Abstract
Mammalian ornithine decarboxylase (ODC) is among the most labile of cellular proteins, with a half-life of usually less than an hour. Like other short-lived proteins ODC is degraded by the 26S proteasome. Its degradation is not triggered by ubiquitination, but is stimulated by the binding of an inducible protein, antizyme. Truncations and mutations in the C terminus of mammalian ODC have been shown to prevent the rapid turnover of the enzyme, demonstrating the presence of a degradation signal in this region. Moreover, ODCs from the trypanosomatid parasites Trypanosoma brucei and Leishmania donovani, which lack this C-terminal domain, are metabolically stable, and recombination of T. brucei ODC with the C terminus of mammalian ODC confers a short half-life to the fusion protein when expressed in mammalian cells. In the present study we have cloned and sequenced the ODC gene from the trypanosomatid Crithidia fasciculata. To our knowledge, this is the first protozoan shown to have an ODC with a rapid turnover. The sequence analysis revealed a high homology between C. fasciculata ODC and L. donovani ODC, despite the difference in stability. We demonstrate that C. fasciculata ODC has a very rapid turnover even when expressed in mammalian cells. Moreover, ODC from C. fasciculata is shown to lack the C-terminal degradation domain of mammalian ODC. Our findings indicate that C. fasciculata ODC contains unique signals, targeting the enzyme for rapid degradation not only in the parasite but also in mammalian cells.
Keywords: Crithidia fasciculata, protein turnover, polyamines, PEST region
Ornithine decarboxylase (ODC), which catalyzes the first step in the polyamine biosynthetic pathway, is an enzyme regulated by several unique mechanisms (1–3). The extensive regulation of ODC most likely reflects the importance of the polyamines in the cell. The polyamines are ubiquitous in nature and have been shown to be essential for a variety of cellular processes ranging from control of the membrane potential to cell growth (3–5). Thus ODC is a potential target for therapeutic agents against cancer and various parasitic diseases (3, 6). A remarkable finding is that humans afflicted with African sleeping sickness refractory to other antitrypanosomal drugs can be cured by treatment with an ODC inhibitor, 2-difluoromethylornithine (DFMO) (6, 7).
Mammalian ODC is regulated at a multitude of levels, including transcriptional, translational, and posttranslational (1). The polyamines exert a strong and rapid feedback control of ODC synthesis and degradation, providing the cell with an efficient mechanism for the regulation of polyamine levels (1–3). An important characteristic of mammalian ODC is its very rapid turnover (2). In fact the biological half-life of ODC, which may be as short as a few minutes, is one of the shortest known for a mammalian enzyme. The turnover of ODC is partly regulated by polyamines (2). An excess of polyamines stimulates the degradation of ODC through the induction of a specific protein, antizyme, that binds strongly to ODC and targets it for degradation (2, 8, 9). The polyamines induce the synthesis of antizyme through a unique mechanism involving a frameshift during translation of the antizyme mRNA (10, 11). Also the degradation of ODC occurs through an extraordinary mechanism, involving an ATP-dependent but ubiquitin-independent process catalyzed by the 26S proteolytic complex called the proteasome (12, 13). This is the first example of a non-ubiquitinated protein being degraded by the 26S proteasome.
Molecular analysis of the ODC protein has revealed sequences important for the short half-life of the enzyme. Truncation of the C-terminal part of mouse ODC converts the enzyme into a stable protein (14). Furthermore, mammalian ODC, like many other proteins with short half-lives, contains so-called PEST regions (15) that are rich in proline (P), glutamic acid (E), aspartic acid (D), serine (S) and threonine (T). Mammalian ODC has two PEST regions, one of which is located within the C terminus of the protein (14). ODC from Trypanosoma brucei, the causative agent of African sleeping sickness, is a stable protein in the parasite (16) as well as when expressed in mammalian cells (17, 18). Compared with mammalian ODC, T. brucei ODC is truncated at the C terminus and therefore lacks one of the PEST sequences (17, 18). Recombining T. brucei ODC with the C terminus of mammalian ODC confers a short half-life to the fusion protein expressed in mammalian cells (17, 18). Also the ODC from Leishmania donovani, the causative agent of visceral leishmaniasis, lacks the sequence corresponding to the C terminus of mammalian ODC and is a stable protein in the parasite (19). Other parasites that express an ODC with a slow turnover rate are Leishmania mexicana (20) and the malaria parasite Plasmodium falciparum (21). However, the amino acid sequences of these ODCs are not yet known.
Recently, it was demonstrated that the nonpathogenic trypanosomatid Crithidia fasciculata contains an ODC that has a fast turnover (22). The protozoan flagellates of the genus Crithidia are monogenetic trypanosomatid parasites that colonize the digestive tract of infected flies. Inhibition of protein synthesis in the parasite, using cycloheximide, resulted in a rapid loss of ODC activity with a half-life of about 30 min (22). In addition, the polyamine-mediated repression of ODC, usually found in organisms expressing a short-lived ODC, was absent in this parasite (22). To our knowledge, this is the first demonstration of a metabolically unstable ODC in a trypanosomatid. Because this experimental system may conceal information important for the understanding of the molecular mechanisms involved in the rapid turnover as well as the feedback control of ODC, we have isolated and sequenced the ODC gene from C. fasciculata. The deduced amino acid sequence of C. fasciculata ODC was almost 70% identical to that of L. donovani ODC (19). We demonstrate that the protein has a very fast turnover even when expressed in mammalian cells. However, the fact that C. fasciculata ODC lacks the part corresponding to the C terminus of mammalian ODC may be taken to denote that other signals mediate the fast turnover of the protein.
