJW6039), or DRS1 (pJW3015) were grown in SD-Ura medium at 28°C and serial dilutions (103 to 10 cells) plotted on plates of the same medium, which were incubated for 3 days at 37°C (left panel). Isolated colonies of each transformant were replica printed to plates containing 10 mM 3AT and to minimally supplemented SD plates and incubated for 3 days at 28°C (right panels). (B) Allele-specific suppression of the nop7 and drs1 mutations by the hcRPL33A plasmid. Mutant drs1-1 and nop7-A strains bearing an hcRPL33A plasmid (pPM13), empty vector (pR8426), or high-copy-number plasmids bearing DRS1 (pJW3015) or NOP7 (pJW6039) were grown as described for panel A and plotted on SC-Ura plates, which were incubated for 2 days at 28°C or for 5 days at 18°C. (C) Dosage suppressors of the Slg− phenotype of the rpl33a-G76R mutant. Transformants of Hm337 (rpl33a-G76R) carrying an empty vector (pRS426), a single-copy plasmid bearing RPL33A (pPM2), or high-copy-number plasmids bearing RPL33A (pPM13), PAB1 (pAS425), IMT4 (p2635), or the three subunits of eIF2 plus IMT4 (hc-TC; p3000) were streaked for single colonies on minimally supplemented SD medium and incubated at 28°C (3 days) or 37°C (4 days). (D) The Gcd− phenotype of rpl33a-G76R is partially suppressed by the hcIMT4 plasmid and the hc-TC, but not by the hcPAB1 plasmid. Isolated colonies of the same transformants as those described in panel C were replica printed to plates containing 10 mM 3AT and to SD plates and incubated for 3 days at 28°C.
Furthermore, we identified the gene encoding the poly(A) binding protein (PAB1) as a dosage suppressor of the Slg− phenotype of the rpl33a-G76R and ΔA mutants (Fig. 6C and data not shown). However, the hcPAB1 strain did not suppress the 3ATR/Gcd− phenotypes of the rpl33a-G76R and ΔA mutants (Fig. 6D and data not shown). It also did not suppress the Slg− phenotypes conferred by mutations that reduce the assembly of the TC, including the gcd11-505 mutation in the γ subunit of eIF2, the gcd1-501 mutation in the γ subunit of eIF2B (the GEF for eIF2), and the gcd14-2 mutation in the catalytic subunit of the methyltransferase that modifies position 58 in tRNAiMet (data not shown). Accordingly, we considered the possibility that the hcPAB1 plasmid only reduces the deficiency in ribosomal subunits in the rpl33a-G76R and ΔA mutants. In agreement with this idea, we found that PAB1 overexpression leads to increased levels of both 25S and 18S rRNAs at 28°C in rpl33a-G76R cells, leaving a moderate deficit in 25S rRNA versus 18S rRNA steady-state levels (see Fig. S1A and B in the supplemental material). Thus, the hcPAB1 plasmid should increase the amounts of 60S and 40S subunits, without eliminating a relative deficiency in 60S subunits, in rpl33a-G76R cells. Importantly, hcPAB1 did not reduce significantly the abundance of half-mers observed in the polysome profiles of the rpl33a-G76R mutant (see Fig. S1C in the supplemental material). The last finding, together with the failure of the hcPAB1 plasmid to reduce the Gcd− phenotype, indicates that a strong subunit joining defect persists in rpl33a-G76R cells overexpressing PAB1 despite the elevated levels of ribosomal subunits. This reinforces our conclusion that rpl33a-G76R confers a specific defect in subunit joining apart from its effect in reducing 60S-subunit levels.
