Abstract
CD8+ T lymphocytes (TCD8+) play an important role in cellular immune responses. TCD8+ recognize MHC class I molecules complexed to peptides of 8 to 10 residues derived largely from cytosolic proteins. Proteins are generally thought to be fragmented in the cytoplasm and delivered to nascent class I molecules in the endoplasmic reticulum (ER) by a peptide transporter encoded by the MHC. To explore the extent to which TCD8+ induction in vivo is limited by proteolysis or peptide transport into the ER, mice were immunized with recombinant vaccinia viruses containing mini-genes encoding antigenic peptides (bypassing the need for proteolysis), or these peptides with a NH2-terminal ER insertion sequence (bypassing the requirements for both proteolysis and transport). Additionally, mice were immunized with recombinant vaccinia viruses encoding rapidly degraded fragments of proteins. We report that limitations in induction of TCD8+ responses vary among Ags: for some, full length proteins are as immunogenic as other forms tested; for others, maximal responses are induced by peptides or by peptides targeted to the ER. Most importantly, in every circumstance examined, targeting peptides to the ER never diminished, and in some cases greatly enhanced, the TCD8+ immune response and provide an important alternative strategy in the design of live viral or naked DNA vaccines for the treatment of cancer and infectious diseases.
CD8+ T lymphocytes (TCD8+)2 play an important role in the eradication of many intracellular pathogens and can destroy cancer cells. The Specificity of the interaction of TCD8+ with a target cell is provided chiefly by an interaction of TCRs on the surfaces of lymphocytes with MHC class I molecules, located on the surfaces of target cells. MHC class I molecules are themselves specialized receptors that bind peptide Ags, which are generally 8 to 10 residues in length.
Most of the peptides that are presented by class I molecules originate in the cytosol. Transport into the endoplasmic reticulum (ER), the site of peptide association with class I molecules, involves two gene products of the MHC region, designated TAP-1 and TAP-2 (1-4). The TAP transporters are required for the efficient presentation of most, but not all, Ags (5, 6). An alternate, TAP-independent pathway for entry into the endoplasmic reticulum is via ER insertion signal sequences, encoded in the 5′ ends of mRNAs (the NH2 termini of precursor peptides) that target peptides for co-translational insertion into the ER (7-10).
Peptides are likely to be generated by the proteolysis of intracellular proteins or glycoproteins (11-13). The mechanisms involved in the targeting of protein Ags for proteolysis and presentation as peptides by class I molecules are largely unknown. It has been argued that ubiquitin-dependent targeting of proteins for proteolytic degradation by the proteasome plays a role in generating peptides for binding to MHC class I (14). There is now considerable evidence that a subset of proteasomes is involved in peptide generation, specifically those proteasomes containing two MHC gene products termed LMP2 and -7 (15-25).
Basal levels of the transcription of genes encoding class I molecules, TAP and LMP2 and -7, are greatly enhanced by exposure of most cells to IFN-γ. It is unknown, however, whether proteolysis, peptide delivery, or both of these processes are limiting in vivo in the generation of TCD8+ responses. Because it is possible to express minigenes preceded by ER insertion signal sequences, much of the Ag-processing machinery can be bypassed. Indeed, the targeting of 8- or 9-amino acid long peptides to the ER, where they are presumed to be liberated from the targeting sequence by signal peptidase, occurs in cells deficient in TAP and LMP2 and -7 (26-28). This targeting in vitro is highly efficient, despite the presumably random diffusion of these peptides to class I molecules. Note that in normal cells, class I molecules are tethered to TAP and are likely to directly receive freshly transported peptides (29, 30).
In the present study, we test the limiting factors in the induction of TCD8+ responses using recombinant vaccinia viruses (rVV) expressing either full length proteins, protein fragments with enhanced rates of proteolysis, or 8- or 9-residue antigenic peptides with NH2-terminal extensions consisting of methionine (needed for initiation of protein synthesis) or sequences that target the peptide to the ER. Implications for the rational design of recombinant vaccines are described.
Materials and Methods
Viruses
The rVV used in this study are listed in Table I. All of the rVV contain foreign genes under control of the VV P7.5 promoter inserted into the VV thymidine kinase gene by homologous recombination, resulting in the generation of thymidine kinase-negative progeny as described previously (31). rVV expressing full length proteins from influenza virus A/Puerto Rico/8/34 (PR8) (viruses lettered A–F) have been described previously (32) and were produced using a vector (pGS60) that does not contain the β-galactosidase gene. The β-galactosidase gene is present in the later generation vector (pSC11) (31) used to produce all of the other recombinants (this facilitates identification of rVV). To eliminate the possibility that β-galactosidase expression influenced the immunogenicity of influenza virus nucleoprotein (NP), a new rVV expressing the full length NP gene was constructed using the pSC11 plasmid (VV-NP). VV-NP1–168, W-NPM147–155, and VV-NPM296–495 were produced by inserting PCR- generated genes produced from a full length NP template into the pSC11. rVV-encoding mini-gene products were produced using synthetic oligonucleotides as described previously (28). Note that in all viruses expressing mini-genes, the start codon (ATG) is preceded by Kozak’s consensus sequence (CCACC) for translation (33). rVV encoding full length OVA or P1A were produced using cDNAs encoding the genes. All rVV were propagated in 143/B cells (ATCC, CRL 8303) and used as crude cell lysates. The amount of infectious virus in stocks was determined by plaque titration using BS-C-1 cells (ATCC, CCL 26). rVV used in a single experiment were titered together to maximize the accuracy of the relative titers. PR8 was propagated in the allantoic cavity of embryonated chicken eggs.
Table I.
Recombinant gene products produced by rVV
| Designation | Sequence of Gene Product |
|---|---|
| A. V-69 | Full length NP from PR8 (32) |
| B. VV-HA | Full length hemagglutinin from PR8 (32) |
| C. VV-NS1 | Full length nonstructural 1 from PR8 (32) |
| D. VV-NA | Full length neuraminidase from PR8 (32) |
| E. VV-PB2 | Full length basic polymerase from PR8 (32) |
| F. VV-PA | Full length acidic polymerase from PR8 (32) |
| G. VV-NP | Full length NP from PR8 (74) |
| H. VV-NPM147–155 | MTYORTRALV |
| I. VV-ESNP147_155 | MRYMILGLLALAAVCSAATYORTRALV |
| J. VV-lSNP147–155 | MTNILCLLOIALLLCFSTTALSTYORTRALV |
| K. VV.NPM50–57 | MSDYEGRLI |
| L. VV-ESNP50–57 | MRYMILGLLALAAVCSAASDYEGRLI |
| M. VV-NP1–168 | Fragment of NP from PR8 |
| N. VV-NPM147–315 | Initiating methionine preceding fragment of NP from PR8 |
| O. VV-NPM296–495 | Initiating Met preceding fragment of NP from PR8 (74) |
| P. VV-OVA | Full length OVA gene (75) |
| Q. VV-0VAM2S7–264 | MSINNFEKL (76) |
| R. VV-ESOVA257–264 | MRYMILGLLALAAVCSAASIINFEKL |
| S. VV-P1A | Full length P1A gene (37) |
| T. VV-P1AM35–43 | MLPYLGWLVF (36) |
| U. VV-ESP1A35–43 | MRYMILGLLALAAVCSAALPYLGWLVF |
| V. VV-NPM147–155ES | MTYORTRALVRYMILGLLALAAVCSA |
(Italics) Foreign initiating methionine; (underlined), naturally processed determinant; (bold) E3/19K signal sequence; (bold underlined) IFN-β signal sequence; (underlined italic), additional Ala resulting from inclusion of a restriction site in the expression vector.
