Abstract
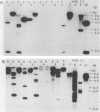
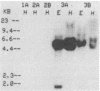
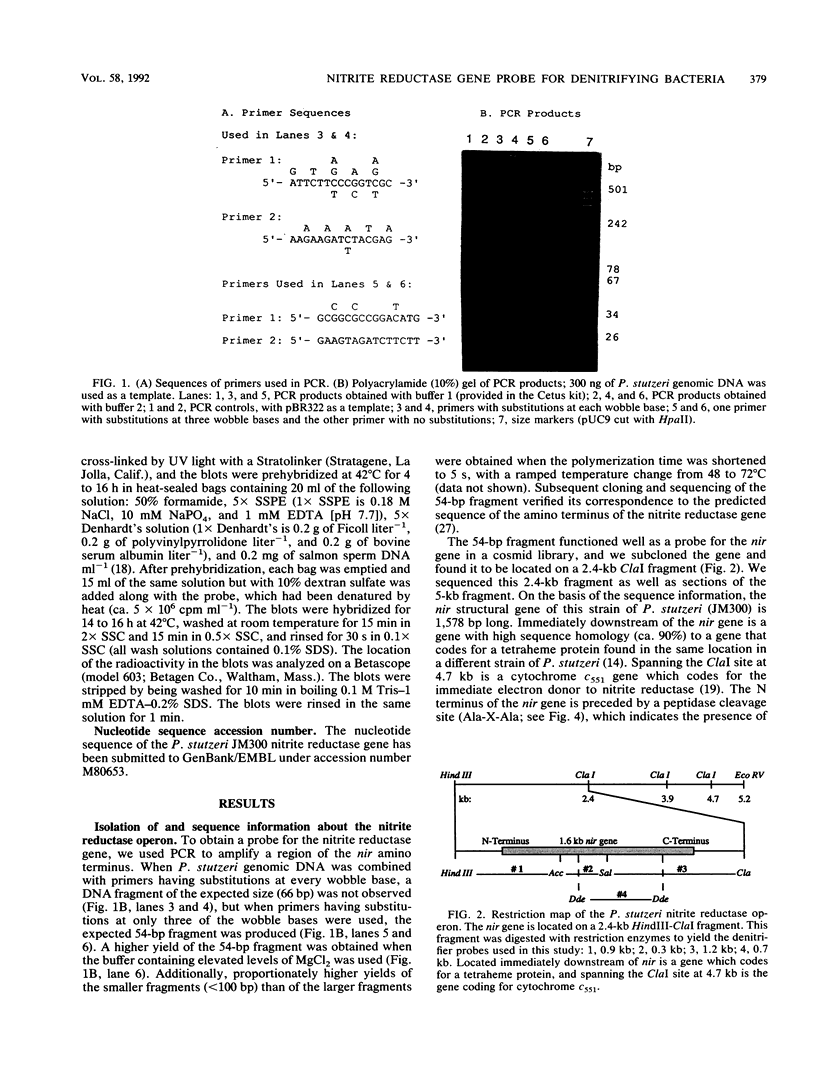
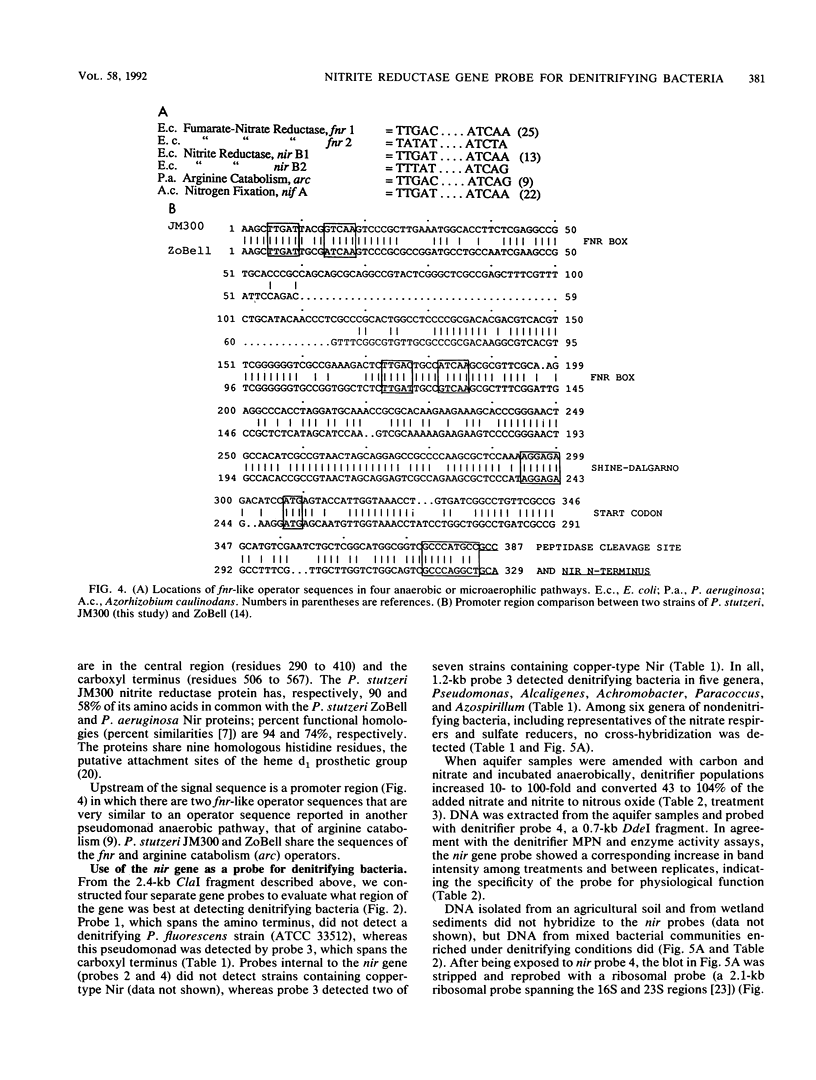
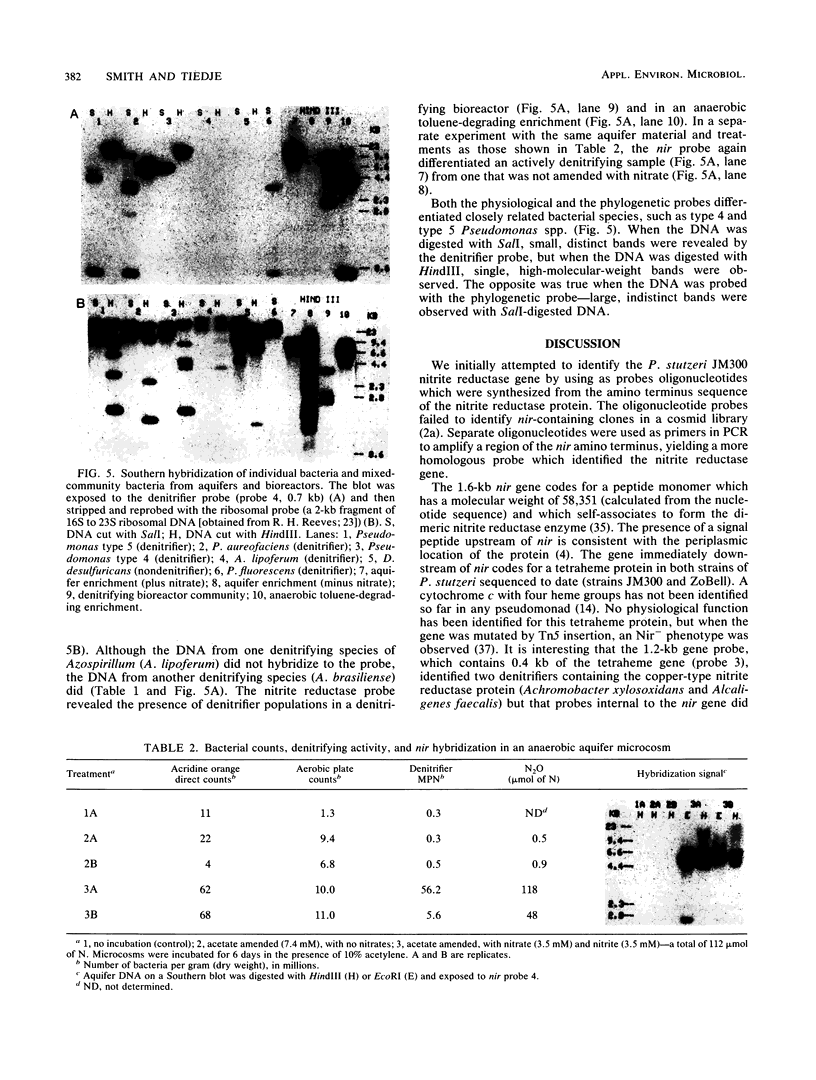
The dissimilatory nitrite reductase gene (nir) from denitrifying bacterium Pseudomonas stutzeri JM300 was isolated and sequenced. In agreement with recent sequence information from another strain of P. stutzeri (strain ZoBell), strain JM300 nir is the first gene in an operon and is followed immediately by a gene which codes for a tetraheme protein; 2.5 kb downstream from the nitrite reductase carboxyl terminus is the cytochrome c551 gene. P. stutzeri JM300 nir is 67% homologous to P. aeruginosa nir and 88% homologous to P. stutzeri ZoBell nir. Within the nitrite reductase promoter region is an fnr-like operator very similar to an operator upstream of a separate anaerobic pathway, that for arginine catabolism in P. aeruginosa. The denitrification genes in P. stutzeri thus may be under the same regulatory control as that found for other anaerobic pathways of pseudomonads. We have generated gene probes from restriction fragments within the nitrite reductase operon to evaluate their usefulness in ecology studies of denitrification. Probes generated from the carboxyl terminus region hybridized to denitrifying bacteria from five separate genera and did not cross-hybridize to any nondenitrifying bacteria among six genera tested. The denitrifier probes were successful in detecting denitrifying bacteria from samples such as a bioreactor consortium, aquifer microcosms, and denitrifying toluene-degrading enrichments. The probes also were used to reveal restriction fragment length polymorphism patterns indicating the diversity of denitrifiers present in these mixed communities.