Abstract
Tear lipocalin (TL) may stabilize the lipid layer of tears through a molten globule state triggered by low pH. EPR spectroscopy with site directed spin labeling, revealed the side chain mobility of residues on the G-strand of TL in a molten globule state; the G-strand retains β-sheet structure. All of the side chains of G strand residues become more loosely packed, especially residues 96–99. In contrast, the highly mobile side chain of residue 95 on the F-G loop, becomes tightly packed. ANS binding to TL in a molten globule state reestablishes tight packing around side chains that are oriented both inside and outside of the barrel. Unlike RBP and BLG; TL has no disulfide bond between G and H strands. It is likely that the central β-sheet in the molten globule state of lipocalins is stabilized by its interactions with the main α-helix, rather than the interstrand disulfide bond.
Keywords: Molten globule, Tear lipocalin, Lipocalin-1, ANS, EPR, β-lactoglobulin, Side-directed spin labeling
Introduction
Molten globule states of proteins, that have features in common with protein-folding intermediates [1–3], are suggested to be involved in such functions as the insertion of proteins into membranes [4], the release of bound ligands [1, 5, 6] and aggregation including amyloid fibril formation [7].
Molten globule states of proteins are characterized by having native-like secondary structure that lacks most of the specific tertiary interactions [1]. Three experimental criteria have been suggested to confirm native-molten globule transitions in proteins. Compared to the native state, a protein in the molten globule state should exhibit (1) small changes in far-UV CD spectrum (intact secondary structure), (2) significant loss in near-UV CD spectrum (loss of tertiary structure) and (3) enhancement of ANS fluorescence upon binding (exposure of hydrophobic sites).
Despite the large body of data available on molten globule states of proteins, site-specific information is limited. Nuclear magnetic resonance (NMR) spectroscopy and a molecular dynamic simulation have been employed to study molten globule states of some proteins [8–11]. The partial loss of secondary structure as well as side chain packing is site specific. Molten globule states of both RBP and equine BLG, which are members of lipocalin family, have similar features. Residues in the second β-sheet, also known as central β-sheet, (strands F, G, H and part of A) of the barrel retain native-like β-sheet structure in the molten globule state. The F,G, and H strands of BLG form hydrogen bonds early in a folding process and it has been hypothesized that the G and H strand strands act as folding initiation sites; residues on these strands have been catergorized as more stable with greater unfolding midpoints in urea [10]. Molten globule states of these RBP and BLG also reveal that the major α-helices are packed against the central β-sheets as in the native structures [9–11]. Both BLG and RBP also have interstrand disulfide bonds that join the G and H strand, which hypothetically restrain unfolding and facilitate retention of β structure in the lipocalins. Some lipocalins, including TL, the G and H strands are not linked by a disulfide bridge. The molten globule state of TL provides an opportunity to obtain information on the G strand in the absence of the potentially constraining disulfide bond.
Tear lipocalin (TL), as a member of the lipocalin family of proteins, exhibits cup shaped ligand binding fold within a continuously hydrogen-bonded β-barrel that is formed by eight antiparallel β-strands [12]. There is a single disulfide bridge linking the C terminal end with the CD loop. The solution structure of TL was resolved by site directed tryptophan fluorescence and revealed a capacious cavity that confers promiscuity in ligand binding [13]. These findings were verified by crystallography of TL [14].
Numerous putative functions, most of which are linked to the binding of various classes of ligand, have been suggested for TL [15–23]. One postulate is that TL stabilizes and modulates lipid in the tear film through a molten globule state triggered by low pH at the aqueous-lipid interface [5]. TL undergoes a native to molten globule transition at pH 3.0 that has been documented by changes in tertiary structure of the protein and ANS fluorescence enhancement [5].