Synthetic peptides
Synthetic peptides corresponding to PR8 NP residues 147–155 (TYQR TRALV) (34), PR8 NP residues 50–57 (SDYEGRLI) (35), P815A residues 35–43 (LPYLGWLVF) (36, 37), and OVA residues 257–264 (SI INFEKL) (38) were synthesized, HPLC-purified, and verified by amino acid analysis by Peptide Technologies (Washington, DC) and the Biologic Resources Branch, NIAID (Bethesda, MD).
Immunization
The mice used were 8- to 10-wk-old female BALB/c (for H-2d PR8 experiments), DBA/2n (for H-2d P815A experiments), C3H and CBA/j (for H-2k PR8 NP experiments), and C57BL/6n (for H-2b PR8 NP experiments) and were obtained from the Animal Production Colonies, Frederick Cancer Research and Development Facility, NCI, NIH (Frederick, MD). Mice were injected i.v. with 5 × 106 plaque-forming units (PFU) of rVV, except in the dose titration experiments where the dose is specified. Six days later, or in the case of secondary immune responses 30 days later, spleens were removed and dispersed to single cell suspensions in Iscove’s modified DMEM medium with 7.5% heat-inactivated FCS using a Pyrex Ten Broeck homogenizer (Corning, New York). For secondary TCD8+ cultures, splenocytes were harvested, dispersed, and cultured for 6 days in media containing RPMI 1640 supplemented with 1 μM of the peptide specsed , 10% FCS, 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate (all from Biofluids, Rockville, MD), 5 × 10−5 M 2-E (Aldrich Chemical Co., Milwaukee, WI), and 0.03% (100 mM) glutamine (NIH media unit, Bethesda, MD).
51Cr releaser assays
Cytolytic assays were performed as described previously (39). In brief, P815 cells (H-2d) (ATCC, TIB64), L929 cells (H-2k) (ATCC, CCL l), or clones of murine methylcholanthrine-induced sarcoma lines MCA 205 (H-2b) (40) and MCA 102 (H-2b) (41) generated in our laboratory, and the N-nitroso-N-methylurethane-induced murine colon tumor line CT-26 (H-2d) (42), were used as targets as specified, and were pulsed with synthetic peptides where specified during 51Cr labeling. Target cells were infected at a multiplicity of 10 PFU/cell with VV or with 100 μl of PR8-infectious allantoic fluid/106 cells for 1 h before labeling for 1h with 51Cr. Target cells were incubated with splenocytes for 8 h at 37°C at the E:T ratio indicated. The amount of released 51Cr was determined by gamma counting, and the percent specific lysis was calculated as follows: [(experimental cpm − spontaneous cpm)/(maximal cpm − spontaneous cpm)] × 100. Lytic units were calculated as described previously (43).
Metabolic radiolabeling experiments
Pulse-chase experiments were performed as described previously (44). In brief, 2.4 × 107 P815 cells were infected for 2 h with rVV at 10 PFU/cell and then incubated in methionine-free RPMI for 30 min at 37°C, which was then supplemented with 250 μCi of 35S-labeled Met (Amersham, Arlington Heights, IL) for 10 min. After washing with ice-cold PBS, cells were either incubated at 0°C (pulse) or chased for up to 90 min at 37°C in warmed medium containing 2 mg/ml unlabeled methionine. Detergent extracts from radiolabeled cells were adjusted to contain equal amounts of acid-precipitable 35S-labeled Met in equal volumes and incubated with protein A Sepharose preloaded with the NP-specific mAb H16-L10 (45). After washing, affinity-purified material was analyzed by SDS-PAGE using a 12.5% polyacrylamide gel and the buffer system of Laemmli (46). Gels were fixed and dried, then imaged using a phosphorimager (Molecular Dynamics, Sunnyvale, CA).
Results
Immunization with rVV-expressing influenza virus proteins
To examine whether Ag processing is limiting in the induction of primary TCD8+, BALB/c mice were immunized with rVV-expressing full length influenza virus proteins. The rVV studied are listed in Table I. Each of the full length PR8 proteins studied is known to be recognized by BALB/c TCD8+ induced by priming and in vitro stimulation with influenza virus (47). Six days after immunization, splenocytes were harvested and tested for their ability to lyse 51Cr-labeled P815 cells (H-2d) infected with influenza virus. None of the splenocyte populations lysed influenza virus-infected cells above values obtained using uninfected cells (Fig. 1). By contrast, each of the populations efficiently lysed VV-infected cells, demonstrating that mice were infected by the various rVV. The absence of lysis of influenza virus-infected cells is not due to low levels of viral gene expression, because the same target cells were efficiently lysed by influenza virus-specific TCD8+ raised by secondary in vitro stimulation (48) with influenza virus (not shown).
FIGURE 1.
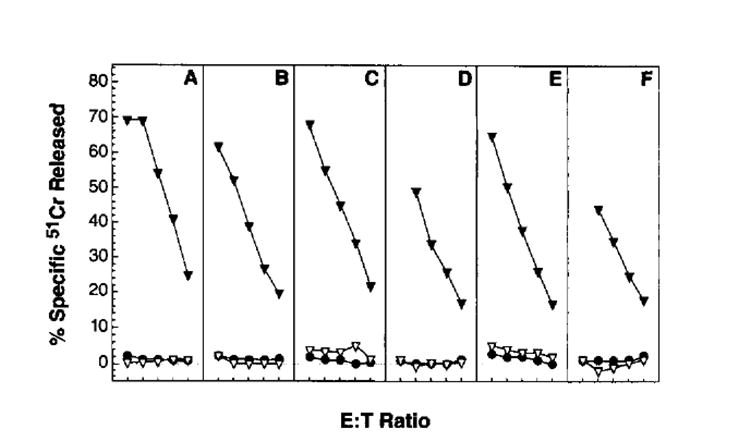
Immunization with rVV expressing full length influenza virus proteins. BALB/c mice were injected i.v. with 5 × 106 PFU of rVV expressing the following full length influenza (PR8) virus gene products: nonstructural 1 (A); neuraminidase (B); hemagglutinin (C); nucleoprotein (D); basic polymerase (E); and acidic polymerase (F). Six days later, splenocytes were harvested and tested in a 51Cr release assay at 200:1, 100:1, 50:1, 25:1, and 12:l E:T ratios using untreated P815 cells (●), VV-infected P815 cells (▼), or PR8- infected P815 cells (▽). The experiment was repeated with similar results.
To explore the limiting factors in the induction of primary TCD8+, we focused on the NP-specific response. TCD8+ from BALB/c mice predominantly (and possibly exclusively) recognize residues 147–155 of NP in conjunction with Kd (34, 49). The functions of rVV-expressing residues 147–155 preceded by either a Met (needed for initiation of translation) or ER insertion sequences derived from either IFN-β or an adenovirus glycoprotein have been studied in vitro (27, 28, 50). To determine whether enhanced generation of the immunodominant fragment of NP, or whether transport into the ER could boost immunogenicity in vivo, mice were immunized with these rVV i.v., and primary immune responses were studied on day 6 after immunization. As seen in Figure 2, VV-NP failed to elicit a NP-specific primary response. VV-NPM147–155 induced a primary NP-specific TCD8+ response similar in magnitude to the VV-specific response. Bypassing TAP by targeting peptide to the ER using either the adenovirus or IFN signal sequence (VV-ESNP147–155 or VV-ISNP147–155) also gave excellent primary TCD8+ responses, but these responses were not consistently better than those obtained with the VV-NPM147–155. Appending the leader sequence to the COOH terminus of the minimal determinant (VV-NPM147–155ES) diminished its immunogenicity, demonstrating that the orientation of the leader sequence is critical to its function. This may be because of its failure to translocate co-translationally the peptide into the ER, or it may result from the leader sequence’s poor cleavage by signal peptidase. Note that in some experiments splenocytes from immunized animals were observed to specifically lyse influenza virus-infected cells at levels roughly similar to those of peptide-pulsed target cells (not shown). All of the recombinants tested elicited VV-specific responses of similar magnitude, demonstrating that each rVV was capable of eliciting TCD8+ responses. The primary TCD8+ responses against fragments of NP as shown in Figure 2, F and G, are discussed below.