MATERIALS AND METHODS
Materials.
l-[1-14C]ornithine (57 mCi/mmol; 1 Ci = 37 GBq) was obtained from New England Nuclear. [35S]dCTP (1000 Ci/mmol) was purchased from Amersham. The eukaryotic expression vector (pSVL) containing the simian virus 40 late promoter and origin of replication was obtained from Pharmacia. The λGEM-11 genomic library from C. fasciculata was a kind gift from D. S. Ray (23). All restriction enzymes, DNA ligase, Taq DNA polymerase, and avian myeloblastosis virus reverse transcriptase were obtained from Boehringer Mannheim. Genomic clones of T. brucei ODC and L. donovani ODC were generously provided by C. C. Wang (16) and B. Ullman (19), respectively.
Cell Culture.
A C. fasciculata laboratory strain (ATCC 11745) was cultured at 28°C in a defined medium (HOSMEM II) (24). The cultures were supplemented with 50 units of penicillin and 50 μg/ml of streptomycin. African green monkey COS-7 cells (ATCC CRL 1651) were grown at 37°C in DMEM containing 10% fetal calf serum, nonessential amino acids, and antibiotics (50 units of penicillin and 50 μg/ml of streptomycin) in the presence of 5% CO2/95% air.
Isolation of Nucleic Acids.
Genomic DNA (25) and total RNA (26) were isolated from C. fasciculata according to published methods.
Isolation of an ODC DNA Fragment from C. fasciculata Using PCR.
A DNA probe for the isolation of the ODC gene from C. fasciculata was obtained by amplifying an ODC-specific DNA fragment using degenerate primers from conserved regions of ODC. The sense primer 5′-GGCGAATTCRTNTWYGCNAAYCCNTGYAA-3′ and the antisense primer 5′-GGCGAATTCACNCCRTCRTTNACRTARTA-3′ corresponded to the conserved regions containing the amino acids (V/I)(Y/F)ANPCK and YYVNDG(V/L), respectively. The PCR was carried out in the presence of 10 mM Tris·HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM of each dNTP, 100 pmol of each oligonucleotide primer, and 2.5 units of Taq DNA polymerase in a volume of 100 μl. The program used was as follows: 95°C for 15 s, 45°C for 30 s, and 75°C for 2 min (35 cycles) followed by a final extension step for 5 min at 75°C. Analysis of the PCR products on a 1% agarose gel gave a single band of about 700 bp which was in agreement with the expected size of the ODC fragment. The fragment was subcloned into pBluescript KS+ (Stratagene) and partially sequenced using the dideoxy chain termination method.
Isolation of the C. fasciculata ODC Gene.
A λGEM-11 C. fasciculata genomic library (23) was screened for ODC positive sequences using the cloned PCR product labeled with [32P]dCTP. The plaque-forming units were blotted onto Hybond N filters (Amersham) and prehybridized at 42°C in 6× SSPE (20× SSPE = 3 M NaCl/0.2 M NaH2PO4/20 mM EDTA, pH 7.4), 0.5% SDS, 10% dextran, 50% formamide and 1 mg/ml DNA for 3–4 h. The 32P-labeled PCR product was then added and the filters were hybridized overnight. The filters were washed for 2 × 5 min and 2 h at room temperature in 2× SSC (20× SSC = 3 M NaCl/0.3 M sodium citrate, pH 7.0) and 0.1% SDS followed by two high stringency washes for 30 min at 65°C in 0.1× SSC and 0.1% SDS. Positive plaques were further purified by new rounds of plating and screening. One of the clones was selected for further analysis.
Subcloning and Sequencing of the C. fasciculata ODC Gene.
The isolated bacteriophage DNA was digested with SfiI or SacI followed by fractionation on a 0.8% agarose gel. The SfiI digest showed a 12-kb insert and the SacI gave three specific bands of about 4.5, 4.3, and 2.3 kb. The SacI fragments were blotted onto a Hybond N membrane and hybridized to the cloned PCR product as described. The 4.5-kb fragment was shown to hybridize to the 32P-labeled PCR product. The isolated fragment was then subcloned into pBluescript KS+ and both strands were sequenced using the dideoxy chain termination method with the T7 sequencing kit from Pharmacia. Additional sequencing was done with an Applied Biosystems model 373A sequencer using the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems). This fragment was shown to contain the complete ORF of C. fasciculata ODC as well as long stretches of the 3′ and 5′ untranslated regions, including the 5′ splice acceptor site.
Southern and Northern Blot Analyses.
