Abstract
Protein TrwC is the conjugative relaxase responsible for DNA processing in plasmid R388 bacterial conjugation. TrwC has two catalytic tyrosines, Y18 and Y26, both able to carry out cleavage reactions using unmodified oligonucleotide substrates. Suicide substrates containing a 3′-S-phosphorothiolate linkage at the cleavage site displaced TrwC reaction towards covalent adducts and thereby enabled intermediate steps in relaxase reactions to be investigated. Two distinct covalent TrwC–oligonucleotide complexes could be separated from noncovalently bound protein by SDS–PAGE. As observed by mass spectrometry, one complex contained a single, cleaved oligonucleotide bound to Y18, whereas the other contained two cleaved oligonucleotides, bound to Y18 and Y26. Analysis of the cleavage reaction using suicide substrates and Y18F or Y26F mutants showed that efficient Y26 cleavage only occurs after Y18 cleavage. Strand-transfer reactions carried out with the isolated Y18–DNA complex allowed the assignment of specific roles to each tyrosine. Thus, only Y18 was used for initiation. Y26 was specifically used in the second transesterification that leads to strand transfer, thus catalyzing the termination reaction that occurs in the recipient cell.
Keywords: 3′-S-phosphorothiolate-containing oligonucleotides, bacterial conjugation, relaxase, transesterification
Introduction
Bacterial conjugation is responsible for the horizontal spread of adaptive genes, including antibiotic resistance and virulence genes. According to the accepted steps of bacterial conjugative DNA processing, conjugation is initiated by cleavage of a specific phosphodiester bond (the nic site) in the donor supercoiled DNA (for a review, see Zechner et al, 2000; Llosa and de la Cruz, 2005). This reaction is catalyzed by a sequence-specific DNA-strand transferase, the relaxase, which remains covalently attached to the 5′-terminus of nic. Subsequent DNA strand displacement through rolling circle replication produces the T-strand, which is transferred to the recipient cell. The unwinding reaction is terminated by a second transesterification reaction, also catalyzed by the relaxase, which leads to DNA circularization. Protein TrwC is the encoded relaxase of plasmid R388 (Avila et al, 1988), responsible for the initiation and termination reactions of DNA processing during conjugation (Grandoso et al, 2000). TrwC is a 103 kDa protein with two domains. The N-terminal relaxase domain (amino acids 1–300) catalyzes cleavage and DNA strand transfer in vitro using either oligonucleotides or supercoiled plasmid DNA. The C-terminal helicase domain (amino acids 300–966) is responsible for a 5′–3′ DNA helicase activity (Llosa et al, 1996). A segment located between amino acids 300 and 600 of this domain enhances oriT-dependent recombination (Cesar et al, 2006).
Two types of conjugative relaxases have been categorized depending upon the number of catalytic tyrosines within the active site: relaxases with one active tyrosine and relaxases with two active tyrosines. Relaxases belonging to the MOBP and MOBQ groups, exemplified by MobA, the relaxase of plasmid RSF1010 (Scherzinger et al, 1993; Monzingo et al, 2007), and TraI, the relaxase of plasmid RP4 (Pansegrau and Lanka, 1996; Francia et al, 2004), respectively, have a single active tyrosine (Francia et al, 2004). Relaxases belonging to the MOBF group exemplified by TrwC, the relaxase of plasmid R388 (Llosa et al, 1995, 1996; Grandoso et al, 2000), TraI relaxase of plasmid F (Datta et al, 2003; Street et al, 2003; Larkin et al, 2005; Matson and Ragonese, 2005; Williams and Schildbach, 2006) and TraH relaxase of plasmid pKM101 (Byrd et al, 2002) contain two catalytic tyrosines (Francia et al, 2004). Relaxases can specifically cleave single-stranded oligonucleotides containing their respective nic site sequences in vitro, so that the 5′-end of the cleaved product becomes covalently bound to the protein via the catalytic tyrosine. This bound single-stranded DNA can then be transferred to an appropriate acceptor oligonucleotide by a second DNA strand-transfer reaction (reviewed by Lanka and Wilkins, 1995). To investigate the ability of the Y-relaxase RP4_TraI to function in ‘second cleavage', single-stranded oligonucleotides containing nic were immobilized at their 3′-termini on magnetic beads and cleaved by TraI (Pansegrau and Lanka, 1996). The resulting covalent TraI–oligonucleotide adducts were active in the joining reaction, but unable to cleave oligonucleotides containing an intact nic. This result indicated that second cleavage probably requires a second TraI monomer, as the monomer engaged in complex formation was unable to terminate the reaction. Both TrwC catalytic tyrosines (Y18 and Y26) are able to carry out cleavage and joining reactions involving covalent complexes (Grandoso et al, 2000). Different atomic structures of noncovalent complexes of the TrwC relaxase protein with its cognate DNA substrate have been solved (Guasch et al, 2003; Boer et al, 2006). The structures were useful to explain the catalytic roles of the metal cations and also of the two active-site tyrosines. A model of action was proposed for TrwC (Guasch et al, 2003; Boer et al, 2006), in which two DNA exit pathways from the active pocket are used at different steps in conjugative DNA-processing.
In vitro analysis of cleavage reactions using standard oligonucleotides suffer from the drawback that the resulting products represent an equilibrium between the kinetics of cleavage and religation. 5′-S-phosphorothiolate-linked oligonucleotides are well established as suicide substrates for several enzymes (Burgin and Nash, 1995). When there is an equilibrium between cleaved and ligated DNA, substitution of the –OH group responsible for the attack in the ligation step for an –SH group was proven to displace the equilibrium towards covalent oligonucleotide–enzyme adducts. Such oligonucleotides have been used to study the mechanism of site-specific integrases (Burgin and Nash, 1995), recombinases (Ghosh et al, 2005) and topoisomerases (Krogh and Shuman, 2000). In this article, we describe for the first time, the application of 3′-S-phosphorothiolate-linked oligonucleotides as suicide substrates for the study of intermediate steps in TrwC DNA processing. Results obtained allow us to propose a distinct role for each catalytic tyrosine as well as for the IR2 hairpin (inverted repeat located 5′ to nic). The covalent complexes thus formed are also useful for studying the conjugation termination reaction, as they mimic the state of the relaxase when it enters the recipient cell.