FIGURE 2.
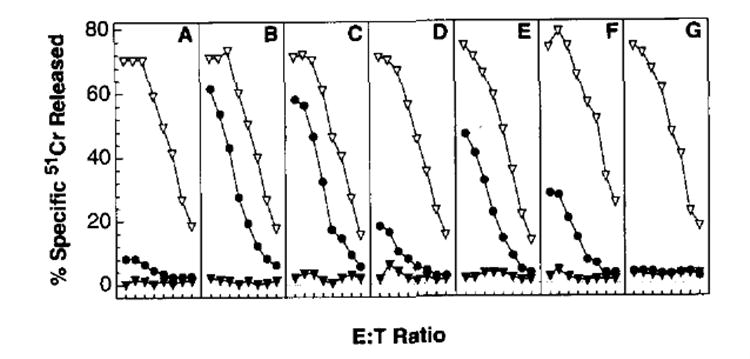
Immunogenicity of rVV-encoding oligopeptides. BALB/c mice were injected i.v. with 5 × 106 PFU of VV-NP (A), VV-NPM147–155 (B), VV-ESNP147–155 (C), VV-NPM147–155ES (D), VV-ISNP147–155 (E), VV-NP1–16, (F), and VV-NPM296–496 (G). Six days later, splenocytes from immunized mice were tested in a 51Cr release assay at E:T ratios of 600:1, 300:1, 150:1, 75:1, 37:1, 19:1, 9:1, and 5:1. Note that high E:T ratios were included to give the full spectrum of lysis. The target cells used in this experiment were untreated P815 cells (▼), P815 cells pulsed with synthetic peptide corresponding to NP residues 147–155 (●) or VV-infected P815 (▽). This experiment was repeated three times with similar results.
Kinetics of the TCD8+ response against rVV-expressing oligonucleotides
To determine whether our findings of the enhanced immunogenicity of the VV-NPM147–155 and VV-ESNP147–155 were attributable simply to a difference in kinetics of the TCD8+ responses to different forms of the Ag, we studied the TCD8+ responses between days 1and 21 after infection with various rVV. Primary NP-specific responses induced by VV-NPM147–155 and VV-ESNP147–155 were maximal between days 5 and 8, with peak responses usually occurring on day 6. Primary responses to VV also peaked on day 6 in most experiments. Importantly, NP-specific responses to VV-NP were minimal throughout the 3-wk trial. One representative experiment showing the kinetics from days 4 through 10 is shown in Figure 3. The addition of a leader sequence had a marginal, although not decisive, effect on the potency of the TCD8+ that was elicited.
FIGURE 3.
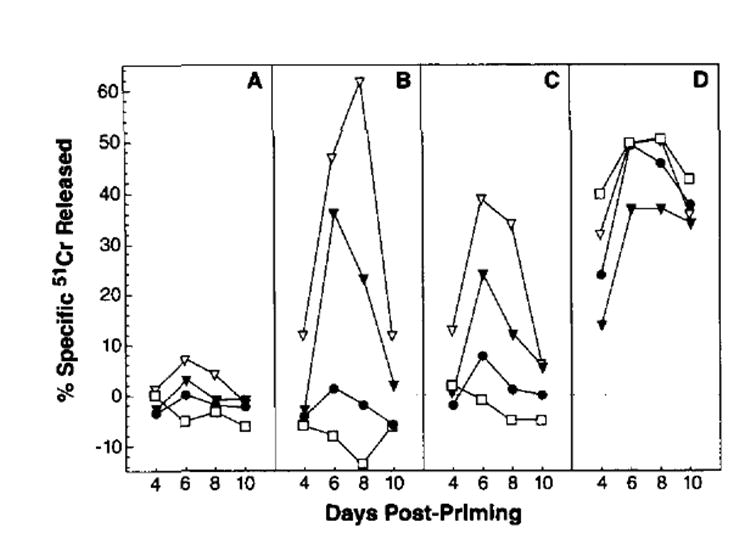
Kinetics of the TCD8+ response against rVV. Splenocytes derived from mice immunized i.v. with 5 × 106 PFU Of VV-NP (●), VV-NP147–155 (▼), VV-ES NP147–155 (▽) or the control VV-ESOVA257–264 (□) were tested for CTL activity against untreated P815 cells (A), P815 cells pulsed with NP147–155 synthetic peptide (B), PR8-infected P815 cells (C), or VV-infected P815 cells (D). Mice were immunized sequentially to allow response to be measured in the same assay. The E:T ratio shown is 100:1. Similar results were obtained in four independent experiments.
Thus, TCD8+ elicited by rVV expressing the NP minigene products followed kinetics typical of primary responses. These experiments also showed that the poor immunogenicity of full length NP was not simply a question of a delayed response. Taken together, these findings demonstrate, first, that processing and presentation of the immunodominant NP147–155 peptide from the full length protein are limiting in vivo. Second, these data suggest that TAP-mediated transport of the VV-NPM147–155 was not severely limiting in the elicitation of a TCD8+ primary immune response, because addition of an ER insertion signal sequence did not consistently enhance immune responses.
The effect of enhanced degradation of NP on immunogenicity
Because it appeared that the proteolytic generation of NP147–155 was limiting in the elicitation of primary TCD8+ responses, we sought to test the immunogenicity of a shorter lived fragment of NP (residues 1–168). As seen in Figure 4, mAb-reactive material present in detergent extracts of VV-NP1–168-infected cells pulse-radiolabeled with [35S]methionine and chased for up to 90 min was degraded with a calculated t1/2 of 27 min, whereas no degradation of full length NP was detected over the 90-min chase period. Mice immunized with VV-NP1–168 (Fig. 2F), but not VV-NPM296–496, which does not contain the H-2 Kd-restricted epitope (Fig. 2G), mounted an anti-NP147–155 peptide-specific TCD8+ response at levels between those elicited with VV-NP and VV-NPM147–155.
FIGURE 4.