Finally, we found that overexpressing tRNAiMet from a high-copy-number plasmid containing the IMT4 gene (hcIMT4) suppressed the 3ATR phenotypes of rpl33a-G76R and ΔA cells but not the corresponding Slg− phenotypes. Similar results were obtained for a high-copy-number plasmid encoding the three subunits of eIF2 in addition to IMT4 to achieve overexpression of the TC (Fig. 6C and D). To explain these results, we propose that the delay in subunit joining at the uORF4 AUG codon in rpl33a mutants leads to the dissociation of tRNAiMet from the 40S subunit and allows the resumption of scanning from uORF4 to GCN4 as the basis for the Gcd− phenotypes of these mutants. In this view, raising the concentration of tRNAiMet would increase the occupancy of the TC on the stalled 40S subunits and thereby prevent their leaky scanning of the uORF4 AUG codon. As this would not correct the defect in ribosome biogenesis and subunit joining defects in these mutants, this model can explain why the hcIMT4 plasmid does not suppress the Slg− phenotypes of the rpl33a-G76R and ΔA mutants.
DISCUSSION
In this report, we have shown that the rpl33a-G76R mutation in RPL33A leads to a defect in 60S-ribosomal-subunit accumulation and constitutive derepression of GCN4 translation. It was shown previously that reducing the concentration of 60S subunits by deleting RPL16B (currently named RPL11B) produced a moderate Gcd− phenotype, partially derepressing the expression of GCN4 and its target gene HIS4 (23). Thus, it was possible that rpl33a-G76R confers a Gcd− phenotype simply by decreasing rpL33A abundance or preventing its incorporation into 60S subunits, with attendant reduction in the 60S-subunit concentration. However, rpl33a-G76R confers a much stronger Gcd− phenotype than does the deletion of RPL33A, producing greater resistance to 3AT and more-extensive derepression of GCN4-lacZ and HIS4-lacZ expression. Importantly, the stronger Gcd− phenotype of rpl33a-G76R was evident at 28°C, at which temperature it produces a less severe reduction in 60S-subunit accumulation than does ΔA. Furthermore, rpl33a-G76R produces a greater reduction in the rate of methionine incorporation than does ΔA at 28°C. Together, these findings suggest that rpl33a-G76R alters the function of rpL33A in a way that impairs translation initiation and GCN4 translational control more dramatically than does a simple reduction in rpL33A abundance. Given that the 60S subunit does not participate in translation initiation until the subunit joining step, it is likely that rpl33a-G76R alters the 60S subunit in a way that impedes the joining reaction. We propose that subunits containing the rpl33a-G76R product are impaired for subunit joining and that they compete with the functional subunits containing rpL33B. (The partial dominance of its Gcd− phenotype shows that competition by defective subunits containing rpl33a-G76R is evident even in the presence of WT rpL33A in heterozygous diploids.) The absence of rpL33A in the ΔA mutant and the attendant reduction in 40S-subunit levels also reduce the rate of subunit joining, but in this case there is no competition for the functional subunits.
We found that overexpressing tRNAiMet, alone or together with eIF2 (TC), partially suppresses the Gcd− phenotype of the rpl33a-G76R mutation. Such suppression also occurs with gcd mutations in subunits of eIF2 or eIF2B that derepress GCN4 by reducing TC formation (38). This has likewise been observed for mutations in initiation factors that impair the rate of TC recruitment, e.g., eIF1A (21). All of the latter mutations are thought to produce Gcd− phenotypes by reducing the rate of TC recruitment to 40S subunits scanning the GCN4 mRNA leader after they complete the translation of uORF1, increasing the proportion of 40S subunits that reach uORF4 without rebinding the TC and, consequently, bypassing uORF4 and reinitiating downstream at GCN4 instead. Boosting TC levels in these mutants restores efficient reinitiation at uORF2 to -4 and thereby prevents reinitiation at GCN4 (38).