Genomic DNA (10 μg) from C. fasciculata was digested with various restriction enzymes. The resulting DNA fragments were separated on a 0.8% agarose gel, transferred to Hybond N (Amersham) membranes, and hybridized to the 32P-labeled PCR product as described. For identification of the C. fasciculata ODC transcript 20 μg of total RNA was separated on a 1% agarose gel containing 2.2 M formaldehyde and blotted onto a Hybond N membrane. The membrane was then hybridized to the 32P-labeled PCR product.
Mapping of the 5′ Splice Acceptor Site of the ODC Transcript.
The 5′ end of the ODC mRNA was determined by amplification and sequencing of the 5′ terminus of the mature ODC transcript. The reverse transcription was performed at 42°C for 60 min in the presence of 300 ng random hexanucleotides, 125 nM of each dNTP, 20 units human placenta ribonuclease inhibitor, 50 μg total RNA from C. fasciculata, and 50 units of avian myeloblastosis virus reverse transcriptase in a total volume of 20 μl. To amplify the 5′ end of the C. fasciculata ODC cDNA, 2.0 μl of the reverse transcriptase reaction was added to a PCR containing 10 mM Tris·HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM of each dNTP, 20 pmol of each oligonucleotide primer, and 2.6 units of Taq DNA polymerase/pwo DNA polymerase (expand high fidelity PCR system, Boehringer Mannheim). The sense primer, 5′-CCATCTAGAGCTATATAAGTATCAGTTTCTGTACTT-3′, contained 27 nt of the 39-nt mini-exon that is trans-spliced onto the 5′ end of the transcript (27, 28). The antisense primer, 5′-CGCACCATCGGCAACTCGTGCC-3′, contained a nucleotide sequence from the coding region of the C. fasciculata ODC gene. The PCR program included a primary denaturation for 5 min at 95°C and 30 cycles at 96°C, 45°C, and 72°C for 15, 30, and 60 s, respectively, followed by a final extension at 72°C for 5 min. The PCR product was then further amplified by nested PCR using two new antisense oligonucleotides, 5′-TCTGGATGTACACGTTGGAGAGG-3′ and 5′-GGCAGAGAAGCTCATCCCG-3′. The PCR resulted in a product of about 900 bp, which was subcloned into the pCR II vector using a TA cloning kit (Invitrogen) and then sequenced as described above.
Expression of ODCs from C. fasciculata, T. brucei, and L. donovani in COS Cells.
As has been demonstrated for many other transcripts from trypanosomatids, the translational initiation sequences of ODCs from C. fasciculata (present study), T. brucei (29) and L. donovani (19) do not comply with the Kozak consensus sequence (30, 31). Thus, to be able to express these ODCs in a mammalian cell we modified the context of the initiation codon to CCACCATG by PCR. The modified ODC DNAs were then subcloned into the mammalian expression vector pSVL.
For transfection, COS cells were harvested during exponential growth and resuspended in 0.8 ml fresh growth medium at a density of 10 × 106 cells/ml. The transfection was performed by electroporation. The cells were mixed with 15 μg of DNA and then pulsed with 0.3 kV at 250 μF. Following a 5-min recovery period at room temperature the cells were seeded in fresh medium at a density of 28,000 cells/cm2. The cells were analyzed 2 days after transfection. The turnover of ODC was determined by following the decay of ODC activity after cycloheximide treatment (50 μg/ml).
ODC Assay.
The COS cells were sonicated in ice-cold 0.1 M Tris·HCl (pH 7.5) containing 0.1 mM EDTA and 2.5 mM dithiothreitol, and the debris was sedimented by centrifugation at 30,000 × g for 20 min. The expression of C. fasciculata ODC in the COS cells, which was much less than that of T. brucei ODC or L. donovani ODC, was determined after precipitation of the endogenous COS ODC using an excess of a specific antibody against mammalian ODC (32). The ODC activity was determined by measuring the release of 14CO2 from l-[1-14C]ornithine in the presence of saturating levels of pyridoxal 5′-phosphate (0.1 mM) and l-ornithine (0.5 mM). Protein concentrations were determined by the method of Bradford (33).
Computer Analysis.
The sequence analyses were performed with the gap/bestfit comparison program and the multiple sequence alignment program pileup of the Wisconsin package, Genetics Computer Group.
RESULTS
Isolation of the ODC Gene from a C. fasciculata Genomic Library.
A suitable probe for screening of the C. fasciculata λGEM-11 genomic library was obtained by amplifying an ODC-specific DNA fragment from C. fasciculata by PCR using degenerate oligonucleotides based on conserved amino acid sequences in ODC molecules from various eukaryotes. We obtained a 689-bp PCR product that was subcloned and partly sequenced. The sequence revealed a high homology to the ODC gene from L. donovani (results not shown), strongly indicating that the PCR product contained a sequence from the C. fasciculata ODC gene. This PCR product was then used to screen the genomic library. Out of 50,000 screened plaques about 10 were positive. One of the positive plaques, containing a clone with a ≈12-kb insert, was used for further analysis. A 4.5-kb restriction fragment of the clone was selected for subcloning and sequencing.
Sequencing of the ODC Gene from C. fasciculata.