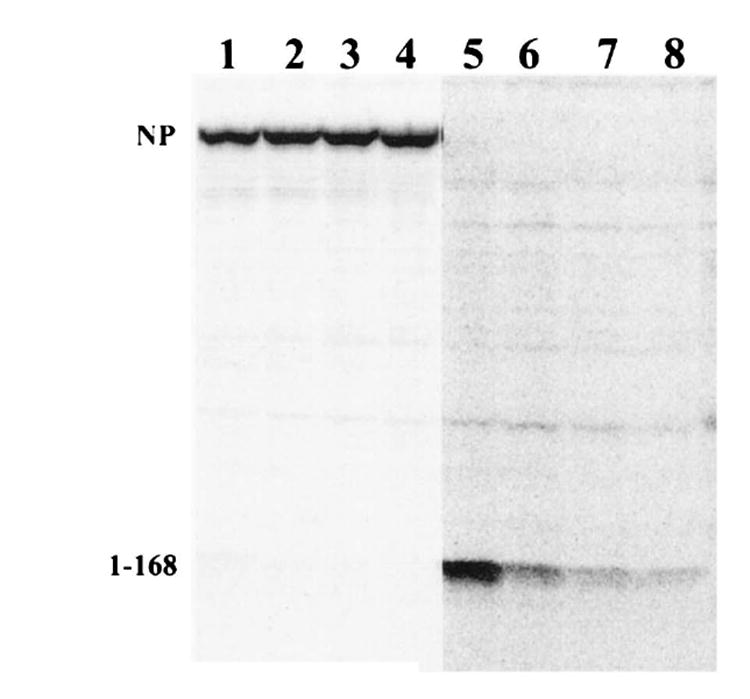
Metabolic stability of NP and NP1–168. L929 cells infected with either VV-NP (lanes 1–4) or with VV-NP1–168 (lanes 5–8) were metabolically radiolabeled with [35S]methionine and chased for 0 min (lanes 1 and 5), 30 min (lanes 2 and 6), 60 min (lanes 3 and 7), or 90 min (lanes 4 and 8). Immunoreactive NP present in detergent extracts were analyzed by SDS-PACE and visualized and quantitated by a Molecular Dynamics phosphorimager. There was no measurable decrease in the amount of full length NP recovered during the 90-min chase. The decrease in NP1–168 recovered showed that it had t1/2 of degradation of 27 min calculated by linear regression using the formula t1/2 = −ln∣(Nt/No)ln∣2 × t, where Nt is the amount recovered in the chase, No is the amount recovered in the pulse, and t is the time of the chase.
Accelerated proteolysis because of misfolding or incomplete folding would not necessarily feed peptide fragments into the Ag-processing pathway. In fact, such proteolytic fragments may not be efficient precursors for antigenic peptides at all (especially if the protein fragment is generated predominantly by exopeptidase activity, generating, instead, free amino acids, dipeptides, or tripeptides). However, assuming that NP1–168 is also more rapidly degraded in APCs in vivo than it is in vitro, these findings suggest that the enhanced degradation of NP increases the production of antigenic peptides, but not to the levels attained by directly synthesizing the peptide. Alternatively, it is possible that the removal of residues 169–495 enhances peptide production from NP1–168 by altering its intracellular trafficking or by providing more suitable flanking sequences for liberation of the antigenic peptide by the relevant protease(s).
Comparison of secondary TCD8+ responses to NP
Although VV-NP does not elicit primary TCDB+ measurable in the 51Cr release assays performed, it is known to prime for a secondary TCD8+ response when splenocytes are rested after primary immunization, then restimulated in vitro with peptide-pulsed or virus-infected stimulators (40). To compare the efficacy of rVV priming of secondary TCD8+ responses, splenocytes were removed from mice immunized 30 days earlier, stimulated in vitro for 7 days with the NP147–155 synthetic peptide, and tested for lytic activity against target cells sensitized with the same peptide. As shown in Figure 5, VV-NP and VV-ESNP147–155, were found to have similar priming potencies at the lowest doses (some minimal priming was obtained using as little as 500 PFU/mouse). However, at virus dosages of 5 × 104 or greater, mice primed with VV-ESNP147–155 produced splenocyte cultures with approximately 10 times greater activity when compared in lytic units than cultures prepared from mice primed with VV-NP.
FIGURE 5.
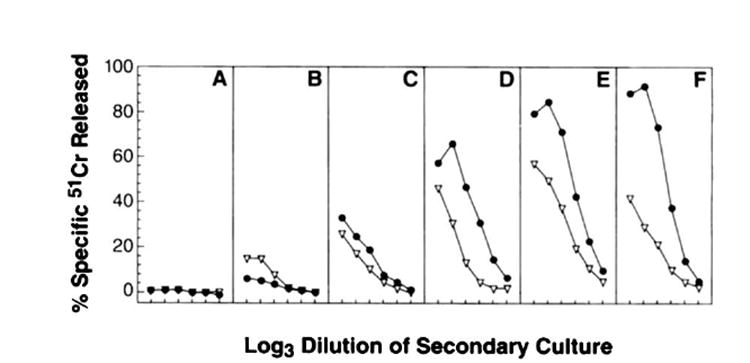
Comparison of secondary TCD8+ responses to NP. BALB/c mice were primed with either VV-NP (▽) or VV-ES NP147–155 (●) at the following PFU/mouse: 5 × 101 (A); 5 × 102 (B); 5 × 103 (C); 5 × 104 (D); 5 × 105 (E); and 5 × 106 (F). Thirty days later, splenocytes were removed and stimulated in vitro for 7 days with the NP147–155 synthetic peptide. Cultured effector cells were then tested for lytic activity against target cells sensitized with NP147–155. The experiment was repeated with similar results.
Extension of findings to other mouse strains and other model Ags
These results provided clear evidence that the ability of rVV to induce NP-specific responses in BALB/c mice was greatly enhanced by expression of a preprocessed peptide. Similar findings were obtained in DBA/2n mice, which are also H-2d (data not shown). Moreover, we found that a rVV expressing the immunodominant NP peptide (residues 50–57) presented by H-2K k (35) was more immunogenic in CBA/j (Fig. 6) or C3H mice (not shown) than a rVV expressing the full length protein. Appendage of the E3/19K signal sequence to the peptide had a mariginal effect on its immunogenicity. Interestingly, NP-specific responses elicited by the rVV expressing the rapidly degraded 1–168 fragment were of similar magnitude as those elicited by the rVV-expressing full length NP, indicating that increased proteolysis does not guarantee increased production of the immunodominant antigenic peptide.
FIGURE 6.
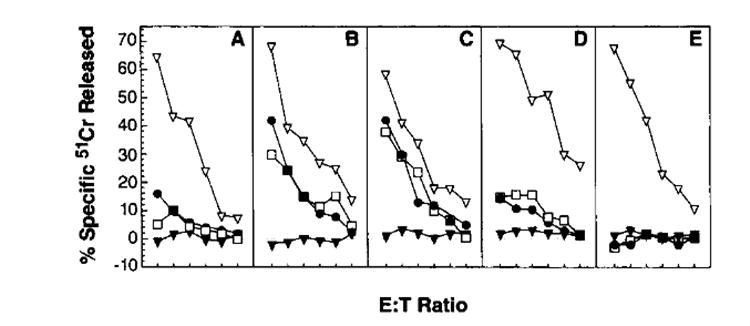
Primary TCD8+ responses to rVV coding for forms of NP. CBA/j mice were immunized with 5 × 106 PFU/mouse of the following constructs: VV-NP (A), VV-NPM50–57 (B), VV-ESNP50–57 (C), VV-NP1–168 (D), and VVNP147–315 (E). Six days after immunization, splenocytes were tested for their ability to lyse target cells at an E:T ratio of 200:1, 100:1, 50:1, 25:1, 12:1, and 6:1. Target cells were uninfected L929 (H-2k) (▼), L929 pulsed with synthetic peptide NP50–57 of PR8 (SDYEGRLI) (●), PR8-infected L929 cells (□), or VV- infected L929 cells (▽). This experiment was repeated with similar results.