There is a lack of site-specific information regarding structural consequences of ANS binding to proteins in molten globule states. Here, based on recent developments in understanding nitroxide side chain motion in β-sheet proteins [24], site directed spin labeling (SDSL) has been employed to extract site-specific information on dynamics of the G-strand of TL in a molten globule state with and without bound ANS. The results reported here, based on changes of dynamic modes of nitroxide side chain for G-strand residues, clearly reveal that in a molten globule state, the G-strand of TL retains β-sheet structure. However, packing of side chains, specifically for residues 95–96, are significantly altered. ANS binding to TL in a molten globule state reestablish tight packing around side chains that are oriented both inside and outside of the barrel.
Materials and methods
Materials
MTSL, (1-oxyl-2,2,5,5-tetramethyl-3-pyrroline-3-methyl) methanethiosulfonate, was obtained from Toronto Research Chemicals, Inc, Toronto, Ontario. ANS (8-anilino-1-naphthalenesulfonic acid) was purchased from Sigma. E. Coli, BL 21 (DE3) cells and pET 20b were obtained from Novagene. Oligonucleotide primers were obtained from Universal DNA, Inc. PCR II was obtained from Invitrogen, San Diego, CA. HiTrap columns were obtained from Pharmacia Biotech Inc., Piscataway, New Jersey. Gas-permeable TPX capillaries were obtained from Wilmad Glass Co. Inc., Buena, NJ.
Site-Directed Mutagenesis and Plasmid Construction
The TL cDNA in PCR II previously synthesized [25], was used as a template to clone the TL gene spanning bases 115–592 of the previously published sequence [20] into pET 20b with flanking restriction sites for NdeI and BamHI as previously described [26]. For single Cys mutants of TL, C101L was chosen as the template for additional mutant cDNAs because it showed similar structural features and binding characteristics for spin labeled lipids [27]. Eight additional mutant cDNAs were constructed, in which corresponding amino acid residues in the G strand of TL were substituted sequentially to cysteine. Amino acid 95 corresponds to aspartic acid, bases 397–399 of Redl [20].
Expression and Purification of Mutant Proteins
The mutant plasmids were transformed in E. Coli, BL 21 (DE3), cells were cultured, and protein was expressed according to the manufacturer’s protocol (Novagene), and purified as previously described [6, 13].
Spin labeling of TL Mutants
Mutant and wild type TLs, 100 μM, were incubated in 5-molar excess of MTSL overnight at 4°C. Unreacted spin labels were removed with a HiTrap column.
EPR Measurements and Fitting of EPR spectra
Electron paramagnetic resonance spectra were recorded at X-band with a Varian E-109 spectrometer fitted with two-loop one-gap resonator [28]. For measurements, the samples of about 5μl of spin-labeled protein in various conditions (generally 50–100 μM) were loaded into Pyrex capillaries (0.84 o.d. × 0.64 i.d.; VitroCom Inc., Mountain Lakes, NJ). The microwave power was 2mW incident, and the modulation amplitude was 1G. 10 mM sodium phosphate and 30 mM sodium citrate buffers were used for pH 7.3 and pH 3.0, respectively. All measurements were conducted at room temperature. To obtain ANS –protein complexes, spin labeled mutant proteins were incubated with 3-molar excess of ANS.
EPR spectra were fit to the MOMD model of Freed and co-workers using the program NLSL as previously described for CRBP [24, 29]. The apparent mean correlation time was calculated as τ= 1/6D, where D is the rotational diffusion coefficient.
Results and Discussion
All single Cys mutant and wild-type proteins used in this study have been characterized previously [27]. For G-strand residues of TL, the effects of mutations on secondary structure and binding characteristics are minimal.
The line shapes of EPR spectra reflect the dynamic modes of the nitroxide side chain that can be modulated by protein backbone fluctuation, tertiary interaction, etc [24, 30–32]. Each EPR spectrum along the sequence provides site-specific dynamic information. It has been shown that the peak-to-peak intensity and line width of the central line of the EPR spectra correlate with dynamic and accessibility parameters. For β-sheet proteins, the sequence pattern of these site specific parameters along the β-strand exhibits characteristic alternating periodicity [24, 27]. The nitroxide side chain in β-sheets that is located in a tertiary interaction with a side chain of a neighboring strand gives rise to additional dynamic modes, ranging from weakly ordered to immobilized. The dynamic modes of the nitroxide side chain, which interacts with nearest-neighbor side chains oriented in parallel on the flanking strands, depend on the type of neighboring residue, whether the neighbor is hydrogen bonded, and the twisting of the β strand [24].