The nucleotide sequence of the C. fasciculata ODC clone was found to contain a single long ORF of 2163 bp encoding a protein of 721 aa with a calculated molecular mass of 78,913 Da (Fig. 1). The deduced amino acid sequence showed a high homology with ODCs from other eukaryotes (Fig. 2). In addition to the ORF, ≈1100 and ≈350 nt of the 5′ and 3′ flanking regions, respectively, were sequenced from the clone. Analysis of the codon usage revealed that the ORF of C. fasciculata ODC, like those of other genes from C. fasciculata and L. donovani, contains an over-representation of G or C at the third codon position (35). About 85% of the codons used contain a G or C residue as the third nucleotide in the C. fasciculata ODC gene.
Figure 1.
Nucleotide and amino acid sequences of the C. fasciculata ODC gene and its predicted protein product. The 2163 nt ORF is numbered starting with the A in the first ATG. The 721 aa are numbered with the initiating methionine as number 1. The sites of the two degenerate PCR primers used for obtaining the ODC-specific probe are underlined. Nucleotides 5′ to the splice acceptor site are printed in lowercase letters.
Figure 2.
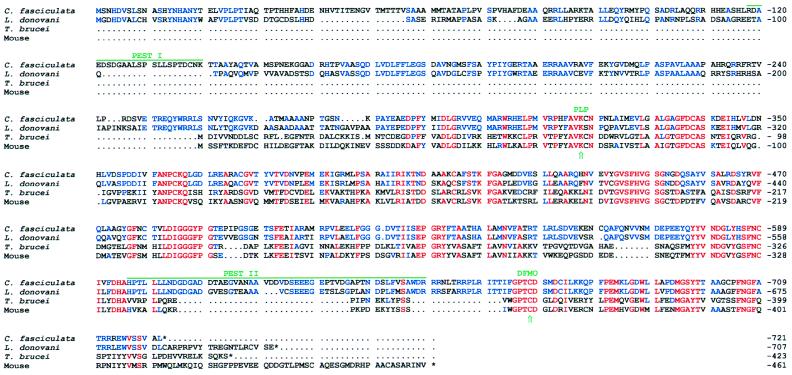
Comparison of the amino acid sequences of ODC proteins from C. fasciculata, T. brucei, L. donovani, and mouse. The deduced amino acid sequences were compared using the multiple sequence alignment program pileup. Amino acids that are identical in all of the species are printed in red. Amino acids that are not identical in all four species but identical in C. fasciculata and L. donovani are printed in blue. The binding sites for the cofactor pyridoxal 5′-phosphate and the irreversible inhibitor DFMO (34) are indicated. The PEST regions in C. fasciculata ODC are indicated by green lines.
The predicted amino acid sequence of C. fasciculata ODC is longer than that of ODC from other species, including L. donovani. ODCs from C. fasciculata and L. donovani (19) consist of 721 and 707 aa, respectively, whereas the enzyme from mammals (36), yeast (37), T. brucei (16, 29), Neurospora crassa (38), Xenopus laevis (39), and the nematode Panarellus redividus (40) contains between 423 and 484 aa. Comparison of the amino acid sequence for ODC from C. fasciculata with those of other eukaryotic species reveals several conserved regions (Fig. 2). The sequence PHFAVKCN, which resembles a consensus sequence of PXXAVKC(N), probably contains the lysine (K) to which the pyridoxal 5′-phosphate cofactor binds (34). The sequence GPTCD is conserved in all eukaryotic ODCs sequenced so far and contains the cysteine (C) that is likely to be the major binding site of the irreversible inhibitor DFMO (34). The deduced amino acid sequence of C. fasciculata ODC contains a ≈250 aa N-terminal extension, which is absent in the mouse ODC. The amino acid sequence of C. fasciculata ODC shows a marked homology with that of L. donovani ODC (Fig. 2). Optimal alignment gives a 69% identity between the two sequences. The homologies between the amino acid sequence of C. fasciculata ODC and those of mouse, yeast, and T. brucei ODC are much lower, with identity scores of 37–40%.
The C terminus of C. fasciculata ODC is shorter than those of ODCs from L. donovani and T. brucei (Fig. 2). Compared with mouse ODC, the C. fasciculata ODC lacks 48 of the C-terminal amino acids whereas the ODCs from L. donovani (19) and T. brucei (16) lack 28 and 36 aa, respectively. This region of the mammalian ODC has been demonstrated to be essential for the extremely fast turnover of the protein (14), which may explain the finding that ODCs from L. donovani (19) and T. brucei (16) are proteins with a very slow turnover.
Analysis of the ODC Gene Locus in C. fasciculata.