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coyne M. S., Arunakumari A., Averill B. A., Tiedje J. M. Immunological identification and distribution of dissimilatory heme cd1 and nonheme copper nitrite reductases in denitrifying bacteria. Appl Environ Microbiol. 1989 Nov;55(11):2924–2931. doi: 10.1128/aem.55.11.2924-2931.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne M. S., Arunakumari A., Pankratz H. S., Tiedje J. M. Localization of the cytochrome cd1 and copper nitrite reductases in denitrifying bacteria. J Bacteriol. 1990 May;172(5):2558–2562. doi: 10.1128/jb.172.5.2558-2562.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criddle C. S., DeWitt J. T., Grbić-Galić D., McCarty P. L. Transformation of carbon tetrachloride by Pseudomonas sp. strain KC under denitrification conditions. Appl Environ Microbiol. 1990 Nov;56(11):3240–3246. doi: 10.1128/aem.56.11.3240-3246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M., Gamper M., Zimmermann A., Haas D. Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J Bacteriol. 1991 Mar;173(5):1598–1606. doi: 10.1128/jb.173.5.1598-1606.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T. N., Betlach M. R., Tiedje J. M. Numerically dominant denitrifying bacteria from world soils. Appl Environ Microbiol. 1977 Apr;33(4):926–939. doi: 10.1128/aem.33.4.926-939.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Jayaraman P. S., Peakman T. C., Busby S. J., Quincey R. V., Cole J. A. Location and sequence of the promoter of the gene for the NADH-dependent nitrite reductase of Escherichia coli and its regulation by oxygen, the Fnr protein and nitrite. J Mol Biol. 1987 Aug 20;196(4):781–788. doi: 10.1016/0022-2836(87)90404-9. [DOI] [PubMed] [Google Scholar]
- Jüngst A., Wakabayashi S., Matsubara H., Zumft W. G. The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett. 1991 Feb 25;279(2):205–209. doi: 10.1016/0014-5793(91)80150-2. [DOI] [PubMed] [Google Scholar]