EPR spectra of spin-labeled apo-Cys mutants in various conditions are shown in Figure 1. These spectra are very similar to that of holo-forms that have been studied previously [27]. For some sites, two dynamic modes, indicated as α and β in Figure 1, can be easily observed. For β-sheets, the detection of two dynamic modes is indicative of nearest-neighbor interactions. Secondary structural content of G-strand residues can be determined from the plots of the spectral parameters, which correlate with side chain mobility and exposure, versus residue number (Fig. 2). The residues 96–101 exhibit the alternating periodicity, which is characteristic of β-sheets. This feature is very consistent with the solution model and crystal structure of TL resolved by site-directed tryptophan fluorescence (SDTF) and x-ray crystallography, respectively [13, 14].
Figure 1.
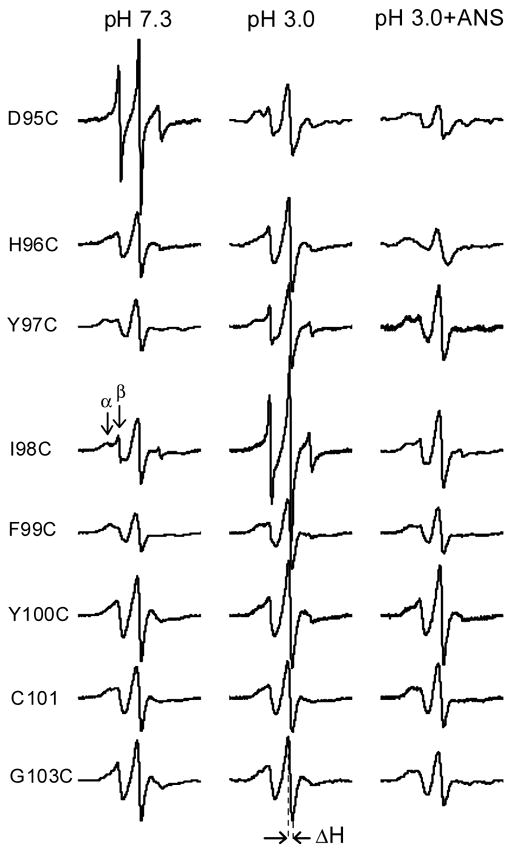
EPR spectra for spin labeled G-strand residues of apo-TL at various conditions. For direct comparison of intensities, the spectra are normalized to represent the same number of spins. Scan width is 100 G for all spectra. α and β represent the motionally restricted and mobile components, respectively. The line width of the central resonance line, ΔH, is denoted by the arrows
Figure 2.
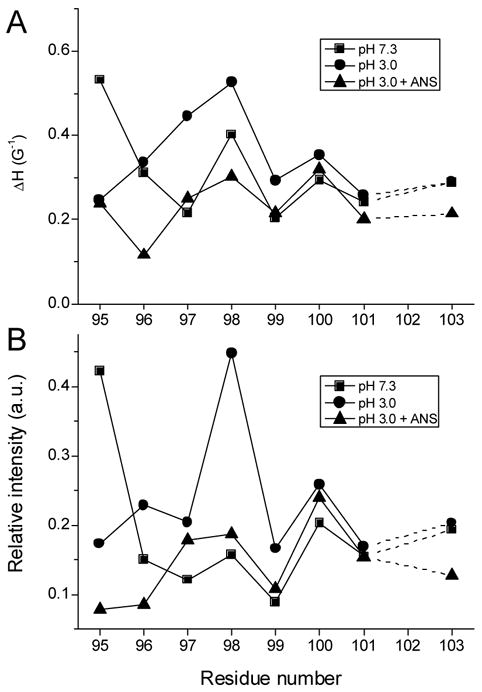
Plots of ΔH−1 (A) and relative intensity of central lines (B) in normalized EPR spectra of G-strand residues at various conditions.