It is unlikely that the rpl33a-G76R or ΔA mutations derepress GCN4 translation by a mechanism involving impaired TC loading. First, the mutations affect the 60S subunit, which should not be present during the process of scanning from uORF1 to uORF4. Second, the mutations derepress GCN4-lacZ fusions containing either all four uORFs or uORF4 alone by similar degrees. Gcd− mutations that impair TC assembly or TC loading on 40S subunits have little effect on GCN4 expression in the presence of uORF4 alone, because their effects on TC recruitment are greatly magnified by the reinitiation process that occurs after uORF1 translation (38). Because the rpl33a mutations clearly affect 40S- to 60S-sububnit joining, giving rise to abundant half-mer polysomes, the simplest interpretation of their Gcd− phenotypes is that they allow 40S subunits that reach uORF4 as fully assembled PICs (i.e., containing the TC) to bypass uORF4 because of a defect in subunit joining. The delay in subunit joining could allow a fraction of these PICs to abort reinititation at uORF4, resume scanning, and reinitiate further downstream at GCN4. This aberrant form of leaky scanning would also occur at the elongated version of uORF1, which cannot support reinitiation at GCN4, explaining why rpl33a-G76R decreases the inhibitory effect of this element, as well as solitary uORF4, on GCN4 translation.
One way to account for this aberrant leaky scanning is to propose that a defect in subunit joining at the uORF4 AUG codon increases the probability of dissociation of Met-tRNAiMet from the P site of the 40S subunit, which then allows the resumption of scanning downstream to GCN4. This would occur following hydrolysis of GTP by the TC and the release of eIF2-GDP from the PIC. Increasing the concentration of Met-tRNAiMet would decrease its rate of disassembly from the PIC by mass action, enabling the PIC to remain intact long enough for the subunit joining reaction (impaired by rpl33a-G76R) to occur at the uORF4 AUG codon. Whereas overexpressing tRNAiMet could prevent Met-tRNAiMet loss at uORF4 (and thus reverse the Gcd− phenotype), it would not correct the low rate of subunit joining conferred by rpl33a-G76R and ΔA. Furthermore, overexpressing Met-tRNAiMet should not suppress the ribosome biogenesis defects of these mutants. The last stipulations of our model can explain why overexpressing Met-tRNAiMet fails to suppress the Slg− phenotypes of the rpl33a mutants.
It is noteworthy that ΔA differs from rpl33a-G76R in preventing full derepression of HIS4-lacZ expression in GCN2 GCN3 cells under amino acid starvation conditions (last two lines of Table 3). In this respect, ΔA resembles the Δrpl16b mutation described previously (23) in conferring moderate Gcd− and Gcn− phenotypes. We have proposed that the Gcn− phenotype of the Δrpl16b mutation results from leaky scanning of the GCN4 AUG codon under starvation conditions, whereas leaky scanning of uORF4 elicits the Gcd− phenotype of this mutation under nonstarvation conditions (23). Leaky scanning of uORF1 could also contribute to the Gcn− phenotype because translation of uORF1 is required for reinitiation at GCN4 under starvation conditions. To explain why rpl33a-G76R produces only a strong Gcd− phenotype, we suggest that its more severe subunit joining defect than that of ΔA or Δrpl16b results in such a high level of leaky scanning at uORF2 to -4 that these defects overpower the opposing effects of leaky scanning at uORF1 and GCN4, to yield a net derepression of GCN4 translation (and HIS4-lacZ expression) that is comparable to the WT-induced level.
It is interesting that the Slg− phenotypes of rpl33a-G76R and ΔA mutants are partially suppressed by overexpression of PAB1. Because PAB1 has been implicated in 60S-subunit joining in yeast (61), we considered the possibility that its overexpression corrects the defect in subunit joining imposed by rpl33a-G76R and ΔA. However, this possibility is at odds with the fact that the hcPAB1 plasmid does not significantly diminish half-mer formation in rpl33a-G76R cells, nor does it suppress the Gcd− phenotypes of either rpl33a mutant. The hcPAB1 plasmid also does not reduce the Slg− or Gcn− phenotypes of a deletion of the gene encoding the subunit joining factor eIF5B (Δfun12) (data not shown). Thus, we conclude that PAB1 overexpression does not rescue defective subunit joining. Interestingly, the hcPAB1 plasmid substantially increased both 25S and 18S rRNA levels (see Fig. S1A and B in the supplemental material) without fully correcting the reduced 25S/18S rRNA ratio in rpl33a-G76R cells. Hence, we think that the hcPAB1 plasmid suppresses the Slg− phenotype of rpl33a-G76R cells primarily by boosting ribosomal-subunit levels but without completely eliminating subunit imbalance. More work is required to determine whether PAB1 overexpression diminishes defects in subunit biogenesis when rpL33A is either mutated or completely absent or whether it stabilizes defective subunits in the rpl33a-G76R mutants.