- Knowles R. Denitrification. Microbiol Rev. 1982 Mar;46(1):43–70. doi: 10.1128/mr.46.1.43-70.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn E. P., Zeyer J., Eicher P., Schwarzenbach R. P. Anaerobic degradation of alkylated benzenes in denitrifying laboratory aquifer columns. Appl Environ Microbiol. 1988 Feb;54(2):490–496. doi: 10.1128/aem.54.2.490-496.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. F., DeMoss J. A. Location of sequences in the nar promoter of Escherichia coli required for regulation by Fnr and NarL. J Biol Chem. 1988 Sep 25;263(27):13700–13705. [PubMed] [Google Scholar]
- Nordling M., Young S., Karlsson B. G., Lundberg L. G. The structural gene for cytochrome c551 from Pseudomonas aeruginosa. The nucleotide sequence shows a location downstream of the nitrite reductase gene. FEBS Lett. 1990 Jan 1;259(2):230–232. doi: 10.1016/0014-5793(90)80015-b. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Ratet P., Pawlowski K., Schell J., de Bruijn F. J. The Azorhizobium caulinodans nitrogen-fixation regulatory gene, nifA, is controlled by the cellular nitrogen and oxygen status. Mol Microbiol. 1989 Jun;3(6):825–838. doi: 10.1111/j.1365-2958.1989.tb00231.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw D. J., Guest J. R. Nucleotide sequence of the fnr gene and primary structure of the Enr protein of Escherichia coli. Nucleic Acids Res. 1982 Oct 11;10(19):6119–6130. doi: 10.1093/nar/10.19.6119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestrini M. C., Galeotti C. L., Gervais M., Schininà E., Barra D., Bossa F., Brunori M. Nitrite reductase from Pseudomonas aeruginosa: sequence of the gene and the protein. FEBS Lett. 1989 Aug 28;254(1-2):33–38. doi: 10.1016/0014-5793(89)81004-x. [DOI] [PubMed] [Google Scholar]