In extending our findings to other Ags, we observed two situations in C57BL/6nmice in which rVV-encoding full length proteins and the corresponding peptides were similarly immunogenic. In the first case, we used rVV expressing either full length OVA (VV-OVA) or the immunodominant Kb-restricted peptide (residues 257–264) (51) preceded by Met or the E3/19K signal sequence (27). As shown in Figure 7, we consistently found that VV-OVA induced detectable primary responses that were only slightly less vigorous than those elicited by the peptide-producing rVV. In the second example, C57BL/6n mice responded vigorously to a rVV expressing the nucleocapsid protein of vesicular stomatitis virus, and responses were not enhanced by the expression of the immunodominant peptide (residues 57–64) (52) preceded by Met or the E3/19K signal sequences (data not shown).
FIGURE 7.
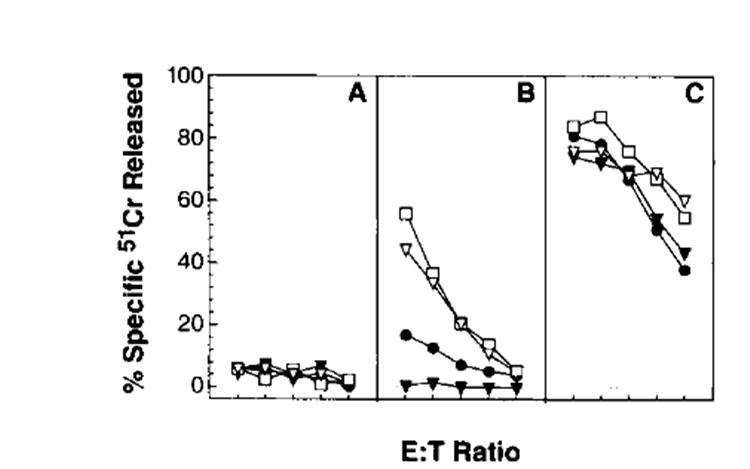
Primary TCD8+ responses to rVV-encoding forms of OVA. C57BL/6n mice were immunized with 5 × 106 PFU/mouse of the following constructs: VV-OVA (●), VV-OVAM257–264 (▽), VV-ESOVA257–264 (□) and, as a control, VV-ESNPM147–155 (▼). Six days after immunization, splenocytes were tested for their ability to lyse target cells at an E:T ratio of 100:1, 50:1, 25:1, 12:1, and 6:1. Target cells were WP6, a clone of the MCA 205 methylcholanthrine-induced sarcoma (H-2b) (A), WP6-pulsed with the Kb-restricted OVA257–264 peptide, SIINFEKL (B), or WP6-infected with a nonrecombinant vaccinia virus (CR19 of WR) (C). This experiment was repeated with similar results.
Immunogenicity of a nonmutated “self” protein Ag
The induction of Ld-restricted, P1A tumor Ag-specific TCD8+ responses in DBA/2n mice by rVV provided a more stringent test of the immunogenicity of rVV expressing different forms of antigenic peptides. The P815A Ag is encoded by the P1A gene, which is a normal, nonmutated cellular protein, is expressed by at least one unrelated, syngeneic mast cell line (36, 37), and is only weakly immunogenic.
rVV expressing full length PlA, the minimal antigenic determinant preceded by Met, or the minimal determinant preceded by theE3/19K ER insertion sequence were all able to sensitize target cells for lysis by P1A-specific TCD8+ (Irvine,1995, This issue). Presumably because of poor immunogenicity, none of the rVV induced measurable primary TCD8+ responses. We thus examined their ability to prime for secondary in vitro responses to the synthetic peptide P815A35–43 (LPYLGWLVF), 30 days after a priming with vaccinia virus. No P1A-specific responses were found after stimulation of splenocytes from mice immunized with a control rVV. By contrast, responses were observed using splenocytes from mice primed with VV-P1A. Interestingly, this response was much more vigorous than that induced by priming with VV-PlAM35–43. (Fig. 8). The most vigorous response was obtained when the ES sequence was added to the NH2 terminus (VV-ESPlA35–43).
FIGURE 8.
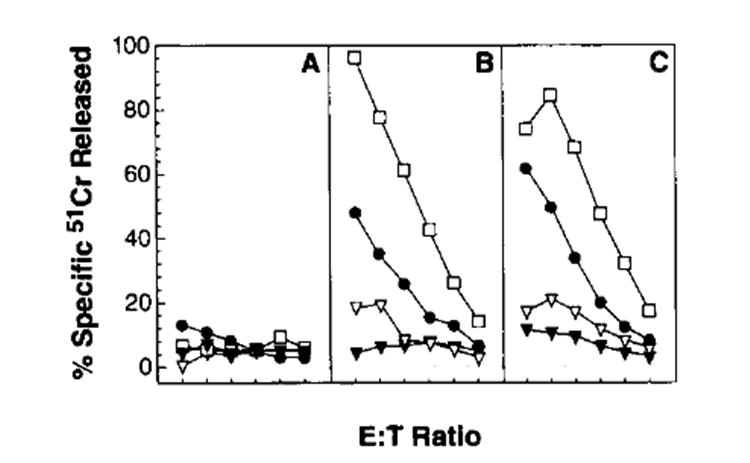
Secondary TCD8+ responses to rVV-encoding forms of P1A. DBA/2n mice were immunized with the following constructs: VV-P1A (●) VV-P1AM35–43 (▽), VV-ESPlA35–43(□) and, as a control, VV-NPM147-155 (▼). Thirty days later, splenocytes were harvested, dispersed, and stimulated in vitro for 7 days with the P815A35–43 synthetic peptide. Cultured effector cells were then tested for lytic activity against target cells at an E:T ratio of 100:1, 50:1, 25:1, 12:1, 6:1, and 3:1. Target cells were the N-nitroso-N-methylurethane-induced murine colon tumor line CT-26 (H-2d) (A); CT-26-pulsed with the P81535–43 peptide presented by Ld (LPYLGWLVF) (B); and the P815 mastocytoma, which is recognized in this experiment without being pulsed with peptide (because it naturally expresses P815A (C). Positive control vaccinia virus-infected target cells were not used because the effectors were secondarily stimulated splenocytes. This experiment was repeated with similar results.
Discussion
We have found that the immunogenicity of Ags recognized by TCD8+, normally expressed as segments of a viral or cellular proteins, can be enhanced when expressed by rVV as minimal determinants. Because transcription of mini-genes and longer genes in infected cells is under the control of the p7.5 W promoter, it is likely that similar amounts of mRNA encoding the various polypeptides are produced. If we assume that all of the rVV we have used produce the potential sources of antigenic peptides at equimolar levels in APCs in vivo, then it is likely that peptides with a single additional NH2-terminal Met or ER insertion signal sequence are more efficient sources of antigenic peptides than full length proteins in vivo. However, lacking suitable Abs reactive with mini-gene products, we unfortunately have no method of comparing the rate of minigene product synthesis with that of longer gene products.
The production of minimal antigenic determinants that translocate into the ER in a TAP-independent fashion, putting the right sized fragment into the right intracellular compartment, has considerable intuitive appeal, but also has a number of theoretical disadvantages. In the natural setting, peptides destined to be complexed with class I molecules may originate with proteolytic structures that interact with TAP, or that transfers the peptides to a carrier that delivers the peptides to TAP (53), and TAP in turn interacts with class I molecules (29, 30). The peptides encoded by our leader sequence plus mini-gene constructs are unlikely to follow this intracellular trafficking pattern, because TAP is not involved (27), but we know nothing more about their topographic relationship with the Ag-processing machinery. If our peptides are produced into the cytosol, some portion of them could undergo proteolysis because of the proteolytic nature of this compartment. By this reasoning, peptides processed naturally by a proteolytic pathway may be more efficiently complexed with class I molecules. Thus, there is no a priori reason why mini-gene constructs should be superior to constructs encoding full length proteins. The data nevertheless assert themselves: in no situation thus far examined is a rVV expressing an ER-targeted oligopeptide less immunogenic than rVV expressing the original full length protein or other forms of the protein.