It was shown previously that the hcPAB1 plasmid is a dosage suppressor of the temperature-sensitivity phenotypes conferred by the rpc31-236 and rpc10-30 mutations in Pol III subunits that substantially reduce the steady-state levels of tRNAiMet (57). However, we found that the hcPAB1 plasmid did not suppress the growth defects of mutations in eIF2, eIF2B, or the m1A58 methyltransferase for tRNAiMet. As noted above, the Slg− and Gcd− phenotypes of these mutants are suppressed by overexpressing tRNAiMet (8; data not shown), consistent with their defects in tRNAiMet synthesis or TC assembly. Thus, it is unclear whether suppression of the growth defects of rpc31-236 and rpc10-30 mutants by the hcPAB1 plasmid involves ameliorating a defect in TC assembly.
The idea that the rpl33a-G76R mutation interferes with subunit joining is consistent with several indications that rpL33 is required for a correct interaction of the 60S subunit with the 40S subunit. Cross-linking data suggest that L35a, the rat homolog of yeast rpL33, is located near the A and P sites of the 60S subunit (68). Moreover, L35a binds in vitro to initiator tRNA and elongator tRNAs (69). Finally, rpL33 was found to interact with 60S protein rpL2A (26), which participates in a conserved protein-rRNA intersubunit bridge at the 40S-subunit-60S-subunit interface (63). The rpl33a-G76R mutation introduces an extra positively charged residue into the rigid loop that belongs to a putative RNA-binding domain in rpL33, and we found recently that replacement of Gly-76 with Lys or the nearby Gly-69 with Arg also yields strong Gcd− phenotypes (unpublished observations). Hence, it is tempting to propose that these mutations lead to an inappropriate interaction of rpL33 with Met-tRNAiMet in the P site or a segment of rRNA that somehow interferes with subunit joining.
Our findings indicate that rpL33 also plays an important role in ribosome biogenesis. Shifting the rpl33a-G76R mutant to 37°C leads to a progressive decrease in the ratio of the 60S subunit to the 40S subunit, indicating a relatively greater effect on 60S- versus 40S-subunit accumulation. This could result from impairment of 60S biogenesis, decreased stability of preexisting 60S subunits, or both. A defect in 60S subunit biogenesis is indicated by our Northern analysis of precursor and mature rRNAs after shifting the rpl33a-G76R mutant to 37°C. We observed an accumulation of the 35S precursor and a decrease in 27S and 20S pre-rRNAs, indicating a reduction in the rate of 35S cleavage at the A0, A1, and A2 sites in the 90S preribosomal particle that generates the pre-40S and pre-60S particles. There is also formation of aberrant <25S and 22.5S species that result from cleavage at the C2 or D processing sites in the absence of A0, A1, and A2 cleavage. Thus, the rpl33a-G76R mutation reduces the rate of processing events in the beginning of the pathway required for both 40S and 60S subunit biogenesis. Consistent with this, the amounts of mature 25S and 18S rRNA relative to total RNA decrease in the mutant at the restrictive temperature. The rpl33a-G76R mutation also increases the ratio of the 27SA2 precursor to mature 25S rRNA, suggesting an impairment of processing steps in the pre-60S particle. By contrast, the ratio of the 20S precursor to mature 18S rRNA is not elevated in the mutant, indicating efficient maturation and export of the 40S subunit.
Thus, our results strongly suggest that rpL33A is a component of pre-60S particles and contributes to one or more critical steps in the maturation of 60S subunits. Consistent with this idea, there is mass spectrometry data supporting the presence of rpL33 in early pre-60S particles (20, 51). Second, rpL33 has been shown to copurify with NOP7, a nucleolar protein essential for the processing and maturation of 27S pre-rRNA and for 60S subunit biogenesis (33). Third, we found that overexpression of WT rpL33A confers allele-specific suppression of mutations in NOP7 and DRS1, encoding other nonribosomal components of pre-60S particles.