TL undergoes a native to molten globule transition at pH 3.0 [5]. Comparison of EPR spectra of G-strand residues at pH 7.3 to that of pH 3.0 is very informative (Fig. 1). First, it is clear from the alternating periodicity that β structure persists in this strand. This is consistent with molecular dynamic simulations for RBP that show retention in β structure in the central sheet but disruption in strands E and F through loss of hydrogen bonds [9]. Second, nitroxide side chain mobilities of all residues, except 95, are increased at pH 3.0, albeit to different degrees. Residues 95 and 98 show dramatic, but opposite changes. The nitroxide at position 98 becomes extremely mobile, indicative of very loose packing around this residue. Unexpectedly, the nitroxide at position 95, which is very mobile at pH 7.3, becomes more immobilized in the molten globule transition. At pH 3.0, this nitroxide side chain shows two dynamic modes, which arise from tertiary interactions. The spectral parameters for the G-strand (96–103) retain alternate periodicity, although the entire plot for these residues is shifted to higher values indicating more mobility and exposure (Fig. 2). Taken together the data indicate that the G-strand retains its β-sheet structure in a molten globule state, but tertiary interactions involving G-strand residues are decreased, consistent with the spectral line shape (Fig. 1). Residues 95 and 98 show the biggest changes in the spectral parameters (Figure 2.) in the transition from pH 7.3 to pH 3.0.
Although ANS is one of the most widely used fluorescent probes, information regarding the structural consequences of ANS binding to proteins, particularly in molten globule state, is limited. Fluorescence enhancement upon binding to proteins can result from ion pairing interactions, hydrophobic interactions, restricted mobility or any combination of these factors [33–36]. EPR spectra of spin labeled G-strand mutants complexed with ANS in molten globule state are shown in Figure 1. It is evident that ANS binding to the proteins in the molten globule state restored tight packing for all nitroxide side chains. In some cases, the nitroxide side chains, particularly at positions 95, 96, 101, 103, become more immobile at pH 3.0 than at pH 7.3 without bound ANS (Fig. 1). The entire plot of the spectral parameters for the mutant proteins in the molten globule state complexed with ANS shifted to lower values compared to that observed for pH 3.0. Hence the outcome of ANS binding in influencing the dynamics of the protein is site specific but there is clearly a global character. ANS induces more tight packing for residues with side chains oriented internally as well as externally.
For each site on the G-strand, tertiary interactions are derived from side chains of residues from flanked β-strands F and H. Therefore, EPR spectra of G-strand residues also reflect the status of the F and H strands. Hence, ANS binding to the protein in molten globule state restores tertiary interaction throughout the strand residues rather than for one or two residues (exposed or buried), that could be expected for a local binding site.
Residue I98 is located at the solvent-exposed site and most sensitive to the protein conformational state (Fig. 1, 2). Therefore, EPR spectra of nitroxide side chain at position 98 were analyzed in more detail with respect to local structural and dynamic properties. It has been shown that the rotamer state of side chains in β-sheets is determined by steric interactions with nearest neighbors [24]. The residue I98 has two nearest neighbors, hydrogen-bonded (HB) I89 and non-hydrogen-bonded (NHB) L115, theCα-Cα distance of which are 5.0 Å and 4.6 Å, respectively (Fig 3A). The nearest neighbor interaction between the residues is modulated by extent of twisting. I98, compared to Y100, resides in a more twisted site of G-strand (Fig. 3 B, C). The EPR spectra for residue I98 reveal more asymmetric nearest neighbor interactions compared to the spectra for Y100 (Fig. 1). Indeed, the EPR spectra for the nitroxide side chain at position 98 show two components, the mobility of which differ significantly from each other ( 3.0 ns and 10.0 ns) (Fig. 4). In the apo-protein, the interaction is more asymmetric. The correlation time of the first component decreases from 3.0 ns to 0.8ns. The relative population of this component is also decreased, from 24 % to 6%. The nitroxide at position 98 in the molten globule state has significantly weaker nearest neighbor interactions compared to that of holo-and apostate of the protein. The correlation time of the second component decreases from 10.2 ns to 4.2 ns, while the relative population of the component decreases from 94% to 75% (Fig. 4). The interaction of the apo-protein with ANS in the molten globule state almost completely restores both the population and the lifetime of the second component. It is noteworthy that the EPR spectra of the nitroxide at position 98 is very similar to that reported for E114 in another β-sheet protein CRBP [24]. The residues 98 of TL and 114 of CRBP are located in β-strands, which have very similar twisting (Fig. 3C). The working model suggested by others appears generally applicable for β-sheet proteins and a powerful tool for monitoring conformational changes in various environments [24].