Results
Protein TrwC forms two distinct covalent complexes with oligonucleotide substrates
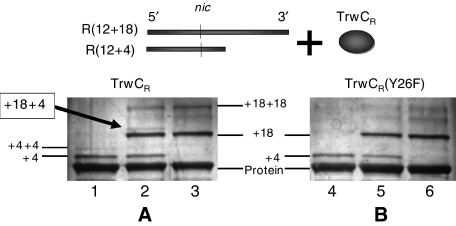
TrwC and related relaxases carry out cleavage and joining reactions involving phosphodiester bonds. The cleavage reaction is not the result of phosphodiester hydrolysis, but of transesterification by an active-site tyrosyl residue, producing a protein–DNA covalent intermediate with the 5′-phosphate end bound to the protein and a free 3′-OH terminus. Strand ligation occurs in a second transesterification event, when a 3′-OH (from the same or a different DNA) cleaves the phosphotyrosyl bond and the covalent intermediate is resolved. In this work, we used TrwC N-terminal relaxase domain (called TrwCR(wt) hereafter) to analyze the cleavage reaction by the two tyrosines involved: Y18 and Y26. When purified, TrwCR(wt) protein reacted with oligonucleotides containing the R388 nic site, TrwCR(wt)/DNA covalent complexes could be separated from free protein and from noncovalently bound complexes by SDS–PAGE, as shown in Figure 1. According to band mobility in these gels, two different covalent complexes were distinguished. For instance, SDS–PAGE analysis of reaction mixtures of TrwCR-DNA with oligonucleotide R(12+4) (lane 1) revealed two bands with reduced mobility (shown as ‘+4' and ‘+4+4', respectively). The ‘+4' band represented roughly 10% of the total protein, whereas the ‘+4+4' band represented less than 1% of the total protein and were attributed to the formation of complexes formed between the protein and either one or two tetranucleotide fragments. Neither band was observed when EDTA was added to the reaction mixture (data not shown), indicating a divalent metal requirement for the reaction. Furthermore, the samples were subject to SDS treatment and boiling before electrophoresis thereby destroying any noncovalent complexes. In light of these results, we assume that TrwCR(wt) cleaves oligonucleotides in the presence of Mg2+ ions and the two product bands represent covalent complexes produced by attachment of one or both TrwC catalytic tyrosines to the 5′-end of the cleaved oligonucleotide 3′-moiety.
Figure 1.
Formation of DNA–TrwCR covalent complexes. (A) Silver-stained SDS–PAGE gel of TrwCR(wt) incubated with different oligonucleotide substrates: R(12+4) (lane 1); R(12+18) (lane 3); a mixture of R(12+4) and R(12+18) (lane 2). The new band that appears in lane 2 corresponds to the formation of a ternary complex (with bound 4-mer and 18-mer oligonucleotide moieties (see text). (B) Same as in (A), except that reactions were carried out using TrwCR(Y26F).
Incubation of TrwCR(wt) with oligonucleotide R(12+18) resulted in two bands corresponding to complexes of higher MW than those obtained with oligonucleotide R(12+4) (lane 3; shown as ‘+18' and ‘+18+18'). When TrwCR(wt) was incubated with a mixture of both oligonucleotides (R(12+4) and R(12+18)), an additional fifth complex appeared (lane 2; labelled ‘+4,+18'). According to its mobility, we assume the new complex is formed when each of the active tyrosines in TrwCR(wt) reacts with a different oligonucleotide.
When the mutant relaxase TrwCR(Y26F) was substituted for TrwCR(wt), only lower molecular weight covalent complexes were observed with R(12+4) or R(12+18) (lanes 4 and 6; ‘+4' and ‘+18', respectively on Figure 1B). Moreover, if R(12+4) and R(12+18) oligonucleotides were added together to reaction mixtures containing TrwCR(Y26F), both single complexes (‘+4' and ‘+18') were observed, but none of the three ternary complexes formed by TrwCR(wt) were detected (lane 5). Similar results were obtained when using TrwCR(Y18F) (data not shown). In summary, it appears that each catalytic tyrosine is equally able to form single complexes. On the other hand, covalent complexes containing two bound oligonucleotides were only obtained when the protein contained both active tyrosines, although only a small amount of the second complex was obtained.
Oligonucleotides containing internucleotide 3′-S-phosphorothiolate linkages displace the TrwC cleavage reaction equilibrium towards covalent adducts
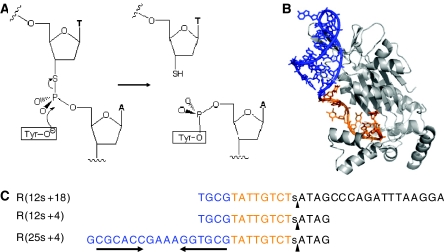
A mechanistic analysis of the relaxase cleavage reaction has been hindered by the fact that the observed cleavage reaction represents an equilibrium between the kinetics of cleavage and religation. To displace the reaction equilibrium toward covalent complexes, 3′-S-phosphorothiolate oligonucleotides were synthesized. These isostructural and isopolar analogs were prepared so that the 3′-bridging oxygen group involved in the ligation step was substituted for a 3′-SH group. The 3′-sulfhydryl group is a soft nucleophile unable to initiate transesterification at the tyrosine-phosphate diester hard electrophilic center. Therefore, the 3′-sulfhydryl group cannot carry out the reverse/ligation step (Figure 2).
Figure 2.
Cleavage reaction of phosphorothiolate oligonucleotides. (A) Scheme of TrwC cleavage reaction. In the normal reaction, the hydroxyl group of the catalytic tyrosil residue is a nucleophile that attacks the phosphate group in the nic site. As a consequence, the phosphodiester bond is broken and a free 3′-OH group is generated. When the oxygen in the scissile P–O bond is replaced by sulfur, the resulting cleavage product ends in a 3′-SH group. The religation reaction that reverses the cleavage reaction when normal oligonucleotides are used is thus inhibited with phosphorothiolate oligonucleotides. As a consequence, the equilibrium is displaced towards oligonucleotide–protein covalent complex formation. (B) X-Ray structure of TrwCR in complex with DNA (pdb:1OMH). The color scheme of the DNA reflects the double- (blue) or single-stranded (ochre) DNA portions of the bound oligonucleotide, and parallels the colors of the sequences in (C). (C) Nucleotide sequences of oligonucleotides R(12s+18), R(12s+4) and R(25s+4), used in this study. The position of the phosphorothiolate modification is shown by a lower case s within the sequences. The cleavage sites are represented by black vertical arrowheads. The 25 nucleotide-long 5′-moiety of R(25s+4) contains an inverted repeat sequence (horizontal arrows) linked via a GAAA loop that results in the double stranded DNA colored blue in (B).