Considering that the mutation of RPL33A strongly impairs A0-A1-A2 processing, it could be proposed that rpL33 also resides in the 90S preribosome and directly influences these early processing events. However, it is important to note that the inhibition of late steps in the 60S pre-rRNA processing pathway seems to indirectly affect early steps in the processing of 35S pre-rRNA (73). Also, the possibility that processing defects could be secondary consequences of reduced translation of r-proteins or other ribosome biogenesis factors in the rpl33 mutant backgrounds cannot be excluded. Consistent with the first idea, rpL33 was found in the 90S preribosome described in the 1970s (66) and was classified as an early assembled r-protein (41, 42). Moreover, recent mass spectrometry data revealed an association of rpL33 with PWP2, a conserved 90S preribosomal component essential for proper endonucleolytic cleavage of 35S rRNA at A0-A1-A2 (26). It should be noted, however, that other large preribosomal particles sedimenting at ∼90S may correspond to the earliest pre-60S particles (67), including the highly complex nucleolar transitional pre-40S-60S particles (reviewed in reference 64). The NSA3-associating pre-rRNP complex (51) is an example of a highly complex nucleolar transitional pre-40S -60S particle, and rpL33 was found associated with NSA3/CIC1 (26). Nevertheless, whereas PWP2 is present in the earliest nucleolar pre-90S particle, it was not detected in the NSA3-associated pre-rRNP complex. Thus, the association with PWP2 favors the idea that rpL33 might reside in the classical 90S preribosome.
The pre-rRNA processing phenotypes of rpl mutants were not systematically investigated, as was recently done for rps mutants (22). Whereas the absence or functional inactivation of r-protein L5, L11, L25, L28, or L30 leads to an accumulation of 27S pre-rRNAs, we observed decreased steady-state levels of 27S and 20S pre-rRNAs, with little accumulation of 23S pre-RNA in rpl33a-G76R cells at 37°C. The delay in processing at A0-A3 leads to low levels of the 23S species, and subsequent cleavage at C2 or D produces <25S and 22.5S species in rpl33a-G76R cells. The <25S and 22.5S species were described previously (i.e., in references 9, 22, 40, and 52) but not observed in the rpl mutants analyzed so far. Hence, it appears that rpL33 makes a distinctive contribution to multiple pre-rRNA processing reactions.
Supplementary Material
Acknowledgments
We thank Dionisio Martín-Zanca and Rosa Esteban for helpful suggestions regarding this work and Thomas Dever for critical reading of the manuscript. We also thank John L. Woolford for the nop7 and drs1 mutants and for the hcNOP7 and hcDRS1 plasmids, Alan Sachs for the hcPAB1 plasmid, and Javier de las Rivas for help with the 3D model of rpL33.
P.M.-M. has been supported by fellowship AP2000-4183 granted by the Spanish Ministerio de Educación y Ciencia (MEC). M.T. was supported by MEC grants BMC 2001-1676 and PTR-1995-1010.