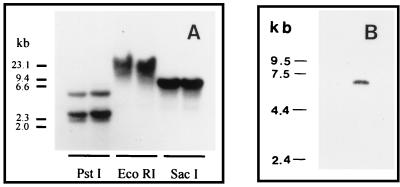
Several of the protozoan parasites have some of their genes organized in tandemly repeated arrays (41, 42). To achieve information on the organization of the ODC gene locus in C. fasciculata we analyzed genomic DNA using Southern blot analysis. The DNA was digested with restriction enzymes that either did not cut at all (EcoRI), or cut only once (PstI and SacI), within the ODC gene. After blotting, the restriction fragments were hybridized to the labeled 689-bp PCR product that was used for the isolation of the ODC gene. As shown in Fig. 3, PstI, which cuts within the sequence corresponding to the probe, gave rise to three fragments; one ≈4 kb and the others ≈2 kb each. The 4-kb fragment probably resulted from incomplete cutting of the DNA, because the amount of this fragment varied greatly between experiments. SacI (which does not cut within the region covered by the probe) and EcoRI both gave rise to a single band of two different sizes (≈6 and ≈15 kb, respectively). Taken together these results suggest that the ODC gene is not arranged in tandemly repeated units but may be present as a single copy in the genome of C. fasciculata. However, a titration of the gene copy number is needed for ultimate confirmation.
Figure 3.
Southern (A) and Northern (B) blot analyses of DNA and RNA from C. fasciculata. (A) Genomic DNA from C. fasciculata was digested with PstI, EcoRI, and SacI and fractionated on a 0.8% agarose gel. The digest was blotted to a Hybond N membrane and hybridized to a 32P-labeled C. fasciculata ODC-specific PCR probe. The size markers were HindIII-digested λ DNA. (B) Total RNA from C. fasciculata was fractionated on a 1% agarose gel containing 2.2 M formaldehyde, blotted to a Hybond N membrane, and then hybridized to a 32P-labeled C. fasciculata ODC-specific PCR probe. Mammalian ribosomal RNA and a commercial RNA ladder (GIBCO) were used as size markers.
Analysis of the ODC mRNA in C. fasciculata.
Northern blot analysis of total RNA from C. fasciculata revealed a single transcript of ≈6.5 kb (Fig. 3). The maturation of mRNAs in C. fasciculata, like in other trypanosomatids, involves a trans-splicing event in which a 39-nt mini-exon is added to the 5′ end of the transcript (27, 28). To determine the site for this splicing event we amplified and sequenced a region of C. fasciculata ODC cDNA corresponding to the 5′ terminus of the mature ODC transcript. One of the primers used for amplification contained part of the 39-nt mini-exon and the other one contained a sequence from the ORF. The PCR gave rise to one major amplification product of about 900 bp in size. Sequencing revealed that the site for the trans-splicing of the mini-exon to the ODC transcript was located at nt −301 from the predicted translation initiation site, thus giving rise to a 5′ untranslated region of 340 nt.
Turnover of C. fasciculata ODC Expressed in a Mammalian Expression System.
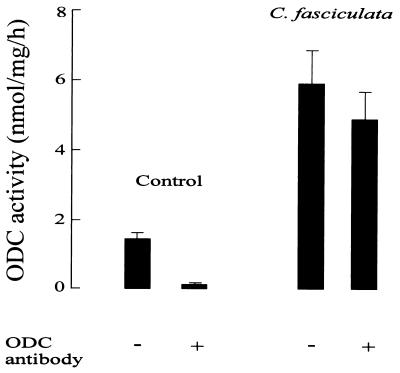
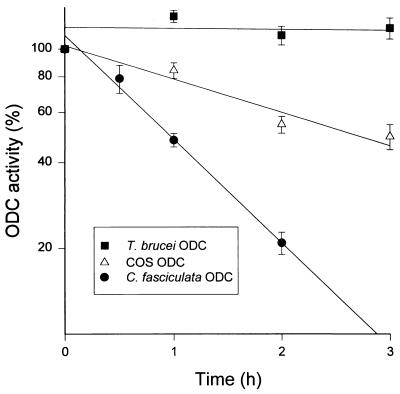
C. fasciculata is the only protozoan known to have an ODC that is turning over rapidly (22). However, as revealed in the present study, C. fasciculata ODC lacks the region corresponding to the C terminus of mammalian ODC, which has been shown to be essential for the fast turnover of the enzyme (9, 14, 17, 18). This finding indicates that the “C-terminal rule” of ODC, in which the rapid turnover of the enzyme is dependent on the presence of a specific C-terminal region, is not generally applicable. It is conceivable, though, that the rule is only valid in mammalian cells and thus, when expressed in such a system, C. fasciculata ODC would be metabolically stable. To determine the turnover of C. fasciculata ODC in a mammalian cell we subcloned the ODC gene from C. fasciculata into the mammalian expression vector pSVL and then expressed it transiently in COS cells. To discriminate between endogenous COS ODC and expressed C. fasciculata ODC, we used a specific antibody against mammalian ODC (32). This antibody does not react with C. fasciculata ODC but only precipitates the COS ODC. As shown in Fig. 4, transfection of COS cells with the expression vector containing the ODC gene from C. fasciculata gave rise to ODC activity in the COS cells that was not precipitated by the ODC antibody, demonstrating the accuracy of the clone and the efficiency of the system. The turnover of C. fasciculata ODC in the COS cells was determined by measuring the decay of nonprecipitable ODC activity after inhibition of protein synthesis by cycloheximide. As seen in Fig. 5, C. fasciculata ODC turns over rapidly, with a half-life of about 1 h. In comparison, ODCs from T. brucei (Fig. 5) and L. donovani (not shown) were found to be stable proteins when expressed in the COS cells. The half-life of the endogenous COS ODC was estimated to be about 3 h.