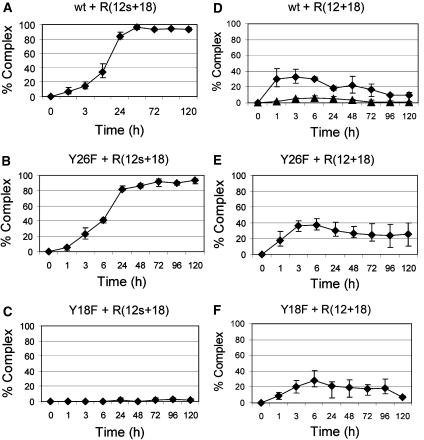
We synthesized three oligonucleotides (R(12s+18), R(12s+4) and R(25s+4); Figure 2) containing a 3′-S-phosphorothiolate internucleotide linkage at the R388 nic site as explained in Materials and methods. First, the kinetics of covalent complex formation by TrwCR(wt), TrwCR(Y26F) or TrwCR(Y18F) when using either R(12+18) or R(12s+18) were compared. Kinetic curves were calculated by plotting the percentage oligonucleotide recovered as a covalent complex (calculated from coomassie stained SDS–PAGE gels such as that in Figure 1) versus time. Figure 3 shows the results obtained. When TrwCR(wt) was incubated with R(12s+18), the yield of complex formation reached 100% (Figure 3A). This result demonstrates that phosphorothiolate-containing oligonucleotides were impaired in the reverse ligation reaction, thus allowing the cleavage reaction to reach completion. Remarkably, only the ‘+18' complex was observed, in contrast with the two complex bands (‘+18' and ‘+18+18') obtained when TrwCR(wt) was incubated with R(12+18) (Figure 3D). In the latter case, only about 10% of the protein was in the form of a complex in the same period of time (and did not increase with time). However, the kinetics of complex formation (forward reaction) was slower with R(12s+18) than with R(12+18), as judged from the elevation of the curves at early reaction times (Figure 3A versus D). When the same two oligonucleotides were incubated with TrwCR(Y26F), the yields were similar to those obtained with TrwCR(wt), the only significant difference being that no ternary (‘+18+18') complex was produced with R(12s+18), as expected (Figure 3B and E). The most surprising and interesting result in Figure 3 occurred when we looked at cleavage of R(12s+18) when incubated with TrwCR(Y18F). As can be seen in Figure 3C, there was no complex formation at all, even when incubation of this mutant protein with R(12+18) resulted in a standard yield of complex (Figure 3F). Thus, we assume that the Y26 transesterification reaction coordinate involves an extended P−O3′ bond (1.57 Å in the groundstate) in a dissociative transition state, which is mimicked by the elongated P−S3′ bond (1.95 Å) of the phosphorothiolate. Although R(12s+18) may therefore be bound by TrwCR(Y18F), no activation of the phosphorothiolate towards cleavage is observed.
Figure 3.
Kinetics of complex formation with R(12s+18) and R(12+18) oligonucleotides. Cleavage assays were performed with oligonucleotide R(12s+18) (A–C) or R(12+18) (D–F). Reactions used protein TrwCR(wt) (A, D), TrwCR(Y18F) (B, E), or TrwCR(Y26F) (C, F). Samples were collected at different times, up to 120 h. These samples were analyzed by SDS–PAGE, and the amount of covalent complex was calculated from the intensity of the bands. When two series of data are present in the same graph, the single complex is represented by solid diamonds, whereas solid triangles represent the double complex.
The nic IR2 hairpin enhances Y18 activity
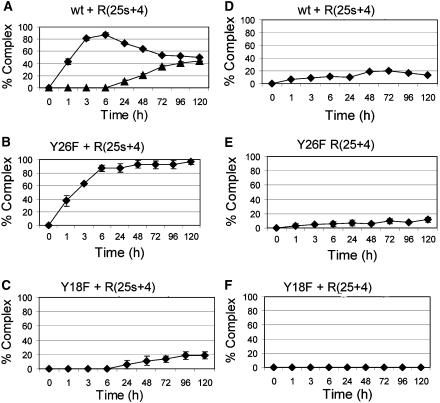
A majority of conjugative plasmid nic sites have an inverted repeat located 5′ to nic (Francia et al, 2004). This inverted repeat was shown to be recognized by the relaxase, as shown for the TrwCR co-crystals in complex with 25- or 27-mer oligonucleotides (Figure 2B, Guasch et al, 2003; Boer et al, 2006). Thus, the kinetics of cleavage was analyzed as above, but using 29-mer oligonucleotides (containing IR2). Results are shown in Figure 4, and can be more easily understood if we analyze the behavior of the relaxase mutants first. As described in the preceding section, Y18, but not Y26, could cleave oligonucleotide R(12s+18). Similarly, Figure 4B shows in fact that TrwCR(Y26F) efficiently cleaves R(25s+4), reaching 100% cleavage in about 6 h. This is a significantly higher rate than that obtained when cleaving R(12s+18), where it took 24 h to reach 100% cleavage. As can be seen, protein TrwCR(Y26F) cleaves R(25+4) to a significantly reduced equilibrium state when compared to R(12+18) (Figure 4E versus Figure 3E). This result has two alternative explanations: either TrwCR(Y26F) is less capable of cleaving hairpin-containing oligonucleotides or the hairpin shifts the equilibrium towards the ligation reaction. The second alternative seems more likely when we look at the results obtained with oligonucleotide R(25s+4) (Figure 4B). It is clear that TrwCR(Y26F) cleaves R(25s+4) at a higher rate than R(12s+18), as judged by the percent cleavage at short times (Figure 4B versus Figure 3B), suggesting that the forward reaction rate is enhanced by the hairpin. As the reaction with the unmodified oligonucleotide R(25+4) remains low (Figure 4E), we must assume that the reverse reaction is also more efficient, probably because of the additional binding strength provided by the hairpin.
Figure 4.
Kinetics of complex formation with R(25s+4) and R(25+4) oligonucleotides. Cleavage assays were carried out as those described in Figure 3, except that oligonucleotides R(25s+4) (A–C) and R(25+4) (D–F) were used.
Mutant protein TrwCR(Y18F) cleaved R(25s+4) although rather slowly (10% cleavage after 24 h), but cleavage of the standard oligonucleotide R(25+4) was not observed (Figure 4C and F). It seems that the presence of IR2 allows a better positioning of the substrate oligonucleotide on the active site (see Discussion).
The IR2 hairpin allows efficient Y26 cleavage only after Y18 cleavage
TrwCR(Y26F) was significantly more active in the cleavage of R(25s+4) than TrwCR(Y18F): >80% versus <5% complex formation after 6 h incubation (Figure 4B and C). TrwCR(wt) was also used with R(25s+4) as substrate (Figure 4A). In this assay, the ‘+4+4' double complex appeared and reached, interestingly, 50% of the cleaved product. The progress of the reaction can be followed in Figure 4A. The ‘+4' complex increased with time during the first 6 h incubation, up to about 80% complex formation. Then, the ‘+4+4' double complex started to accumulate, at the expense of the single complex. It reached a maximum of about 50%, but did not increase further, even after adding an excess oligonucleotide or keeping the reaction at 37°C for 7 days.
The composition of the single and the double complex formed by TrwCR(wt) and oligonucleotide R(25s+4) was analyzed by protease digestion and mass spectrometry (Table I). Trypsin treatment of SDS–PAGE gel slices containing TrwCR(wt) protein generated a peptide peak with a molecular mass of 851.35 m/z, consistent with it being the peptide that contains both tyrosines (TrwC amino acid residues 15–29: AASYYEDGADDYYAK). This peak however was not present when either the ‘+4' or the ‘+4+4' bands in the TrwC(wt):R(25s+4) complexes were analyzed by MS, indicating that, as expected, the Y18–Y26-containing segment was involved in the formation of both complexes. Moreover, the single ‘+4' complex yielded an additional peak at 1480.95 m/z. This peak corresponds to the mass of the peptide containing residues 15–29 plus the tetranucleotide ATAG. The intensity of the peak was reduced 50-fold compared with that of the nonconjugated peptide, a typical observation for MS-analysis of DNA–peptide complexes which can have multiple charged states (and hence m/z ratios). This effect was stronger with the double complex to the point that, in this case, no peak corresponding to the double complex was detected (Table I, column 1: trypsin treatment). Its existence is inferred from the fact that neither the peak corresponding to the peptide alone nor that of the single complex were detected.