Footnotes
Published ahead of print on 4 June 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, C. C., J. Jakovljevic, J. Roman, P. Harnpicharnchai, and J. L. Woolford, Jr. 2002. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA 8:150-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J., L. Phan, R. Cuesta, B. A. Carlson, M. Pak, K. Asano, G. R. Bjork, M. Tamame, and A. G. Hinnebusch. 1998. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev. 12:3650-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asano, K., L. Phan, J. Anderson, and A. G. Hinnebusch. 1998. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 17:18573-18585. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, J., P. Grandi, O. Gadal, T. Lessmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 5.Belk, J. P., F. He, and A. Jacobson. 1999. Overexpression of truncated Nmd3p inhibits protein synthesis in yeast. RNA 5:1055-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, K. A., S. Granneman, A. V. Lee, S. Manickam, and S. J. Baserga. 2006. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases involved in large ribosomal subunit biogenesis. Mol. Cell. Biol. 26:1195-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke, D., D. Dawson, and T. Stearns. 2000. Assay of β-galactosidase in yeast, p. 125. In D. Burke, D. Dawson, and T. Stearns (ed.), Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 8.Calvo, O., R. Cuesta, J. Anderson, N. Gutierrez, M. T. García-Barrio, A. G. Hinnebusch, and M. Tamame. 1999. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4167-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colley, A., J. D. Beggs, D. Tollervey, and D. L. Lafontaine. 2000. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C+D snoRNP U3. Mol. Cell. Biol. 20:7238-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuesta, R., A. G. Hinnebusch, and M. Tamame. 1998. Identification of GCD14 and GCD15, novel genes required for translational repression of GCN4 mRNA in Saccharomyces cerevisiae. Genetics 148:1007-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Cruz, J., D. Kressler, and P. Linder. 2004. Ribosomal subunit assembly, p. 258-285. In M. O. J. Olson (ed.), The nucleolus. Kluwer Academic Publishers, New York, NY.
- 12.Deshmukh, M., Y. F. Tsay, A. G. Paulovich, and J. L. Woolford, Jr. 1993. Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 13:2835-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutschbauer, A. M., D. F. Jaramillo, M. Proctor, J. Kumm, M. E. Hillenmeyer, R. W. Davis, C. Nislow, and G. Giaever. 2005. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169:1915-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. F. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585-596. [DOI] [PubMed] [Google Scholar]
- 15.Dever, T. E., W. Yang, S. Åström, A. S. Byström, and A. G. Hinnebusch. 1995. Modulation of tRNAiMet, eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2·GTP·Met-tRNAiMet ternary complexes. Mol. Cell. Biol. 15:6351-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dresios, J., I. L. Derkatch, S. W. Liebman, and D. Synetos. 2000. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39:7236-7244. [DOI] [PubMed] [Google Scholar]
- 18.Dresios, J., P. Panopoulos, C. P. Frantziou, and D. Synetos. 2001. Yeast ribosomal protein deletion mutants possess altered peptidyltransferase activity and different sensitivity to cycloheximide. Biochemistry 40:8101-8108. [DOI] [PubMed] [Google Scholar]
- 19.Dresios, J., P. Panopoulos, K. Suzuki, and D. Synetos. 2003. A dispensable yeast ribosomal protein optimizes peptidyltransferase activity and affects translocation. J. Biol. Chem. 278:3314-3322. [DOI] [PubMed] [Google Scholar]
- 20.Fatica, A., A. D. Cronshaw, M. Dlakic, and D. Tollervey. 2002. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell 9:341-351. [DOI] [PubMed] [Google Scholar]
- 21.Fekete, C. A., D. J. Applefield, S. A. Blakely, N. Shirokikh, T. Pestova, J. R. Lorsch, and A. G. Hinnebusch. 2005. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J. 24:3588-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira-Cerca, S., G. Poll, P. E. Gleizes, H. Tschochner, and P. Milkereit. 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell 20:263-275. [DOI] [PubMed] [Google Scholar]
- 23.Foiani, M., A. M. Cigan, C. J. Paddon, S. Harashima, and A. G. Hinnebusch. 1991. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:3203-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallagher, J. E., D. A. Dunbar, S. Granneman, B. M. Mitchell, Y. Osheim, A. L. Beyer, and S. J. Baserga. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18:2506-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 26.Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche, M. Marzioch, C. Rau, L. J. Jensen, S. Bastuck, B. Dumpelfeld, A. Edelmann, M. A. Heurtier, V. Hoffman, C. Hoefert, K. Klein, M. Hudak, A. M. Michon, M. Schelder, M. Schirle, M. Remor, T. Rudi, S. Hooper, A. Bauer, T. Bouwmeester, G. Casari, G. Drewes, G. Neubauer, J. M. Rick, B. Kuster, P. Bork, R. B. Russell, and G. Superti-Furga. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631-636. [DOI] [PubMed] [Google Scholar]