Figure 3.
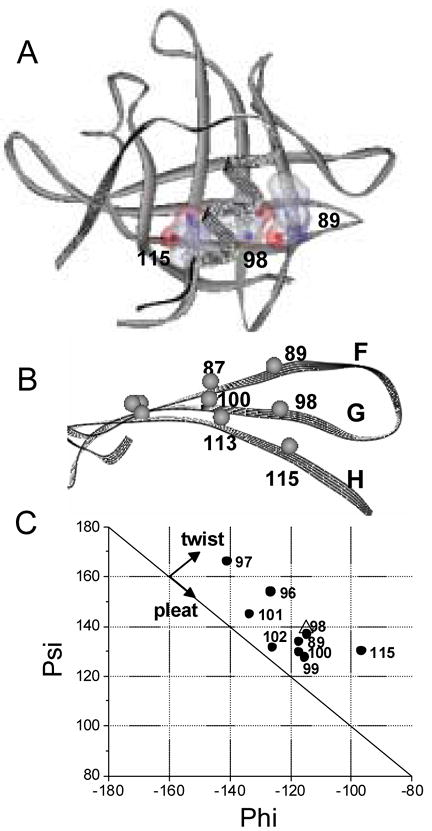
Ribbon model of TL generated from PDB: 1XKI [14] using ViewerLite 5.0 (Accelrys Inc.). (A) The position of the side chain for residue I98 (G-strand) relative to that of the flanking residues I89 (F-strand) and L115 (H-strand). The side chains are shown by combination of “ball and stick” and surface representations. Gray, blue and red balls represent carbon, nitrogen and oxygen atoms, respectively. (B) Side view of G and flanking strands, F and H. Gray balls are the positions of the α-carbons of respective residues. (C) A Ramachandran plot of residues in G and its flanking F (I89), H (L115) strands (solid circles) (PDB: 1XKI). To compare the extent of the β-strand twists, residue E114 (open triangle) of CRBP (PDB: 1CRB [38]) are also added to the plot.
Figure 4.
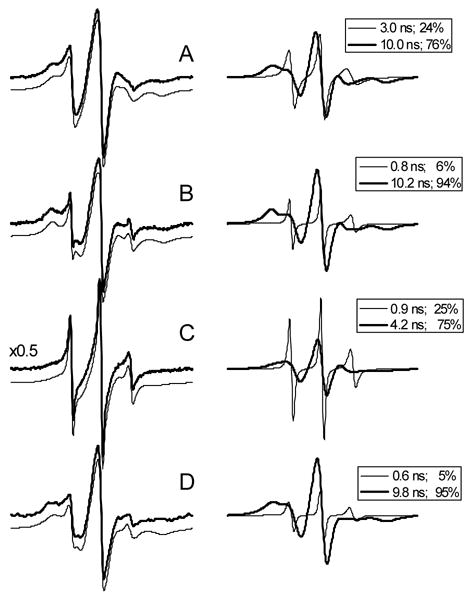
EPR spectra of spin labeled C98 mutant of TL at various conditions. Scan width is 100 G. (A) holoC98 at pH 7.3 [27]; (B) apoC98 at pH 7.3; (C) apoC98 at pH 3.0; (D) apoC98 with ANS (1/3 ratio). Left column: the thick traces are experimental spectra and thin traces are the least-squares best fit to the MOMD model. The best-fit spectra are vertically shifted relative to the experimental spectra for better view. Right column: the thick and thin traces are immobile and mobile components, respectively, from 2-site MOMD model. The percentages represent relative populations of each component.