Table 1.
Mass spectrometry experiments
| Sample | Trypsin | Specific Glu-C | Nonspecific Glu-C | |||
|---|---|---|---|---|---|---|
| Peptide 15–29 aa′ | + Oligonucleotide | Peptide 21–35 aa′ | + Oligonucleotide | Peptide 12–20 aa′ | + Oligonucleotide | |
| TrwCR(wt) | 851.35 m/z | — | 796.3 m/z | — | 515.25 m/z | — |
| ‘+4' complex | — | 1480.95 m/z | 796.3 m/z | — | — | 1144.85 m/z |
| ‘+4+4' complex | — | — | — | — | — | 1144.85 m/z |
The results of mass spectrometry experiments are outlined in this table. The sample analyzed in each case is shown in the left column. Three different digestions were carried out with Trypsin, Glu-C using specific cleavage conditions and Glu-C using nonspecific cleavage conditions, respectively. In each case, the presence/absence of peptide or complex was analyzed. m/z values indicate that the peak corresponding to the theoretical m/z expected for that sample has been found, on the contrary, — indicates that the peak did not appear.
To determine which tyrosine was covalently bound to DNA in each complex, a new protease treatment was carried out with protease GluC. By changing the digestion conditions (Materials and methods) GluC can cleave either Glu specifically, or Glu and Asp nonspecifically. Under stringent conditions, GluC cleaves TrwCR(wt) only once between Y18 and Y26, yielding a peptide containing Y18 (residues 1–20) and a second peptide containing Y26 (residues 21–35). Neither peaks corresponding to the unmodified peptide containing Y18, nor its DNA adduct were observed. In contrast, peptides containing Y26 were found in a peak with 796.3 m/z in both the single complex and the free protein, indicating that this peptide (and therefore, Y26) is not involved in its formation, as this m/z corresponds precisely to the Y26-containing peptide, and not to the oligonucleotide-bound complex (Table I, column 2: specific Glu-C treatment).
There is an Asp residue (Asp11) in the 1–20 peptide containing Y18. As this peptide was not observed in the specific GluC treatment, a nonspecific digestion was performed to find the resulting peptide (amino acid residues 12–20) and study Y18 behavior. In the TrwCR(wt) protein sample, a peak of 515.2 m/z was observed, consistent with our expectations. Using GluC nonspecific digestion with the TrwCR(wt) DNA complexes, a 4-mer-Y18-peptide peak was found (a peak of 1144.2 m/z). This peak was present in the ‘+4' and the ‘+4+4' complexes, indicating that Y18 was involved in formation of both complexes (Table I, column 3: nonspecific Glu-C treatment).
Thus, the observed double complex is consistent with a protein complex with one 4-mer oligonucleotide covalently bound to Y18 and a second 4-mer oligonucleotide bound to Y26. Under these conditions, it is possible to assume that the amount of double complex reflects the amount of Y26 cleavage once that Y18 is already covalently bound to DNA. The efficiency of double complex formation in the native protein was 50%, significantly higher than the 20% of single complex formed by TrwCR(Y18F) (Figure 4A versus C). Therefore, Y26 exhibits significantly enhanced reactivity within the conformation of the covalent complex compared to the free protein.
Strand-transfer reactions catalyzed by TrwC covalent complexes
The covalent complexes isolated in vitro, in which only Y18 is bound to DNA, probably emulate the state of the relaxase when it arrives to the recipient cell. Therefore, analysis of these complexes can be useful to understand how active tyrosines proceed in the termination reaction. We decided to carry out strand-transfer reactions to obtain information about this issue. Strand-transfer reactions between the wild-type complex and an oligonucleotide containing a nic site were carried out and the reactions analyzed by capillary electrophoresis.
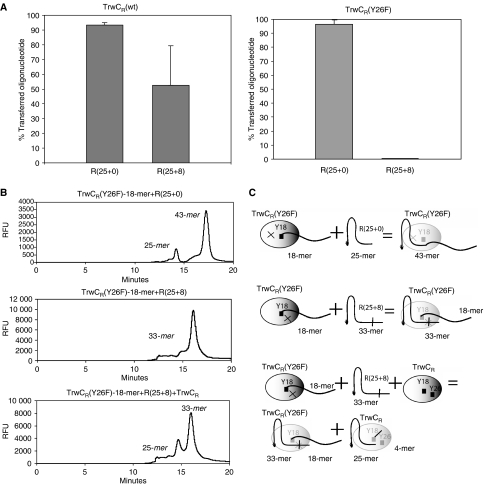
To ensure that unreacted proteins present in samples of the complexes did not interfere with their biochemical analysis, the covalent complexes obtained after 5 days incubation at 37°C were purified as described in Materials and methods. Separation was achieved using complexes of the modified oligonucleotide R(12s+18) with either TrwCR(wt) or TrwCR(Y26F) proteins; TrwCR(Y18F) did not cleave this substrate. Purified covalent complexes thus contained TrwCR(wt) or TrwCR(Y26F) and a covalently bound 18-mer oligonucleotide moiety linked via Y18. Texas Red 5′-labelled oligonucleotides R(25+0) or R(25+8) containing the 25-mer IR2 hairpin sequence were subsequently added to these complexes. The resulting transesterification product would be a 43-mer oligonucleotide in both cases, as a result of ligation of the 25-mer 5′-moiety of the newly added oligonucleotides to the 18-mer nucleotide forming part of the complex.
As shown in Figure 5A, the TrwCR(wt) binary complex catalyzed the strand-transfer reaction with both substrates although reaction with R(25+0) resulted in a higher yield than with R(25+8) (90 versus 50%). When divalent metal ions were sequestered using EDTA, the reaction did not take place (data not shown). Moreover, the reaction using a 5′-Texas Red labelled R(12+0) substrate showed that strand transfer was more efficient with R(25+0) than with R(12+0) (80 versus 20%), underscoring the important role of the IR2 hairpin (see Discussion). When these experiments were repeated with the TrwCR(Y26F) complex, the result with R(25+0) was the same as with TrwCR(wt). In contrast, TrwCR(Y26F) was unable to carry out the strand-transfer reaction with the hairpin-containing oligonucleotide R(25+8) (Figure 5A).
Figure 5.
Strand-transfer reactions catalyzed by binary complexes. Purified TrwCR(Y26F) complex covalently bound to 18-mer oligonucleotide was mixed with Texas Red-labelled oligonucleotides R(25+0) or R(25+8). Reactions were visualized after capillary electrophoresis, where the appearance of the resulting 43-mer oligonucleotide was quantified. (A) Shows the percentage of strand transfer product obtained in each reaction. (B) Three relevant electropherograms are shown. The molecular species involved in the respective reactions are shown schematically in (C). In the first reaction, oligonucleotide R(25+0) was used. The electropherogram shows the appearance of a 43-mer (25+18) transfer product. In the second reaction, oligonucleotide R(25+8) was used. No 43-mer transfer product appears. In the third electropherogram, the first reaction is repeated in the presence of TrwCR(wt). Although TrwCR(wt) can cleave R(25+8), the cleaved oligonucleotide (25+0) appears not to be available for the strand transfer reaction with the binary complex.