One particularly striking finding was that the P1A determinant was less immunogenic as a mini-gene product than as a full length gene product. The underlying mechanism could involve inefficient transport of the peptide by TAP, the destruction or sequestration of the peptide before TAP has a chance to transport to the peptide, or perhaps the most likely explanation for the failure of the mini-gene in the PIA construct is the presence of the initiating methionine. Met-aminopeptidase may fail to cleave in this case, because the second residue (Leu) is unfavorable to cleavage (54). The initiating methionine, in turn, could diminish the transport or binding of the peptide to the Ld molecule.
We noted a wide difference in the immunogenicity of different full length proteins relative to their corresponding mini-gene products. This may reflect differences in the efficiency of cellular proteases in liberating peptides from their longer precursors. The greater susceptibility of NP1–168 to degradation could account for its slightly enhanced immunogenicity relative to full length NP. Other factors involved could include differences in TAP-mediated peptide transport in the ER, peptide affinity for class I molecules, and the sensitivity of triggering primary T cells specific for a given peptide. All of these factors theoretically could be overcome by either overproducing a peptide in the cytosol or by delivering these peptides to the ER in a TAP-independent manner.
Use of minimal determinant constructs may have the advantage of avoiding the potential dangers of immunizing with proteins of known or potential oncogenic function if present in full length, such as ras or p53. Obviously, an oligopeptide is unlikely to retain the same oncogenic potential. Synthetic oligonucleotides expressed by a recombinant vaccinia also have the advantage that they could be rapidly tailor-made in cases in which a variety of mutations can exist, such as mutant p53 or SV40 Ags (55-57).
TCD8+ have been shown to play an important role in the immune response against cancer cells (58-60) and can be grown to large numbers in vitro and transferred adoptively to treat even large tumor burdens of some tumor histologies in both mouse and human (61, 62). Now that several tumor-associated Ags recognized by TCD8+ have been cloned and the relevant antigenic peptides have been identified (36, 37, 63-71), it is now possible to apply the strategy described here to anticancer vaccines. The recent demonstration of the uses of TCD8+ in the restoration of viral immunity to CMV (72), and the possibility that such strategies could be useful for HIV (73), points the way to immunotherapeutic strategies involving ER-targeted peptide vaccines.
Acknowledgments
The technical contributions of D. Jones, B. Palmer, J. Stephens, and A. Taggarse are gratefully acknowledged.
Abbreviations used in this paper
- TCD8+
CD8+ T lymphocytes
- ER
endoplasmic reticulum
- NP
nucleoprotein
- PR8
influenza A/PR/8/34 virus
- rVV
recombinant vaccinia virus
- PFU
plaque-forming units
References
- 1.Deverson E, Gow IR, Coadwell WJ, Monaco JJ, Butcher GW, Howard JC. MHC class II region encoding proteins related to the multidrug resistance family of transmembrane transporters. Nature. 1990;348:738. doi: 10.1038/348738a0. [DOI] [PubMed] [Google Scholar]
- 2.Trowsdale J, Hanson I, Mockridge I, Beck S, Townsend A, Kelly A. Sequences encoded in the class II region of the MHC related to the “ABC” superfamily of transporters. Nature. 1990;348:741. doi: 10.1038/348741a0. [DOI] [PubMed] [Google Scholar]
- 3.Spies T, Bresnahan M, Bahram S, Arnold D, Blanck G, Mellins E, Pious D, DeMars R. A gene in the human major histocompatibility complex class II region controlling the class I antigen presentation pathway. Nature. 1990;348:744. doi: 10.1038/348744a0. [DOI] [PubMed] [Google Scholar]
- 4.Monaco JJ, Cho S, Attaya M. Transport protein genes in the murine MHC: possible implications for antigen processing. Science. 1990;250:1723. doi: 10.1126/science.2270487. [DOI] [PubMed] [Google Scholar]
- 5.Hammond SA, Bollinger RC, Tobery TW, Siliciano RF. Transporter-independent processing of HIV-1 envelope protein for recognition by CD8+ T cells. Nature. 1993;364:158. doi: 10.1038/364158a0. [DOI] [PubMed] [Google Scholar]
- 6.Esquivel F, Yewdell J, Bennink J. RMA/S cells present endogenously synthesized cytosolic proteins to class I-restricted cytotoxic T lymphocytes. J Exp Med. 1992;175:163. doi: 10.1084/jem.175.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartmann E, Sommer T, Prehn S, Gorlich D, Jentsch S, Rapoport TA. Evolutionary conservation of components of the protein translocation complex. Nature. 1994;367:654. doi: 10.1038/367654a0. [DOI] [PubMed] [Google Scholar]
- 8.Nicchitta CV, Blobel G. Lumenal proteins of the mammalian endoplasmic reticulum are required to complete protein translocation. Cell. 1993;73:989. doi: 10.1016/0092-8674(93)90276-v. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann E, Gorlich D, Kostka S, Otto A, Kraft R, Knespel S, Burger E, Rapoport TA, Prehn S. A tetrameric complex of membrane proteins in the endoplasmic reticulum. Eur J Biochem. 1993;214:375. doi: 10.1111/j.1432-1033.1993.tb17933.x. [DOI] [PubMed] [Google Scholar]
- 10.Henderson RA, Michel H, Sakaguchi K, Shabanowitz J, Appella E, Hunt DF, Engelhard VH. HLA-A2.1-associated peptides from a mutant cell line: a second pathway of antigen presentation. Science. 1992;255:1264. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 11.Townsend A, Bastin J, Gould K, Brownlee G, Andrew M, Coupar B, Boyle D, Chan S, Smith G. Defective presentation to class I-restricted cytotoxic T lymphocytes in vaccinia-infected cells is overcome by enhanced degradation of antigen. J Exp Med. 1988;168:1211. doi: 10.1084/jem.168.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Townsend A, Bodmer H. Antigen recognition by class I-restricted T lymphocytes. Annu Rev Immunol. 1989;7:601. doi: 10.1146/annurev.iy.07.040189.003125. [DOI] [PubMed] [Google Scholar]
- 13.Yewdell JW, Bennink JR. Cell biology of antigen processing and presentation to major histocompatibility complex class I molecule-restricted T lymphocytes. Adv Immunol. 1992;52:1. doi: 10.1016/s0065-2776(08)60875-5. [DOI] [PubMed] [Google Scholar]
- 14.Michalek MT, Grant EP, Gramm C, Goldberg AL, Rock KL. A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature. 1993;363:552. doi: 10.1038/363552a0. [DOI] [PubMed] [Google Scholar]
- 15.Fehling HJ, Swat W, Laplace C, Kuhn R, Rajewsky K, Muller U, von Boehmer H. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science. 1994;265:1234. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 16.Akiyama K, Yokota K, Kagawa S, Shimbara N, Tamura T, Akioka H, Nothwang HG, Noda C, Tanaka K, Ichihara A. cDNA cloning and interferon gamma down-regulation of proteasomal subunits X and Y. Science. 1994;265:1231. doi: 10.1126/science.8066462. [DOI] [PubMed] [Google Scholar]