- 27.Grallath, S., J. P. Schwarz, U. M. Bottcher, A. Bracher, F. U. Hartl, and K. Siegers. 2006. L25 functions as a conserved ribosomal docking site shared by nascent chain-associated complex and signal-recognition particle. EMBO Rep. 7:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Grandi, P., V. Rybin, J. Bassler, E. Petfalski, D. Strauss, M. Marzioch, T. Schafer, B. Kuster, H. Tschochner, D. Tollervey, A. C. Gavin, and E. Hurt. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell 10:105-115. [DOI] [PubMed] [Google Scholar]
- 29.Granneman, S., and S. J. Baserga. 2004. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 296:43-50. [DOI] [PubMed] [Google Scholar]
- 30.Granneman, S., M. R. Nandineni, and S. J. Baserga. 2005. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol. Cell. Biol. 25:10352-10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grant, C. M., P. F. Miller, and A. G. Hinnebusch. 1994. Requirements for intercistronic distance and level of eIF-2 activity in reinitiation on GCN4 mRNA vary with the downstream cistron. Mol. Cell. Biol. 14:2616-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harashima, S., and A. G. Hinnebusch. 1986. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 6:3990-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 34.Helser, T. L., R. A. Baan, and A. E. Dahlberg. 1981. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide. Mol. Cell. Biol. 1:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinnebusch, A. G. 1985. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:2349-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinnebusch, A. G. 1988. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 52:248-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinnebusch, A. G. 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J. Biol. Chem. 272:21661-21664. [DOI] [PubMed] [Google Scholar]
- 38.Hinnebusch, A. G. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59:407-450. [DOI] [PubMed] [Google Scholar]
- 39.Hinnebusch, A. G., and G. R. Fink. 1983. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 80:5374-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kressler, D., J. de la Cruz, M. Rojo, and P. Linder. 1998. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1855-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kressler, D., P. Linder, and J. de la Cruz. 1999. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7897-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kruiswijk, T., R. J. Planta, and J. M. Krop. 1978. The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta 517:378-389. [DOI] [PubMed] [Google Scholar]
- 43.Lucchini, G., A. G. Hinnebusch, C. Chen, and G. R. Fink. 1984. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mager, W. H., R. J. Planta, J. P. G. Ballesta, J. C. Lee, K. Mizuta, K. Suzuki, J. R. Warner, and J. L. Woolford, Jr. 1997. A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res. 25:4872-4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meskauskas, A., and J. D. Dinman. 2001. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA 7:1084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meskauskas, A., A. N. Petrov, and J. D. Dinman. 2005. Identification of functionally important amino acids of ribosomal protein L3 by saturation mutagenesis. Mol. Cell. Biol. 25:10863-10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milkereit, P., H. Kuhn, N. Gas, and H. Tschochner. 2003. The pre-ribosomal network. Nucleic Acids Res. 31:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moritz, M., B. A. Pulaski, and J. L. Woolford, Jr. 1991. Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol. Cell. Biol. 11:5681-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müeller P. P., and A. G. Hinnebusch. 1986. Multiple upstream AUG codons mediate translational control of GCN4. Cell 45:201-207. [DOI] [PubMed] [Google Scholar]
- 50.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nissan, T. A., J. Bassler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oeffinger, M., A. Fatica, M. P. Rout, and D. Tollervey. 2007. Yeast Rrp14p is required for ribosomal subunit synthesis and for correct positioning of the mitotic spindle during mitosis. Nucleic Acids Res. 35:1354-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raué, H. A. 2004. Pre-ribosomal RNA processing and assembly in Saccharomyces cerevisiae: the machine that makes the machine, p. 199-222. In M. O. J. Olson (ed.), The nucleolus. Kluwer Academic Publishers, New York, NY.