Conclusion
SDSL enabled the acquisition of site specific dynamic information about the central β sheet in the native-molten globule transition of TL. β structure of the G strand persisted. However, tertiary interactions were mitigated to a variable extent throughout the strand. Residue 98 in the twisted region of the strand showed the greatest loss of tertiary interaction. Despite the absence of a disulfide bridge joining the G-H strands in TL, the central β sheet was stabilized probably by the interaction with the alpha helix as suggested for RBP and BLG [10, 11].
The pH driven molten globule transition is a central theme in the proposed ligand release mechanism for TL [6, 37]. The behavior of specific sites on individual strands of TL in the molten globule state has implications for understanding its mechanism of ligand release and stabilization the lipid layer of the tear film.
Acknowledgments
Supported by U.S. Public Health Service Grants NIH EY-11224 and EY00331 as well as the Edith and Lew Wasserman Endowed Professorship in Ophthalmology.
Abbreviations
- ANS
8-anilino-1-naphthalenesulfonic acid
- BLG
β-lactoglobulin
- CRBP
cellular retinol-binding protein
- EPR
electron paramagnetic resonance
- MOMD
microscopic order/macroscopic disorder
- RBP
retinol-binding protein
- SDSL
side directed spin labeling
- TL
tear lipocalin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bychkova VE, Berni R, Rossi GL, Kutyshenko VP, Ptitsyn OB. Retinol-binding protein is in the molten globule state at low pH. Biochemistry. 1992;31:7566–7571. doi: 10.1021/bi00148a018. [DOI] [PubMed] [Google Scholar]
- 2.Jennings PA, Wright PE. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 1993;262:892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 3.Yao H, Takasawa R, Fukuda K, Shiokawa D, Sadanaga-Akiyoshi F, Ibayashi S, Tanuma S, Uchimura H. DNA fragmentation in ischemic core and penumbra in focal cerebral ischemia in rats. Brain Res Mol Brain Res. 2001;91:112–118. doi: 10.1016/s0169-328x(01)00135-8. [DOI] [PubMed] [Google Scholar]
- 4.van der Goot FG, Gonzalez-Manas JM, Lakey JH, Pattus F. A ‘molten-globule’ membrane-insertion intermediate of the pore-forming domain of colicin A. Nature. 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 5.Gasymov OK, Abduragimov AR, Yusifov TN, Glasgow BJ. Structural changes in human tear lipocalins associated with lipid binding. Biochim Biophys Acta. 1998;1386:145–156. doi: 10.1016/s0167-4838(98)00092-2. [DOI] [PubMed] [Google Scholar]
- 6.Gasymov OK, Abduragimov AR, Yusifov TN, Glasgow BJ. Interstrand loops CD and EF act as pH-dependent gates to regulate fatty acid ligand binding in tear lipocalin. Biochemistry. 2004;43:12894–12904. doi: 10.1021/bi049076o. [DOI] [PubMed] [Google Scholar]
- 7.Khurana R, Gillespie JR, Talapatra A, Minert LJ, Ionescu-Zanetti C, Millett I, Fink AL. Partially folded intermediates as critical precursors of light chain amyloid fibrils and amorphous aggregates. Biochemistry. 2001;40:3525–3535. doi: 10.1021/bi001782b. [DOI] [PubMed] [Google Scholar]
- 8.Kuwajima K. The molten globule state of alpha-lactalbumin. Faseb J. 1996;10:102–109. doi: 10.1096/fasebj.10.1.8566530. [DOI] [PubMed] [Google Scholar]
- 9.Paci E, Greene LH, Jones RM, Smith LJ. Characterization of the molten globule state of retinol-binding protein using a molecular dynamics simulation approach. Febs J. 2005;272:4826–4838. doi: 10.1111/j.1742-4658.2005.04898.x. [DOI] [PubMed] [Google Scholar]