- 17.Zhou X, Momburg F, Liu T, Abdel Motal UM, Jondal M, Hammerling GJ, Ljunggren HG. Presentation of viral antigens restricted by H-2Kb, Db or Kd in proteasome subunit LMP2-and LMP7-deficient cells. Eur J Immunol. 1994;24:1863. doi: 10.1002/eji.1830240822. [DOI] [PubMed] [Google Scholar]
- 18.Fruh K, Gossen M, Wang K, Bujard H, Peterson PA, Yang Y. Displacement of housekeeping proteasome subunits by MHC-encoded LMPs: a newly discovered mechanism for modulating the multicatalytic proteinase complex. EMBO J. 1994;13:3236. doi: 10.1002/j.1460-2075.1994.tb06625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dick LR, Aldrich C, Jameson SC, Moomaw CR, Pramanik BC, Doyle CK, DeMartino GN, Bevan MJ, Forman JM, Slaughter CA. Proteolytic processing of ovalbumin and beta-galactosidase by the proteasome to yield antigenic peptides. J Immunol. 1994;152:3884. [PMC free article] [PubMed] [Google Scholar]
- 20.Boes B, Hengel H, Ruppert T, Multhaup G, Koszinowski UH, Kloetzel PM. Interferon gamma stimulation modulates the proteolytic activity and cleavage site preference of 20S mouse proteasomes. J Exp Med. 1994;179:901. doi: 10.1084/jem.179.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yewdell J, Lapham C, Bacik I, Spies T, Bennink J. MHC-encoded proteasome subunits LMP2 and LMP7 are not required for efficient antigen presentation. J Immunol. 1994;152:1163. [PubMed] [Google Scholar]
- 22.Monaco JJ, McDevitt HO. H-2-linked low molecular weight polypeptide antigens assemble into an unusual macromolecular complex. Nature. 1984;309:797. doi: 10.1038/309797a0. [DOI] [PubMed] [Google Scholar]
- 23.Gaczynska M, Rock KL, Goldberg AL. γ-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365:264. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 24.Driscoll J, Brown MG, Finley D, Monaco JJ. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature. 1993;365:262. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 25.Ortiz-Navarrete V, Seelig A, Gernold M, Frentzel S, Kloetzel PM, Hammerling GJ. Subunit of the “20S” proteasome (multicatalytic proteinase) encoded by the major histocompatibility complex. Nature. 1991;353:662. doi: 10.1038/353662a0. [DOI] [PubMed] [Google Scholar]
- 26.Anderson K, Cresswell P, Gammon M, Hermes J, Williamson A, Zweerink H. Endogenously synthesized peptide with an endoplasmic reticulum signal sequence sensitizes antigen processing mutant cells to class I-restricted cell-mediated lysis. J Exp Med. 1991;174:489. doi: 10.1084/jem.174.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bacik I, Cox JH, Anderson R, Yewdell JW, Bennink JR. TAP (transporter associated with antigen processing)-independent presentation of endogenously synthesized peptides is enhanced by endoplasmic reticulum insertion sequences located at the amino — but not carboxyl — terminus of the peptide. J Immunol. 1994;152:381. [PubMed] [Google Scholar]
- 28.Eisenlohr LC, Bacik I, Bennink JR, Bemstein K, Yewdell JW. Expression of a membrane protease enhances presentation of endogenous antigens to MHC class I-restricted T lymphocytes. Cell. 1992;71:963. doi: 10.1016/0092-8674(92)90392-p. [DOI] [PubMed] [Google Scholar]
- 29.Ortmann B, Androlewicz MJ, Cresswell P. MHC class I/β2-microglobulin complexes associate with TAP transporters before peptide binding. Nature. 1994;368:864. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 30.Suh WK, Cohen-Doyle MF, Fruh K, Wang K, Peterson PA, Williams DB. Interaction of MHC class I molecules with the transporter associated with antigen processing. Science. 1994;264:1322. doi: 10.1126/science.8191286. [DOI] [PubMed] [Google Scholar]
- 31.Chakrabarti S, Brechling K, Moss B. Vaccinia virus expression vector: coexpression of β-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985;5:3403. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith GL, Levin JZ, Palese P, Moss B. Synthesis and cellular location of the ten influenza polypeptides individually expressed by recombinant vaccinia viruses. Virology. 1987;160:336. doi: 10.1016/0042-6822(87)90004-3. [DOI] [PubMed] [Google Scholar]
- 33.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867. [PubMed] [Google Scholar]
- 34.Falk K, Rötzschke O, Deres K, Metzger J, Jung G, Rammensee H-G. Identification of naturally processed viral non-apeptides allows their quantification in infected cells and suggests an allele-specific T cell epitope forecast. J Exp Med. 1991;174:425. doi: 10.1084/jem.174.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gould KG, Scotney H, Brownlee GG. Characterization of two distinct major histocompatibility complex class I Kk-restricted T cell epitopes within the influenza A/PR/8/34 virus hemagglutinin. J Virol. 1991;65:5401. doi: 10.1128/jvi.65.10.5401-5409.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lethe B, Van den Eynde B, Van Pel A, Corradin G, Boon T. Mouse tumor rejection antigens P815A and P815B: two epitopes carried by a single peptide. Eur J Immunol. 1992;22:2283. doi: 10.1002/eji.1830220916. [DOI] [PubMed] [Google Scholar]
- 37.Van den Eynde B, Lethe B, Van Pel A, De Plaen E, Boon T. The gene coding for a major tumor rejection antigen of tumor P815 is identical to the normal gene of syngeneic DBA/2 mice. J Exp Med. 1991;173:1373. doi: 10.1084/jem.173.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falk K, Rötzschke O, Stevanovic S, Jung G, Rammensee H-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 39.Restifo NP, Esquivel F, Kawakami Y, Yewdell JW, Mule JJ, Rosenberg SA, Bennink JR. Identification of human cancers deficient in antigen processing. J Exp Med. 1993;177:265. doi: 10.1084/jem.177.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Restifo NP, Esquivel F, Asher AL, Stötter H, Barth RJ, Bennink JR, Mulé JJ, Yewdell JW, Rosenberg SA. Defective presentation of endogenous antigens by a murine sarcoma: implications for the failure of an antitumor immune response. J Immunol. 1991;147:1453. [PMC free article] [PubMed] [Google Scholar]
- 41.Restifo NP, Spiess PJ, Karp SE, Mule JJ, Rosenberg SA. A nonimmunogenic sarcoma transduced with the cDNA for interferon gamma elicits CD8+ T cells against the wild-type tumor: correlation with antigen presentation capability. J Exp Med. 1992;175:1423. doi: 10.1084/jem.175.6.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brattain MG, Strobel-Stevens J, Fine D, Webb M, Sarrif AM. Establishment of mouse colonic carcinoma cell lines with different metastatic properties. Cancer Res. 1980;40:2142. [PubMed] [Google Scholar]
- 43.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yewdell JW, Frank E, Gerhard W. Expression of influenza A virus internal antigens on the surface of infected P815 cells. J Immunol. 1981;126:1814. [PubMed] [Google Scholar]
- 45.Yewdell JW, Yellen A, Bächi T. Monoclonal antibodies localize events in the folding, assembly, and intracellular transport of the influenza virus hemagglutinin glycoprotein. Cell. 1988;52:843. doi: 10.1016/0092-8674(88)90426-6. [DOI] [PubMed] [Google Scholar]