- 54.Ripmaster, T. L., G. P. Vaughn, and J. L. Woolford, Jr. 1992. A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc. Natl. Acad. Sci. USA 89:11131-11135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rose, M. D., P. Novick, J. H. Thomas, D. Botstein, and G. R. Fink. 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60:237-243. [DOI] [PubMed] [Google Scholar]
- 56.Rotenberg, M. O., M. Moritz, and J. L. Woolford, Jr. 1988. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 2:160-172. [DOI] [PubMed] [Google Scholar]
- 57.Rubbi, L., S. Labarre-Mariotte, S. Chedin, and P. Thuriaux. 1999. Functional characterization of ABC10α, an essential polypeptide shared by all three forms of eukaryotic DNA-dependent RNA polymerases. J. Biol. Chem. 27:31485-31492. [DOI] [PubMed] [Google Scholar]
- 58.Saveanu, C., D. Bienvenu, A. Namane, P. E. Gleizes, N. Gas, A. Jacquier, and M. Fromont-Racine. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20:6475-6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schäfer, T., D. Strauss, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Searfoss, A., T. E. Dever, and R. Wickner. 2001. Linking the 3′ poly(A) tail to the subunit-joining step of the translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5B (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol. 21:4900-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Snyder, D. A., et al. March 2004. posting date. Solution structure of the 50S ribosomal protein L35Ae from Pyrococcus furiosus. Northeast Structural Genomics Consortium target PfR48. http://www.rcsb.org/pdb.
- 63.Spahn, C. M., R. Beckmann, N. Eswar, P. A. Penczek, A. Sali, G. Blobel, and J. Frank. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit-subunit interactions. Cell 107:373-386. [DOI] [PubMed] [Google Scholar]
- 64.Takahashi, N., M. Yanagida, S. Fujiyama, T. Hayano, and T. Isobe. 2003. Proteomic snapshot analyses of preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom. Rev. 22:287-317. [DOI] [PubMed] [Google Scholar]
- 65.Tornow, J., and G. M. Santangelo. 1994. Saccharomyces cerevisiae ribosomal protein L37 is encoded by duplicate genes that are differentially expressed. Curr. Genet. 25:480-487. [DOI] [PubMed] [Google Scholar]
- 66.Trapman, J., J. Retèl, and R. J. Planta. 1975. Ribosomal precursor particles from yeast. Exp. Cell Res. 90:95-104. [DOI] [PubMed] [Google Scholar]
- 67.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 68.Uchiumi, T., M. Kikuchi, K. Terao, and K. Ogata. 1985. Cross-linking study on protein topography of rat liver 60S ribosomal subunits with 2-iminothiolane. J. Biol. Chem. 260:5675-5682. [PubMed] [Google Scholar]
- 69.Ulbrich, N., E. Wool, P. Ackerman, and P. Sigler. 1980. The identification by affinity chromatography of the rat liver ribosomal proteins that bind to elongator and initiator transfer ribonucleic acids. J. Biol. Chem. 255:7010-7016. [PubMed] [Google Scholar]
- 70.Underwood, M. R., and H. M. Fried. 1990. Characterization of nuclear localizing sequences derived from yeast ribosomal protein L29. EMBO J. 9:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Valasek, L., A. A. Mathew, B. S. Shin, K. H. Nielsen, B. Szamecz, and A. G. Hinnebusch. 2003. The yeast eIF3 subunits TIF32/α, NIP1/χ, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 17:786-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Beekvelt, C. A., M. de Graaff-Vincent, A. W. Faber, J. van't Riet, J. Venema, and H. A. Raué. 2001. All three functional domains of the large ribosomal subunit protein L25 are required for both early and late pre-rRNA processing steps in Saccharomyces cerevisiae. Nucleic Acids Res. 29:5001-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 74.Vilardell, J., and J. R. Warner. 1997. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 17:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wach, A. 1996. PCR synthesis of marker cassettes with long-flanking homology regions for disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 76.Wach, A., A. Brachat, R. Pöhlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 77.Warner, J. R., J. Vilardell, and J. H. Sohn. 2001. Economics of ribosome biosynthesis. Cold Spring Harbor Symp. Quant. Biol. 66:567-574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.