- Smith R. L., Duff J. H. Denitrification in a sand and gravel aquifer. Appl Environ Microbiol. 1988 May;54(5):1071–1078. doi: 10.1128/aem.54.5.1071-1078.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V., Goksøyr J., Daae F. L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990 Mar;56(3):782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschech A., Fuchs G. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch Microbiol. 1987 Sep;148(3):213–217. doi: 10.1007/BF00414814. [DOI] [PubMed] [Google Scholar]
- Viebrock A., Zumft W. G. Molecular cloning, heterologous expression, and primary structure of the structural gene for the copper enzyme nitrous oxide reductase from denitrifying Pseudomonas stutzeri. J Bacteriol. 1988 Oct;170(10):4658–4668. doi: 10.1128/jb.170.10.4658-4668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner U., Kemmler J., Weilenmann H. U., Egli T., el-Banna T., Auling G. Isolation and growth of a bacterium able to degrade nitrilotriacetic acid under denitrifying conditions. Biodegradation. 1990;1(1):31–41. doi: 10.1007/BF00117049. [DOI] [PubMed] [Google Scholar]
- Ward D. M., Weller R., Bateson M. M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990 May 3;345(6270):63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- Weeg-Aerssens E., Wu W. S., Ye R. W., Tiedje J. M., Chang C. K. Purification of cytochrome cd1 nitrite reductase from Pseudomonas stutzeri JM300 and reconstitution with native and synthetic heme d1. J Biol Chem. 1991 Apr 25;266(12):7496–7502. [PubMed] [Google Scholar]
- Zumft W. G., Döhler K., Körner H., Löchelt S., Viebrock A., Frunzke K. Defects in cytochrome cd1-dependent nitrite respiration of transposon Tn5-induced mutants from Pseudomonas stutzeri. Arch Microbiol. 1988;149(6):492–498. doi: 10.1007/BF00446750. [DOI] [PubMed] [Google Scholar]