- 10.Greene LH, Wijesinha-Bettoni R, Redfield C. Characterization of the molten globule of human serum retinol-binding protein using NMR spectroscopy. Biochemistry. 2006;45:9475–9484. doi: 10.1021/bi060229c. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi T, Ikeguchi M, Sugai S. Molten globule structure of equine beta-lactoglobulin probed by hydrogen exchange. J Mol Biol. 2000;299:757–770. doi: 10.1006/jmbi.2000.3761. [DOI] [PubMed] [Google Scholar]
- 12.Flower DR. The lipocalin protein family: structure and function. Biochem J. 1996;318( Pt 1):1–14. doi: 10.1042/bj3180001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gasymov OK, Abduragimov AR, Yusifov TN, Glasgow BJ. Site-directed tryptophan fluorescence reveals the solution structure of tear lipocalin: evidence for features that confer promiscuity in ligand binding. Biochemistry. 2001;40:14754–14762. doi: 10.1021/bi0110342. [DOI] [PubMed] [Google Scholar]
- 14.Breustedt DA, Korndorfer IP, Redl B, Skerra A. The 1.8-a crystal structure of human tear lipocalin reveals an extended branched cavity with capacity for multiple ligands. J Biol Chem. 2005;280:484–493. doi: 10.1074/jbc.M410466200. [DOI] [PubMed] [Google Scholar]
- 15.Glasgow BJ, Marshall G, Gasymov OK, Abduragimov AR, Yusifov TN, Knobler CM. Tear lipocalins: potential lipid scavengers for the corneal surface. Invest Ophthalmol Vis Sci. 1999;40:3100–3107. [PubMed] [Google Scholar]
- 16.Gasymov OK, Abduragimov AR, Prasher P, Yusifov TN, Glasgow BJ. Tear lipocalin: evidence for a scavenging function to remove lipids from the human corneal surface. Invest Ophthalmol Vis Sci. 2005;46:3589–3596. doi: 10.1167/iovs.05-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selsted ME, Martinez RJ. Isolation and purification of bactericides from human tears. Exp Eye Res. 1982;34:305–318. doi: 10.1016/0014-4835(82)90079-3. [DOI] [PubMed] [Google Scholar]
- 18.van’t Hof W, Blankenvoorde MF, Veerman EC, Amerongen AV. The salivary lipocalin von Ebner’s gland protein is a cysteine proteinase inhibitor. J Biol Chem. 1997;272:1837–1841. doi: 10.1074/jbc.272.3.1837. [DOI] [PubMed] [Google Scholar]
- 19.Blaker M, Kock K, Ahlers C, Buck F, Schmale H. Molecular cloning of human von Ebner’s gland protein, a member of the lipocalin superfamily highly expressed in lingual salivary glands. Biochim Biophys Acta. 1993;1172:131–137. doi: 10.1016/0167-4781(93)90279-m. [DOI] [PubMed] [Google Scholar]
- 20.Redl B, Holzfeind P, Lottspeich F. cDNA cloning and sequencing reveals human tear prealbumin to be a member of the lipophilic-ligand carrier protein superfamily. J Biol Chem. 1992;267:20282–20287. [PubMed] [Google Scholar]
- 21.Lechner M, Wojnar P, Redl B. Human tear lipocalin acts as an oxidative-stress-induced scavenger of potentially harmful lipid peroxidation products in a cell culture system. Biochem J. 2001;356:129–135. doi: 10.1042/0264-6021:3560129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glasgow BJ, Abduragimov AR, Gassymov OK, Yusifov TN, Ruth EC, Faull KF. Vitamin E associated with the lipocalin fraction of human tears. Adv Exp Med Biol. 2002;506:567–572. doi: 10.1007/978-1-4615-0717-8_79. [DOI] [PubMed] [Google Scholar]
- 23.Yusifov TN, Abduragimov AR, Gasymov OK, Glasgow BJ. Endonuclease activity in lipocalins. Biochem J. 2000;347(Pt 3):815–819. [PMC free article] [PubMed] [Google Scholar]