The TrwCR atomic structure showed that Y26 is located in a mobile loop at the surface of the protein, far from the active site. These data suggest that Y26 could act on a different protein molecule. Thus, we were interested to find out if Y26 could act in trans from the protein supplied, carrying out an intermolecular strand-transfer reaction with the oligonucleotide bound to the complex. To check for this possibility, we performed strand-transfer reaction experiments supplying TrwCR(wt) protein and R(25+8) to the TrwCR(Y26F) complex (Figure 5B). As inferred from the electropherogram, the strand-transfer reaction did not take place between different molecules, even considering that the TrwCR(wt) protein supplied in trans cleaved the R(25+8) oligonucleotide. Note that the 25-mer cleaved product was observed, but not the transferred R(25+18) product (Figure 5B, panel 3). As expected, a similar result was obtained when R(25+8) and TrwCR(Y26F) or TrwCR(Y18F) were added in trans.
Full-length TrwC (TrwCFL), in its (wt) and (Y26F) versions, had previously been shown to catalyze strand transfer reactions with R(14+4) and R(25+8) oligonucleotides (Grandoso et al, 2000). As TrwC is a dimer in solution, whereas TrwCR is a monomer, the possibility existed that Y18 could act in trans in TrwCFL but not in TrwCR. To address this question, we purified covalent complexes containing TrwCFL(wt) or TrwCFL(Y26F) and a covalently-bound 18-mer oligonucleotide. These complexes were obtained by the reaction of TrwCFL(wt) or TrwCFL(Y26F) with the suicide substrate R(12s+18) as described in Materials and methods.
When TrwCFL(wt) binary complex was incubated with stoichiometric amounts of oligonucleotide R(25+0), strand transfer was barely detected (<1%). The result could be a consequence of nonspecific oligonucleotide binding to the helicase domain of TrwCFL(wt) protein, thus reducing the amount of R(25+0) molecules available for reaction with the 18-mer covalently bound oligonucleotide. The inhibitory effect could be reversed by the addition of an excess of an unrelated mixture of oligonucleotides. Then, the strand transfer reaction occurred at a significant rate (14%). When R(25+8) oligonucleotide was used instead, DNA strand transfer was 4%, that is, 70% lower than with R(25+0). When the experiment was repeated, but TrwCFL(Y26F) was used, 11% strand transfer was observed with R(25+0). No strand transfer (<1%) was observed when TrwCFL(Y26F) was incubated with oligonucleotide R(25+8).
Thus, results indicate that the TrwC-catalyzed strand-transfer reaction can only occur in vitro in an intramolecular fashion.
Discussion
Relaxases are proteins with unique biochemical properties, as they catalyze site-specific DNA-transfer reactions among single-stranded DNA molecules. Protein TrwC of plasmid R388 is the prototype of the two-Tyr family of relaxases (see Introduction). Analysis of the ternary complexes shown in Figure 1 and Table I provides incontrovertible evidence that both TrwC residues, Y18 and Y26, give rise to protein–DNA covalent adducts, together or in isolation. Detailed biochemical analysis of two-Tyr relaxases is helped by the fact that the atomic structures of both TraI_F (Datta et al, 2003; Larkin et al, 2005) and TrwC_R388 (Guasch et al, 2003; Boer et al, 2006) proteins, in complex with their DNA substrates, are known. Cleavage and strand-transfer reactions performed by conjugative relaxases have been widely studied (Llosa et al, 1995, 1996; Pansegrau and Lanka, 1996; Moncalian et al, 1997; Becker and Meyer, 2000; Grandoso et al, 2000; Matson and Ragonese, 2005). However, none of the published works were able to analyze cleavage and ligation reactions separately, as the rapid equilibrium between cleavage and joining hindered a detailed biochemical analysis of the process. In particular, identification of the specific role of each catalytic tyrosine could not be addressed. The suicide substrates used in this work allowed us to characterize each of the two distinct transesterification reactions isolating the reactions of cleavage and strand transfer. This approach was useful to gain further insight at the intermediate steps of these reactions. Phosphorothiolate-containing oligonucleotide R(12s+18) enabled cleavage reactions to attain completion. We assume this is due to inhibition of the reverse ligation reaction, which requires transesterification at a hard electrophilic center (the phosphate diester which links the protein and bound oligonucleotide), by the 3′-SH which is a soft nucleophile. Also, nucleophilic attack by Y26 in the initial reaction with R(12s+18) was inhibited. Thus, phosphorothiolate-linked oligonucleotides allowed us to analyze the kinetics of the cleavage reaction, the implication of IR2 in cleavage and to distinguish between cleavages by each of the two active site tyrosines. Besides, they enabled the purification of oligonucleotide–relaxase complexes and subsequent analysis of the second transesterification reaction.
The forward cleavage reaction was analyzed by comparing standard and suicide oligonucleotides containing or not IR2. When using the standard R(25+4) oligonucleotide there was little cleavage on reaching equilibrium. Cleavage was better with R(12+18), where two complexes (binary and ternary) could be observed. Thus, cleavage with standard oligonucleotides reaches equilibrium before much of the oligonucleotide has been cleaved. On the contrary, with suicide oligonucleotide R(12s+18) cleavage reached 100%. With respect to the standard R(12+18), only the binary complex, catalyzed by Y18, was produced. As indicated above, Y26 cannot attack the phosphorothiolate bond, precluding formation of the ternary complex. When oligonucleotide R(25s+4) was used, the forward kinetics was even more rapid and attained completion in 3 h, instead of 24 h. In fact, the ternary complex reached 50% at the expense of the binary complex on equilibrium reached after 72 h. From these results, we conclude that progress of the cleavage reactions with standard oligonucleotides is delayed by efficient reversal of the reactions (re-ligation). In this respect (Williams and Schildbach, 2006), when reporting poor TraI-mediated cleavage of oligonucleotides analogous to R(25+4), assumed that hairpin-containing oligonucleotides were poor substrates for the relaxase. However, they did not rule out the possibility that their observation was a result of efficient religation, which seems the most likely possibility under the light of our results.
A second conclusion from the present work is that Y26 cleaves oligonucleotides efficiently only when they interact with the binary complexes produced by initial ligation to Y18. This result is of the outmost importance, as it implies that Y26 will act specifically after the binary complexes are produced by the initial nicking reaction, but much less efficiently to initiate the cleavage reaction itself. The fact that Y26-mediated cleavage (assumed to be the termination reaction) does not occur on R(12s+18) oligonucleotide, but occurs efficiently with R(25s+4), substantiates the notion that IR2 recognition is required for Y26-mediated cleavage and, hence, for termination. This idea was suggested previously by others (Becker and Meyer, 2000) and by us (Garcillan-Barcia et al, 2007) on the basis of in vivo results. It should be remembered that IR2 is only exposed as a hairpin in the termination reaction (see model in Figure 6). Thus, we assume that, when TrwC binds a IR2 hairpin-containing oligonucleotide, it orientates Y26 towards the catalytic center, but only when Y18 is already covalently bound to DNA after the first cleavage reaction. In fact, Grandoso et al (2000) showed that both TrwCFL(wt) or TrwCFL(Y26F) proteins efficiently cleaved supercoiled oriT-containing DNA in vitro, whereas neither TrwCFL(Y18F) nor TrwCFL(Y18FY26F) were able to cleave such DNA. Thus, our present results nicely explain the perplexing asymmetry observed in the previous work.