- 46.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 47.Bennink JR, Yewdell JW. Recombinant vaccinia viruses as vectors for studying T lymphocyte specificity and function. Curr Top Microbiol Immunol. 1990;163:153. doi: 10.1007/978-3-642-75605-4_6. [DOI] [PubMed] [Google Scholar]
- 48.Bennink JR, Yewdell JW, Smith GL, Moller C, Moss B. Recombinant vaccinia virus primes and stimulates influenza haemagglutinin-specific cytotoxic T cells. Nature. 1984;311:578. doi: 10.1038/311578a0. [DOI] [PubMed] [Google Scholar]
- 49.Bodmer HC, Pemberton RM, Rothbard JB, Askonas BA. Enhanced recognition of a modified peptide antigen by cytotoxic T cells specific for influenza nucleoprotein. Cell. 1988;52:253. doi: 10.1016/0092-8674(88)90514-4. [DOI] [PubMed] [Google Scholar]
- 50.Eisenlohr LC, Yewdell JW, Bennink JR. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxicT lymphocytes. J Exp Med. 1992;175:481. doi: 10.1084/jem.175.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jameson SC, Bevan MJ. Dissection of major histocompatibility complex (MHC) and T cell receptor contact residues in a Kb-restricted ovalbumin peptide and an assessment of the predictive power of MHC-binding motifs. Eur J Immunol. 1992;22:2663. doi: 10.1002/eji.1830221028. [DOI] [PubMed] [Google Scholar]
- 52.Van Bleek GM, Nathenson SG. Isolation ofan endogenously processed immunodominant viral peptide from the class I H-2Kb molecule. Nature. 1990;348:213. doi: 10.1038/348213a0. [DOI] [PubMed] [Google Scholar]
- 53.Srivastava PK. Heat shock proteins in immune response to cancer: the fourth paradigm. Experientia. 1994;50:1054. doi: 10.1007/BF01923461. [DOI] [PubMed] [Google Scholar]
- 54.Moerschell RP, Hosokawa Y, Tsunasawa S, Sherman F. The specificities of yeast methionine aminopeptidase and acetylation of amino-terminal methionine in vivo. Processing of altered iso-1-cytochromes c created by oligonucleotide transformation. J Biol Chem. 1990;265:19638. [PubMed] [Google Scholar]
- 55.Yanuck M, Carbone DP, Pendleton CD, Tsukui T, Winter SF, Minna JD, Berzofsky JA. A mutant p53 tumor suppressor protein is a target for peptide-induced CD8+ cytotoxic T cells. Cancer Res. 1993;53:3257. [PubMed] [Google Scholar]
- 56.Deckhut AM, Tevethia SS. Effect of point mutations in the native simian virus 40 tumor antigen, and in synthetic peptides corresponding to the H-2Db-restricted epitopes, on antigen presentation and recognition by CTL clones. J Immunol. 1992;148:3012. [PubMed] [Google Scholar]
- 57.Lill NL, Tevethia MJ, Hendrickson WG, Tevethia SS. Cytotoxic T lymphocytes (CTL) against a transforming gene product select for transformed cells with point mutations within sequences encoding CTL recognition epitopes. J Exp Med. 1992;176:449. doi: 10.1084/jem.176.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greenberg PD, Cheever MA, Fefer A. H-2 restriction of adoptive immunotherapy of advanced tumors. J Immunol. 1981;126:2100. [PubMed] [Google Scholar]
- 59.Rosenberg SA, Spiess P, Lafreniere R. A new approach to the adoptive immunotherapy of cancer with tumor-infiltrating lymphocytes. Science. 1986;233:1318. doi: 10.1126/science.3489291. [DOI] [PubMed] [Google Scholar]
- 60.Shimizu K, Shen F-W. Role of different T cell set in the rejection of syngeneic chemically induced tumors. J Immunol. 1979;122:1162. [PubMed] [Google Scholar]
- 61.Old LJ. Tumor immunology: the first century. Curr Opin Immunol. 1992;4:603. doi: 10.1016/0952-7915(92)90034-c. [DOI] [PubMed] [Google Scholar]
- 62.Greenberg PD. Adoptive T cell therapy of tumors: mechanisms operative in the recognition and elimination of tumor cells. Adv Immunol. 1991;49:281. doi: 10.1016/s0065-2776(08)60778-6. [DOI] [PubMed] [Google Scholar]
- 63.Brichard V, Van Pel A, Wolfel T, Wolfel C, De Plaen E, Lethe B, Coulie P, Boon T. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1993;178:489. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Traversari C, van der Bruggen P, Luescher IF, Lurquin C, Chomez P, Van Pel A, De Plaen E, Amar-Costesec A, Boon T. A nonapeptide encoded by human gene MAGE-1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J Exp Med. 1992;176:1453. doi: 10.1084/jem.176.5.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boon T, Cerottini J, Van den Eynde B, van der Bruggen P, Van Pel A. Tumor antigens recognized by T lymphocytes. Annu Rev Immunol. 1994;12:337. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- 66.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Rivoltini L, Topalian SL, Miki T, Rosenberg SA. Cloning of the gene coding for a shared human melanoma antigen recognized by autologous T cells infiltrating into tumor. Proc Natl Acad Sci USA. 1994;91:3515. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bakker AB, Schreurs MW, de Boer AJ, Kawakami Y, Rosenberg SA, Adema GJ, Figdor CG. Melanocyte lineage-specific antigen gpl00 is recognized by melanoma-derived tumor-infiltrating lymphocytes. J Exp Med. 1994;179:1005. doi: 10.1084/jem.179.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pardoll DM. Tumour antigens. A new look for the 1990s. Nature. 1994;369:357. doi: 10.1038/369357a0. [DOI] [PubMed] [Google Scholar]
- 69.Houghton AN. Cancer antigens: immune recognition of self and altered self. J Exp Med. 1994;180:1. doi: 10.1084/jem.180.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kawakami Y, Eliyahu S, Delgado CH, Robbins PF, Sakaguchi K, Appella E, Yannelli JR, Adema GJ, Miki T, Rosenberg SA. Identification of a human melanoma antigen recognized by tumor-infiltrating lymphocytes associated with in vivo tumor rejection. Proc Natl Acad Sci USA. 1994;91:6458. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cox AL, Skipper J, Chen Y, Henderson RA, Darrow TL, Shabanowitz J, Engelhard VH, Hunt DF, Slingluff CL., Jr Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994;264:716. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 72.Riddell SR, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science. 1992;257:238. doi: 10.1126/science.1352912. [DOI] [PubMed] [Google Scholar]
- 73.Riddell SR, Gilbert MJ, Greenberg PD. CD8+ cytotoxic T cell therapy of cytomegalovirus and HIV infection. Curr Opin Immunol. 1993;5:484. doi: 10.1016/0952-7915(93)90027-p. [DOI] [PubMed] [Google Scholar]
- 74.Winter G, Fields S. The structure of the gene encoding the nucleoprotein of human influenza virus A/PR/8/34. Virology. 1981;114:423. doi: 10.1016/0042-6822(81)90223-3. [DOI] [PubMed] [Google Scholar]
- 75.McReynolds L, O’Malley BW, Nisbet AD, Fothergill JE, Givol D, Fields S, Robertson M, Brownlee GG. Sequence of chicken ovalbumin mRNA. Nature. 1978;273:723. doi: 10.1038/273723a0. [DOI] [PubMed] [Google Scholar]
- 76.Rotzschke O, Falk K, Stevanovic S, Jung G, Walden P, Rammensee HG. Exact prediction of a natural T cell epitope. Eur J Immunol. 1991;21:2891. doi: 10.1002/eji.1830211136. [DOI] [PubMed] [Google Scholar]