- 24.Lietzow MA, Hubbell WL. Motion of spin label side chains in cellular retinol-binding protein: correlation with structure and nearest–neighbor interactions in an antiparallel beta-sheet. Biochemistry. 2004;43:3137–3151. doi: 10.1021/bi0360962. [DOI] [PubMed] [Google Scholar]
- 25.Glasgow BJ, Heinzmann C, Kojis T, Sparkes RS, Mohandas T, Bateman JB. Assignment of tear lipocalin gene to human chromosome 9q34-9qter. Curr Eye Res. 1993;12:1019–1023. doi: 10.3109/02713689309029229. [DOI] [PubMed] [Google Scholar]
- 26.Gasymov OK, Abduragimov AR, Yusifov TN, Glasgow BJ. Solution structure by site directed tryptophan fluorescence in tear lipocalin. Biochem Biophys Res Commun. 1997;239:191–196. doi: 10.1006/bbrc.1997.7451. [DOI] [PubMed] [Google Scholar]
- 27.Glasgow BJ, Gasymov OK, Abduragimov AR, Yusifov TN, Altenbach C, Hubbell WL. Side chain mobility and ligand interactions of the G strand of tear lipocalins by site-directed spin labeling. Biochemistry. 1999;38:13707–13716. doi: 10.1021/bi9913449. [DOI] [PubMed] [Google Scholar]
- 28.Hubbell WL, Froncisz W, Hyde JS. Continuous and stopped flow EPR spectrometer based on a loop gap resonator. Rev Sci Instrum. 1987;58:1879–1886. [Google Scholar]
- 29.Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-Least-Squares Analysis of Slow-Motion EPR Spectra in One and Two Dimensions Using a Modified Levenberg–Marquardt Algorithm. J Magn Reson Series A. 1996;120:155–189. [Google Scholar]
- 30.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 31.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 32.McHaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35:7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 33.Collini M, D’Alfonso L, Molinari H, Ragona L, Catalano M, Baldini G. Competitive binding of fatty acids and the fluorescent probe 1-8-anilinonaphthalene sulfonate to bovine beta-lactoglobulin. Protein Sci. 2003;12:1596–1603. doi: 10.1110/ps.0304403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matulis D, Baumann CG, Bloomfield VA, Lovrien RE. 1-anilino-8-naphthalene sulfonate as a protein conformational tightening agent. Biopolymers. 1999;49:451–458. doi: 10.1002/(SICI)1097-0282(199905)49:6<451::AID-BIP3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 35.Matulis D, Lovrien R. 1-Anilino-8-naphthalene sulfonate anion-protein binding depends primarily on ion pair formation. Biophys J. 1998;74:422–429. doi: 10.1016/S0006-3495(98)77799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gasymov OK, Glasgow BJ. ANS Fluorescence: Potential to Augment the Identification of the External Binding Sites of Proteins. BBA- Proteins and Proteomics. 2007;1774:403–411. doi: 10.1016/j.bbapap.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasymov OK, Abduragimov AR, Gasimov EO, Yusifov TN, Dooley AN, Glasgow BJ. Tear lipocalin: potential for selective delivery of rifampin. Biochim Biophys Acta. 2004;1688:102–111. doi: 10.1016/j.bbadis.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Cowan SW, Newcomer ME, Jones TA. Crystallographic studies on a family of cellular lipophilic transport proteins. Refinement of P2 myelin protein and the structure determination and refinement of cellular retinol-binding protein in complex with all-trans-retinol. J Mol Biol. 1993;230:1225–1246. doi: 10.1006/jmbi.1993.1238. [DOI] [PubMed] [Google Scholar]