Figure 6.
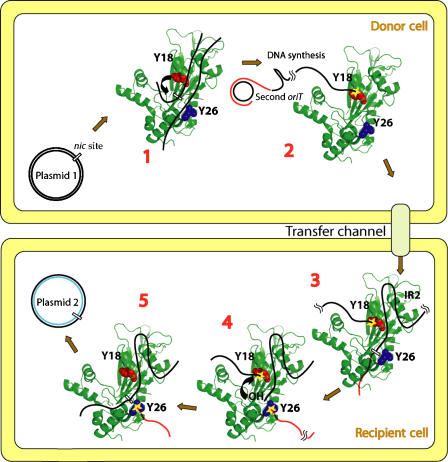
Model for the mechanism of DNA processing during R388 plasmid conjugation. (1) First oriT binding: TrwC (the protein structure is represented as a cartoon, colored in green; Y18 and Y26 residues are shown as spacefill representations in red and blue, respectively) binds supercoiled R388 plasmid DNA (black lines) around the nic site in oriT. (2) Y18 cleavage: TrwC tyrosine Y18 cleaves R388 at nic (curved arrow in (1)). Upon cleavage, TrwC becomes bound to the 5′ end of nic (yellow star). RCR in the donor plasmid DNA (new DNA represented as a red line) displaces the T-strand. TrwC bound to the T-strand pilots the DNA to the recipient cell by the transfer channel. (3) Second oriT binding: TrwC recognizes IR2 when the trailing DNA arrives in the recipient cell and specifically binds nic of the second oriT. (4) Y26 cleavage: TrwC tyrosine Y26 cleaves the second oriT at nic, forming a second phosphotyrosine bond (yellow star bound to red line). A free 3′-OH is generated that attacks the Y18–DNA complex (curved arrow in (4)). (5) Plasmid release: DNA-strand ligation occurs by a second transesterification event, when the newly generated 3′-OH cleaves the phosphotyrosyl bond and the covalent intermediate is resolved. This last step allows recircularization and release of the transferred plasmid DNA.
Thus, Y26 is specifically suit to perform the strand-transfer reaction that constitutes the termination reaction of conjugative DNA processing. To further analyze its role in strand-transfer, we purified binary complexes produced by covalent binding of oligonucleotides to Y18 in TrwCR. Whereas the TrwCR(Y26F)-18-mer complex carried out strand-transfer with R(25+0), but not with R(25+8), the TrwCR(wt)-18-mer complex was able to complete strand-transfer with either of the two oligonucleotides. To be able to engage in strand-transfer, R(25+8) had to be first cleaved to generate the 25-mer containing a free 3′-OH. This second cleavage reaction would only be possible when Y26 was present (as Y18 was already bound to the 18-mer generated by the suicide substrate). When TrwCR(Y18F), TrwCR(Y26F) or TrwCR(wt) were added to the reaction, no transfer was observed. Thus, the attacking Y26 has to come from the same relaxase molecule as the Y18 that performed the first cleavage reaction and is covalently bound to DNA. A similar cis termination requirement was observed in the YY bacteriophage φX174 gene A* protein (Hanai and Wang, 1993). However, Grandoso et al (2000) showed that TrwCFL(Y18F) or TrwCFL(Y26F) were able to perform strand-transfer reactions in vitro with a similar efficiency to that of TrwCFL(wt). Full-length TrwC has been shown to be a dimer in solution, whereas TrwCR is a monomer. Thus, it was possible that dimerization affected the reaction mechanism, perhaps by facilitating an intermolecular re-ligation step using the free Y18 on the second TrwC molecule in the dimer. This explanation was discarded as TrwCFL(wt) or TrwCFL(Y26F) covalent complexes behaved exactly as their TrwCR counterparts in the transfer reaction experiments.
The results obtained in this work nicely explain the previous data with whole-cell experiments. It is known that TrwC relaxase is transported to the conjugative recipient cells as a pilot protein during conjugation (Draper et al, 2005). Besides, the relaxase plays an essential role in the recipient cell, as shown by Garcillan-Barcia et al (2007), who showed that anti-TrwC antibodies expressed in the recipient cells could inhibit R388-mediated conjugation specifically. Importantly, Y26 was a key residue in this function as active anti-conjugation antibodies bound the region of TrwC containing Y26. It was inferred that Y26 was catalyzing the termination reaction in the recipient cell. The in vivo effects of Y26 mutations could even be rescued by expression of the relaxase in recipient cells. All these data taken together allow us to propose a detailed model for the mechanism of DNA processing during conjugation. The model is explained in Figure 6.
Proteins involved in the initiation of RCR in plasmids and phages were classified in two major groups, conjugative relaxases (Mob) and replicases (Rep) (Ilyina and Koonin, 1992; Mendiola and de la Cruz, 1992; Koonin and Ilyina, 1993). The replicase group was subdivided in two superfamilies (SF). SF-I RCR-Rep proteins contain two active-site tyrosines (YY-Rep proteins), whereas SF-II proteins contain just one tyrosine (Y-Rep proteins) (Odegrip and Haggard-Ljungquist, 2001). In the latter case, termination can occur either by transesterification using a tyrosine from a second Rep monomer or by hydrolysis using an activated water molecule. Bacteriophage φX174 gene A* protein (an example of a YY-Rep protein), acts by a flip-flop mechanism in which the two active tyrosines alternate in initiation and termination in an indistinguishable manner (Hanai and Wang, 1993). Whereas the two active tyrosines play equivalent roles in the viral protein, they play specific roles in TrwC-mediated conjugative transfer. Thus, only Y18 is used for initiation, whereas Y26 specifically terminates conjugation. Interestingly, the Rep protein from bacteriophage P2 also displays two nonequivalent tyrosines with alternating roles in initiation and termination, a variant of the flip-flop mechanism (Odegrip and Haggard-Ljungquist, 2001). On the other hand, plasmids that replicate by RCR do not use a flip-flop mechanism. This difference could be a consequence of the different fundamental strategy of plasmid replication versus the infectious nature of phage replication. Thus, plasmid RCR requires a mechanism for copy number control that should avoid the generation of multiple DNA copies from a single initiation event. This is easily accommodated in plasmid RCR by including an irreversible step in the termination reaction. Thus, plasmid pT181 RepC is a Y-Rep protein dimer. One of the monomers catalyzes the initiation reaction, whereas the second monomer catalyzes termination by a second transesterification reaction in which the dimer is rendered inactive by the formation of a DNA–protein complex (Rasooly and Novick, 1993). As a second example, plasmid pC194 Y-Rep replicase RepA acts as a monomer. It initiates RCR by the usual tyrosine-based reaction, but terminates it by hydrolysis of the Tyr–DNA bond by a water molecule activated by a glutamic acid residue in the same protein monomer (Noirot-Gros et al, 1994).