Figure 4.
Expression of C. fasciculata ODC in mammalian cells. COS cells were transfected with pSVL (control) or pSVL containing the ODC gene from C. fasciculata. Two days after transfection the cells were harvested and analyzed for ODC activity. In some samples, ODC activity derived from endogenous COS ODC was immunoprecipitated using an antibody specific against mammalian ODC (mean ± SEM, n = 6).
Figure 5.
Turnover of C. fasciculata ODC and T. brucei ODC expressed in mammalian cells. COS cells were transfected with pSVL or pSVL containing the ODC gene from C. fasciculata or T. brucei. The turnover of ODC in the cells was determined by following the decay of ODC activity after addition of cycloheximide. The C. fasciculata-specific ODC activity was determined after immunoprecipitation of the endogenous COS ODC using an antibody specific against mammalian ODC. The fraction of endogenous ODC in the COS cells expressing the T. brucei ODC was less than 1% (mean ± SEM, n = 3–5).
DISCUSSION
ODC is among the most labile enzymes in the mammalian cell with a half-life often shorter than 1 h (2). The molecular mechanisms as well as the necessary sequences for this rapid degradation are presently being unraveled. It has previously been shown that removal of or mutations in the C terminus of mammalian ODC can transform the protein into a stable one (14, 18, 43). Moreover, when this region of mammalian ODC is fused to stable proteins it confers a rapid turnover rate to these proteins (17, 18, 44). That the C terminus of mammalian ODC is indeed essential for the rapid turnover of the enzyme is also supported by the findings that ODCs from T. brucei (16) and L. donovani (19), which are stable proteins, lack the corresponding region. Based on these results it has been assumed that the C terminus is necessary for a rapid turnover of the protein in mammalian cells (9, 18). However, we now show that the C. fasciculata ODC lacks this region but nevertheless turns over rapidly in the parasite (22) as well as in mammalian cells. Thus, it appears that the function of the C-terminal domain of mammalian ODC in the rapid turnover of the enzyme is taken over by some other part(s) of the C. fasciculata ODC.
Although information on the size of the ODC expressed in C. fasciculata is lacking, the ORF shown in Fig. 1 is most probably the one used in vivo. The following observations support this notion: (i) the predicted initiation codon is the first ATG 3′ to the splice acceptor site, (ii) all reading frames 5′ to this ATG contain multiple stop codons; (iii) there is a codon bias for G or C as the third nucleotide also in the codons encoding the N-terminal part of the enzyme, and (iv) the deduced amino acid sequence corresponding to the sequence between the first and the second ATG of the C. fasciculata ODC gene is highly homologous to the N terminus of the L. donovani ODC.
C. fasciculata ODC shows a relatively high homology to the core region of mammalian ODC (Fig. 3). The main differences are found in the N-terminal and in the C-terminal regions. The N terminus of C. fasciculata ODC is extended by 256 aa as compared with that of mouse ODC. L. donovani ODC also has a long (219 aa) N-terminal extension, whereas the T. brucei ODC does not. The C terminus of mammalian ODC contains one of the two PEST regions of the enzyme (17). The PEST sequences are regions identified in proteins with a rapid turnover (15). Although C. fasciculata ODC lacks the C-terminal PEST region of mammalian ODC, it contains two regions which fulfil the requirements of PEST regions; the regions between aa 118–140 and 596–649. Especially the latter one appears to be a very strong PEST sequence. Interestingly, these parts of the C. fasciculata ODC are missing in the mammalian ODC. L. donovani ODC, which is a stable enzyme (19), lacks the region corresponding to the first PEST sequence of C. fasciculata ODC, and has a much weaker PEST sequence [according to the algorithm described by Rogers et al. (15)] in the region corresponding to the second PEST sequence of C. fasciculata ODC.
Mammalian ODC is degraded by the 26S proteasome (13, 45) in an energy-dependent but ubiquitin-independent process (12). Thus far, ODC and c-Jun (46) are the only proteins known to be degraded by this protease system without being ubiquitinated. Instead, degradation of mammalian ODC is stimulated by a specific protein, named antizyme, that binds very strongly to the enzyme and also inhibits its activity (2, 8, 9, 47). It has been suggested that antizyme acts in place of ubiquitin in ODC degradation (9). The recent finding that fusion of the N terminus of antizyme to other proteins induces their rapid degradation is consistent with this hypothesis (48, 49). The antizyme binding site within mouse ODC has been mapped to the region between aa 117 and 140 (47), which corresponds to aa 368–391 of C. fasciculata ODC. T. brucei ODC, which does not bind antizyme (47), shows a low degree of homology with mouse ODC in this region. Because the homology between C. fasciculata ODC and mouse ODC in this part of the enzyme is even lower, it can be assumed that antizyme does not bind to C. fasciculata ODC. Although it is not known at what stage of evolution that antizyme appeared, it is conceivable that the trypanosomatids, like C. fasciculata, T. brucei, and L. donovani, do not express this protein. As for C. fasciculata, the rapid turnover of its ODC may instead be induced by a different mechanism. Nevertheless, it is clear from the present study that this mechanism is active in the mammalian cell and capable of inducing a rapid degradation of C. fasciculata ODC.