It should be remembered that, as RCR replicases, conjugative relaxases can have either one or two active-site tyrosines (Zechner et al, 2000; Francia et al, 2004). The mechanism shown in this work exemplifies the YY-Mob mechanism. How do we explain the mechanism of termination in Y-relaxases? In the only case analyzed experimentally (relaxase TraI of plasmid RP4), the tyrosine for the second transesterification reaction was made available intermolecularly by a second relaxase molecule (Pansegrau and Lanka, 1996). The second molecule should act either in the donor cell (and thus providing the free hydroxyl group for the attack on the DNA–Tyr bond), or in the recipient cell after conjugative-mediated transport of the relaxase, which has been proven experimentally at least for TrwC (Draper et al, 2005).
The results presented in this work emphasize the asymmetrical nature of the initiation and termination reactions in plasmid R388 conjugation. Thus, it would seem that plasmid conjugation is more similar to phage than to plasmid replication, in that no strict control of termination is required. In fact, the transfer of as many DNA molecules as possible could ensure higher conjugation efficiency (or higher opportunities for recombination of the transferred material). In this respect, bacterial conjugation may be envisioned as an invasive process like phage infection.
Materials and methods
Synthesis of phosphorothiolate oligonucleotides
The phosphorothiolate oligonucleotides used in this work were synthesized by the incorporation of 3′-thiothymidine (X) into the following TrwC substrates at the cleavage site: 5′-TGTGTATTGTCXATAG, R(12s+4); 5′-TGTGTATTGTCXATAGCCCAGATTTAAGGA, R(12s+18); and 5′-GCGCACCGAAAGGTGCGTATTGTCXATAG, R(25s+4). The detailed synthesis of the phosphorothiolate oligonucleotides is described in Supplementary data.
Protein expression and purification
TrwCR(wt), the relaxase domain of TrwC (residues 1–293) and its mutants TrwCR(Y18F) and TrwCR(Y26F) were purified as described previously (Boer et al, 2006). Briefly, pET23a∷trwC-N293 or its mutants were expressed in Escherichia coli strain C43 using a micro-DCU fermentation system (B Braun, Biotech International). Cells were lysed and the lysate centrifuged at 45 000 g for 45 min at 4°C. Protein purification was carried out in two chromatographic steps: P11-phosphocellulose (Whatman) and MonoS (Amersham Pharmacia) column chromatography. The full-length proteins TrwCFL(wt) and TrwCFL(Y26F) were purified as described previously (Grandoso et al, 2000).
Cleavage reactions
Reaction mixtures (20 μl) contained 10 μM TrwCR(wt) (or TrwCR mutants, TrwCFL(wt) or TrwCFL(Y26F)), 25 μM oligonucleotide and 100 μM MgCl2. The mixture was incubated at 37°C for different periods of time. Samples taken from reaction mixtures were electrophoresed by 12% SDS–PAGE and stained with coomassie brilliant blue. Gels were digitally scanned and percent cleavage was calculated from band intensities using Quantity One® software (Bio-Rad).
Strand-transfer reactions
Before DNA strand-transfer reactions, covalent complexes (either TrwCR(wt)-18-mer, TrwCR(Y26F)-18-mer, TrwCFL(wt)-18-mer or TrwCFL(Y26F)-18-mer) were purified by Superdex 75 (Amersham Pharmacia) size-exclusion chromatography. Then, 1 μM of the relevant complex was incubated with 0.5 μM Texas Red-labelled oligonucleotide R(25+0)=(GCGCACCGAAAGGTGCGTATTGTCT), or R(25+8)=(GCGCACCGAAAGGTGCGTATTGTCTATAGCCCA). An unrelated mixture of single-stranded oligonucleotides at a final concentration of 5 μM was also included in the reactions involving TrwCFL. When additional protein was added to the initial reaction to analyze intermolecular reactions, the added protein final concentration was 10 μM. After incubation for 60 min at 37°C, samples were treated with 0.6 mg/ml proteinase K and 0.05% (w/v) SDS for 20 min at 37°C. Reaction products were separated and quantified by capillary electrophoresis as described previously (Boer et al, 2006), using the CE Oligonucleotide Analysis Kit (BioRad) in the capillary system BioFocus®2000 (BioRad). The capillary used was a BioCAP Oligonucleotide Analysis Capillary (30 cm × 75 μm i.d. × 375 μm o.d.). Samples were introduced in the capillary by pressure injection (200 psi/s). Electrophoresis was carried out at 12 kV and 40°C. A laser-induced detector was employed for detection of Texas Red-labelled oligonucleotides. Peak information (migration time, peak area and height) was obtained using the CE Integrator Software (BioRad).
Mass spectrometry
Selected protein bands were excised manually from the gel and subjected to in-gel digestion with Trypsin (Roche) or Glutamic-C (Princeton Separations). Trypsin digestion was performed according to Shevchenko et al (1996) with minor modifications. The gel pieces were swollen in digestion buffer containing 50 mM NH4HCO3 and 12.5 ng/μl proteomics grade trypsin in an ice bath. After 45 min, the supernatant was discarded and 5 μl of 50 mM NH4HCO3 was added to the gel pieces. Digestion proceeded at 37°C overnight. The supernatant was recovered and peptides were extracted twice: first, with 25 mM NH4HCO3 and acetonitrile, and then with 0.1% TFA and acetonitrile. The recovered supernatants and extracted peptides were pooled, dried in a SpeedVac (Thermo Electron) redissolved in 10 μl of 0.1% FA and sonicated for 5 min. For specific Glutamic-C digestion, same procedure was followed but the digestion proceeded at 30°C. Unspecific Glutamic-C digestion was performed using 50 mM sodium phosphate buffer (pH 7.8).
LC-MS spectra were acquired using a Q-Tof micro mass spectrometer (Waters) interfaced with a CapLC capillary chromatograph (Waters). An aliquot (5 μl) of each sample was loaded onto a Symmetry 300 C18 NanoEase Trap precolumn (Waters) and washed with 0.1% FA for 5 min at a flow rate of 20 μl/min. The precolumn was connected to an Atlantis dC18 NanoEase column (75 μm × 150 mm; Waters) equilibrated in 5% acetonitrile and 0.1% FA. A flow splitter was used to decrease the flow rate to 0.2 μl/min and peptides were eluted with a 30 min linear gradient of 10–60% acetonitrile directly onto a NanoEase Emitter (Waters). Obtained spectra were manually analyzed using MassLynx 4.1 software (Waters). The presence or absence of peptides or peptide–DNA complexes in different samples was determined by comparison of the retention time, charge state and signal intensity of the selected ions against the LC-MS profile of the native protein.
Supplementary Material
Supplementary Data
Acknowledgments
We are grateful to Dr Jesus María Arizmendi and Dr Kerman Aloria for mass spectrometry analysis performed in the Proteomics Facility at the University of the Basque Country. This work was financed by Grants BFU2005-03477 from the Spanish Ministry of Education and Science, and EU 6th Framework Programme project SHM-CT-2005-019023 to FC and by the School of Chemistry and Chemical Engineering to JSV and LAC.