In spite of the fact that there is a high homology between the C. fasciculata ODC and the L. donovani ODC (Fig. 2), there is a marked difference in stability between the two proteins. C. fasciculata ODC has a very rapid turnover, whereas L. donovani ODC is a stable protein (19). Due to the high homology between the two proteins it should be possible to make various hybrids, which should provide information on what parts of the protein are essential for this “C-terminal-independent” rapid turnover of C. fasciculata ODC.
Recent experiments carried out with Herpetomonas samuelpessoai have shown that this trypanosomatid parasite, like C. fasciculata, contains a metabolically unstable ODC (C.C., unpublished results). Both C. fasciculata and H. samuelpessoai are monogenetic trypanosomatids parasitizing on a single insect host, whereas T. brucei, L. donovani, L. mexicana, and P. falciparum are digenetic parasites with more complex life cycles involving insects and mammals as alternating hosts. The fact that monogenetic parasites, in contrast to digenetic parasites, appear to express metabolically unstable ODCs may be related to environmental differences affecting the parasite’s need to rapidly control its synthesis of polyamines or their metabolites. Such polyamine metabolites include the unique conjugate of glutathione and spermidine, called trypanothione, which plays an essential role in protecting the parasite against various stressful conditions (50).
Acknowledgments
This study was supported by grants from the Swedish Agency for Research Cooperation with Developing Countries, the Swedish Cancer Society, the Swedish Natural Science Research Council, the Medical Faculty (University of Lund), the United Nations Industrial Development Organization, the University of Buenos Aires (Argentina), the Swedish Society for Medical Research, and the IngaBritt and Arne Lundberg, the Dr. P. Håkansson, the Crafoord, the Wiberg, the Gyllenstiernska Krapperup, and the Gunnar, Arvid and Elisabeth Nilsson foundations.
Footnotes
References
- 1.Heby O, Persson L. Trends Biochem Sci. 1990;15:153–158. doi: 10.1016/0968-0004(90)90216-x. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi S, Murakami Y. Biochem J. 1995;306:1–10. doi: 10.1042/bj3060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marton L J, Pegg A E. Annu Rev Pharmacol Toxicol. 1995;35:55–91. doi: 10.1146/annurev.pa.35.040195.000415. [DOI] [PubMed] [Google Scholar]
- 4.Lopatin A N, Makhina E N, Nichols C G. Nature (London) 1994;372:366–369. doi: 10.1038/372366a0. [DOI] [PubMed] [Google Scholar]
- 5.Ficker E, Taglialatela M, Wible B A, Henley C M, Brown A M. Science. 1994;266:1068–1072. doi: 10.1126/science.7973666. [DOI] [PubMed] [Google Scholar]
- 6.McCann P P, Pegg A E. Pharmacol Ther. 1992;54:195–215. doi: 10.1016/0163-7258(92)90032-u. [DOI] [PubMed] [Google Scholar]
- 7.Fairlamb A H, Henderson G B, Bacchi C J, Cerami A. Mol Biochem Parasitol. 1987;24:185–191. doi: 10.1016/0166-6851(87)90105-8. [DOI] [PubMed] [Google Scholar]
- 8.Murakami Y, Matsufuji S, Miyazaki Y, Hayashi S. J Biol Chem. 1992;267:13138–13141. [PubMed] [Google Scholar]
- 9.Li X, Coffino P. Mol Cell Biol. 1993;13:2377–2383. doi: 10.1128/mcb.13.4.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins J F, Gesteland R F, Hayashi S. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rom E, Kahana C. Proc Natl Acad Sci USA. 1994;91:3959–3963. doi: 10.1073/pnas.91.9.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg-Hasson Y, Bercovich Z, Ciechanover A, Kahana C. Eur J Biochem. 1989;185:469–474. doi: 10.1111/j.1432-1033.1989.tb15138.x. [DOI] [PubMed] [Google Scholar]
- 13.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Nature (London) 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 14.Ghoda L, van Daalen Wetters T, Macrae M, Ascherman D, Coffino P. Science. 1989;243:1493–1495. doi: 10.1126/science.2928784. [DOI] [PubMed] [Google Scholar]
- 15.Rogers S, Wells R, Rechsteiner M. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 16.Phillips M A, Coffino P, Wang C C. J Biol Chem. 1987;262:8721–8727. [PubMed] [Google Scholar]
- 17.Ghoda L, Phillips M A, Bass K E, Wang C C, Coffino P. J Biol Chem. 1990;265:11823–11826. [PubMed] [Google Scholar]
- 18.Ghoda L, Sidney D, Macrae M, Coffino P. Mol Cell Biol. 1992;12:2178–2185. doi: 10.1128/mcb.12.5.2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson S, Adelman J, Ullman B. J Biol Chem. 1992;267:2350–2359. [PubMed] [Google Scholar]
- 20.Sánchez C P, González N S, Algranati I D. Biochem Biophys Res Commun. 1989;161:754–761. doi: 10.1016/0006-291x(89)92664-8. [DOI] [PubMed] [Google Scholar]
- 21.Assaraf Y G, Kahana C, Spira D T, Bachrach U. Exp Parasitol. 1988;67:20–30. doi: 10.1016/0014-4894(88)90004-5. [DOI] [PubMed] [Google Scholar]
- 22.Ceriani C, González N S, Algranati I D. FEBS Lett. 1992;301:261–264. doi: 10.1016/0014-5793(92)80253-d. [DOI] [PubMed] [Google Scholar]
- 23.Pasion S G, Hines J C, Aebersold R, Ray D S. Mol Biochem Parasitol. 1992;50:57–67. doi: 10.1016/0166-6851(92)90244-e. [DOI] [PubMed] [Google Scholar]
- 24.Berens R L, Marr J J. J Parasitol. 1978;64:160. [PubMed] [Google Scholar]
- 25.Miller S A, Dykes D D, Polesky H F. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chomczynski P, Sacchi N. Anal Biochem. 1987;132:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 27.Perry K L, Watkins K P, Agabian N. Proc Natl Acad Sci USA. 1987;84:8190–8194. doi: 10.1073/pnas.84.23.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabriel A, Sisodia S S, Cleveland D W. J Biol Chem. 1987;262:16192–16199. [PubMed] [Google Scholar]
- 29.Kuntz D A, Phillips M A, Moore T D E, Craig S P, III, Bass K E, Wang C C. Mol Biochem Parasitol. 1992;55:95–104. doi: 10.1016/0166-6851(92)90130-c. [DOI] [PubMed] [Google Scholar]
- 30.Kozak M. Nucleic Acids Res. 1981;9:5233–5262. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamauchi K. Nucleic Acids Res. 1991;19:2715–2720. doi: 10.1093/nar/19.10.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Persson L. Acta Chem Scand B. 1982;36:685–688. doi: 10.3891/acta.chem.scand.36b-0685. [DOI] [PubMed] [Google Scholar]
- 33.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Poulin R, Lu L, Ackermann B, Bey P, Pegg A E. J Biol Chem. 1992;267:150–158. [PubMed] [Google Scholar]
- 35.Alvarez F, Robello C, Vignali M. Mol Biol Evol. 1994;11:790–802. doi: 10.1093/oxfordjournals.molbev.a040159. [DOI] [PubMed] [Google Scholar]
- 36.Kahana C, Nathans D. Proc Natl Acad Sci USA. 1985;82:1673–1677. doi: 10.1073/pnas.82.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fonzi W A, Sypherd P S. J Biol Chem. 1987;262:10127–10133. [PubMed] [Google Scholar]
- 38.Williams L J, Barnett G R, Ristow J L, Pitkin J, Perriere M, Davis R H. Mol Cell Biol. 1992;12:347–359. doi: 10.1128/mcb.12.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bassez T, Paris J, Omilli F, Dorel C, Osborne H B. Development (Cambridge, UK) 1990;110:955–962. doi: 10.1242/dev.110.3.955. [DOI] [PubMed] [Google Scholar]
- 40.Von Besser H, Niemann G, Domdey B, Walter R D. Biochem J. 1995;308:635–640. doi: 10.1042/bj3080635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tschudi C, Young A S, Ruben L, Patton C L, Richards F F. Proc Natl Acad Sci USA. 1985;82:3998–4002. doi: 10.1073/pnas.82.12.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, van der Ploeg L H. Mol Cell Biol. 1991;11:3180–3190. doi: 10.1128/mcb.11.6.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenberg Hasson Y, Bercovich Z, Kahana C. Eur J Biochem. 1991;196:647–651. doi: 10.1111/j.1432-1033.1991.tb15861.x. [DOI] [PubMed] [Google Scholar]
- 44.Loetscher P, Pratt G, Rechsteiner M. J Biol Chem. 1991;266:11213–11220. [PubMed] [Google Scholar]
- 45.Bercovich Z, Kahana C. Eur J Biochem. 1993;213:205–210. doi: 10.1111/j.1432-1033.1993.tb17749.x. [DOI] [PubMed] [Google Scholar]
- 46.Jariel Encontre I, Pariat M, Martin F, Carillo S, Salvat C, Piechaczyk M. J Biol Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Coffino P. Mol Cell Biol. 1992;12:3556–3562. doi: 10.1128/mcb.12.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Stebbins B, Hoffman L, Pratt G, Rechsteiner M, Coffino P. J Biol Chem. 1996;271:4441–4446. doi: 10.1074/jbc.271.8.4441. [DOI] [PubMed] [Google Scholar]
- 49.Li X, Coffino P. J Biol Chem. 1996;271:4447–4451. doi: 10.1074/jbc.271.8.4447. [DOI] [PubMed] [Google Scholar]
- 50.Fairlamb A H, Cerami A. Annu Rev Microbiol. 1992;46:695–729. doi: 10.1146/annurev.mi.46.100192.003403. [DOI] [PubMed] [Google Scholar]