References
- Avila P, Grinsted J, de la Cruz F (1988) Analysis of the variable endpoints generated by one-ended transposition of Tn21. J Bacteriol 170: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker EC, Meyer RJ (2000) Recognition of oriT for DNA processing at termination of a round of conjugal transfer. J Mol Biol 300: 1067–1077 [DOI] [PubMed] [Google Scholar]
- Boer R, Russi S, Guasch A, Lucas M, Blanco AG, Perez-Luque R, Coll M, de la Cruz F (2006) Unveiling the molecular mechanism of a conjugative relaxase: the structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J Mol Biol 358: 857–869 [DOI] [PubMed] [Google Scholar]
- Burgin AB Jr, Nash HA (1995) Suicide substrates reveal properties of the homology-dependent steps during integrative recombination of bacteriophage lambda. Curr Biol 5: 1312–1321 [DOI] [PubMed] [Google Scholar]
- Byrd DR, Sampson JK, Ragonese HM, Matson SW (2002) Structure-function analysis of Escherichia coli DNA helicase I reveals non-overlapping transesterase and helicase domains. J Biol Chem 277: 42645–42653 [DOI] [PubMed] [Google Scholar]
- Cesar CE, Machon C, de la Cruz F, Llosa M (2006) A new domain of conjugative relaxase TrwC responsible for efficient oriT-specific recombination on minimal target sequences. Mol Microbiol 62: 984–996 [DOI] [PubMed] [Google Scholar]
- Datta S, Larkin C, Schildbach JF (2003) Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure 11: 1369–1379 [DOI] [PubMed] [Google Scholar]
- Draper O, Cesar CE, Machon C, de la Cruz F, Llosa M (2005) Site-specific recombinase and integrase activities of a conjugative relaxase in recipient cells. Proc Natl Acad Sci USA 102: 16385–16390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia MV, Varsaki A, Garcillan-Barcia MP, Latorre A, Drainas C, de la Cruz F (2004) A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev 28: 79–100 [DOI] [PubMed] [Google Scholar]
- Garcillan-Barcia MP, Jurado P, Gonzalez-Perez B, Moncalian G, Fernandez LA, de la Cruz F (2007) Conjugative transfer can be inhibited by blocking relaxase activity within recipient cells with intrabodies. Mol Microbiol 63: 404–416 [DOI] [PubMed] [Google Scholar]
- Ghosh K, Lau CK, Gupta K, Van Duyne GD (2005) Preferential synapsis of loxP sites drives ordered strand exchange in Cre-loxP site-specific recombination. Nat Chem Biol 1: 275–282 [DOI] [PubMed] [Google Scholar]
- Grandoso G, Avila P, Cayon A, Hernando MA, Llosa M, de la Cruz F (2000) Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J Mol Biol 295: 1163–1172 [DOI] [PubMed] [Google Scholar]
- Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, Gomis-Ruth FX, de la Cruz F, Coll M (2003) Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat Struct Biol 10: 1002–1010 [DOI] [PubMed] [Google Scholar]
- Hanai R, Wang JC (1993) The mechanism of sequence-specific DNA cleavage and strand transfer by phi X174 gene A* protein. J Biol Chem 268: 23830–23836 [PubMed] [Google Scholar]
- Ilyina TV, Koonin EV (1992) Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res 20: 3279–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Ilyina TV (1993) Computer-assisted dissection of rolling circle DNA replication. Biosystems 30: 241–268 [DOI] [PubMed] [Google Scholar]
- Krogh BO, Shuman S (2000) Catalytic mechanism of DNA topoisomerase IB. Mol Cell 5: 1035–1041 [DOI] [PubMed] [Google Scholar]
- Lanka E, Wilkins BM (1995) DNA processing reactions in bacterial conjugation. Annu Rev Biochem 64: 141–169 [DOI] [PubMed] [Google Scholar]
- Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF (2005) Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure 13: 1533–1544 [DOI] [PubMed] [Google Scholar]
- Llosa M, de la Cruz F (2005) Bacterial conjugation: a potential tool for genomic engineering. Res Microbiol 156: 1–6 [DOI] [PubMed] [Google Scholar]
- Llosa M, Grandoso G, de la Cruz F (1995) Nicking activity of TrwC directed against the origin of transfer of the IncW plasmid R388. J Mol Biol 246: 54–62 [DOI] [PubMed] [Google Scholar]
- Llosa M, Grandoso G, Hernando MA, de la Cruz F (1996) Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J Mol Biol 264: 56–67 [DOI] [PubMed] [Google Scholar]
- Matson SW, Ragonese H (2005) The F-plasmid TraI protein contains three functional domains required for conjugative DNA strand transfer. J Bacteriol 187: 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiola MV, de la Cruz F (1992) IS91 transposase is related to the rolling-circle-type replication proteins of the pUB110 family of plasmids. Nucleic Acids Res 20: 3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncalian G, Grandoso G, Llosa M, de la Cruz F (1997) oriT-processing and regulatory roles of TrwA protein in plasmid R388 conjugation. J Mol Biol 270: 188–200 [DOI] [PubMed] [Google Scholar]
- Monzingo AF, Ozburn A, Xia S, Meyer RJ, Robertus JD (2007) The structure of the minimal Relaxase domain of MobA at 2.1 A resolution. J Mol Biol 366: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot-Gros MF, Bidnenko V, Ehrlich SD (1994) Active site of the replication protein of the rolling circle plasmid pC194. EMBO J 13: 4412–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegrip R, Haggard-Ljungquist E (2001) The two active-site tyrosine residues of the a protein play non-equivalent roles during initiation of rolling circle replication of bacteriophage p2. J Mol Biol 308: 147–163 [DOI] [PubMed] [Google Scholar]
- Pansegrau W, Lanka E (1996) Mechanisms of initiation and termination reactions in conjugative DNA processing. Independence of tight substrate binding and catalytic activity of relaxase (TraI) of IncPalpha plasmid RP4. J Biol Chem 271: 13068–13076 [DOI] [PubMed] [Google Scholar]
- Rasooly A, Novick RP (1993) Replication-specific inactivation of the pT181 plasmid initiator protein. Science 262: 1048–1050 [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Kruft V, Otto S (1993) Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur J Biochem 217: 929–938 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Street LM, Harley MJ, Stern JC, Larkin C, Williams SL, Miller DL, Dohm JA, Rodgers ME, Schildbach JF (2003) Subdomain organization and catalytic residues of the F factor TraI relaxase domain. Biochim Biophys Acta 1646: 86–99 [DOI] [PubMed] [Google Scholar]
- Williams SL, Schildbach JF (2006) Examination of an inverted repeat within the F factor origin of transfer: context dependence of F TraI relaxase DNA specificity. Nucleic Acids Res 34: 426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner EL, de la Cruz F, Eisenbrandt R, Grahn AM, Koraimann G, Lanka E, Muth G, Pansegrau W, Thomas CM, Wilkins BM, Zatyka M (2000) Conjugative DNA Transfer Processes. In The Horizontal Gene Pool: Bacterial Plasmids and Gene Spread, Thomas CM (ed), Vol. xxiii, 419 p. London: Harwood Academic Publishers [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data