Abstract
Cyclin-dependent kinases (CDKs) control yeast morphogenesis, although how they regulate the polarity machinery remains unclear. The dimorphic fungus Candida albicans uses Cdc28/Hgc1, a CDK/cyclin complex, to promote persistent actin polarization for hyphal growth. Here, we report that Rga2, a GTPase-activating protein (GAP) of the central polarity regulator Cdc42, undergoes Hgc1-dependent hyperphosphorylation. Using the analog-sensitive Cdc28as mutant, we confirmed that Cdc28 controls Rga2 phosphorylation in vitro and in vivo. Deleting RGA2 produced elongated yeast cells without apparent effect on hyphal morphogenesis. However, deleting it or inactivating its GAP activity restored hyphal growth in hgc1Δ mutants, suggesting that Rga2 represses hyphal development and Cdc28/Hgc1 inactivates it upon hyphal induction. We provide evidence that Cdc28/Hgc1 may act to prevent Rga2 from localizing to hyphal tips, leading to localized Cdc42 activation for hyphal extension. Rga2 also undergoes transient Cdc28-dependent hyperphosphorylation at bud emergence, suggesting that regulating a GAP(s) of Cdc42 by CDKs may play an important role in governing different forms of polarized morphogenesis in yeast. This study reveals a direct molecular link between CDKs and the polarity machinery.
Keywords: Candida albicans, cyclin-dependent kinase, GTPase, GTPase-activating protein, polarized growth
Introduction
The ability to change cell shape and growth pattern is important for fungi to adapt to the constantly changing environment. For example, upon nitrogen starvation, Saccharomyces cerevisiae (Sc) undergoes a growth transition involving drastic changes in cell morphology and the budding pattern, resulting in a filamentous form of growth that allows the organism to grow away from the colony and penetrate solid media to forage for nutrients (Gimeno et al, 1992). Similarly, the human fungal pathogen Candida albicans (Ca) switches from yeast to hyphal growth when exposed to serum or when ingested by macrophages, which is crucial for the penetration of host tissues and escape from phagocytic destruction (Gow et al, 2002).
Several signaling pathways regulate hyphal morphogenesis in C. albicans (Liu, 2001; Berman and Sudbery, 2002; Gow et al, 2002; Kumamoto and Vinces, 2005), among which the cAMP/protein kinase A (PKA) and mitogen-activated protein (MAP) kinase pathways play central roles (Liu, 2001). They activate the transcription factors, Efg1 and Cph1 respectively, leading to the expression of hypha-specific genes (HSGs) responsible for a diverse range of infection-related functions (Liu et al, 1994; Lo et al, 1997; Stoldt et al, 1997; Kumamoto and Vinces, 2005). Among numerous HSGs found so far, the only one essential for hyphal growth is the G1 cyclin-related protein gene HGC1 (Zheng et al, 2004). Hgc1, in association with Cdc28, promotes persistent actin polarization to hyphal tips, and deleting it severely impairs hyphal development (Zheng et al, 2004). However, how Cdc28/Hgc1 regulates the polarity machinery remains entirely unknown.
Current models for S. cerevisiae morphogenesis emphasize a central role of Cdc28 (also known as Cdk1) (Lew and Reed, 1993, 1995). It determines bud shape through association with different cyclins: the G1 cyclins Cln1 and Cln2 promote bud emergence and apical elongation, whereas the mitotic cyclins Clb1 and Clb2 stimulate isotropic bud expansion. A primary function of the G1 CDK at the beginning of the cell cycle is to activate the polarity machinery for bud formation (Moffat and Andrews, 2004). Potential G1 CDK substrates include regulators of the Rho GTPase Cdc42 and its effectors and associated proteins. Cdc24 is the only guanine-nucleotide-exchange factor (GEF) that positively regulates Cdc42, while several GTPase-activating proteins (GAPs), including Rga1, Rga2 and Bem3, negatively regulate it (Smith et al, 2002; Park and Bi, 2007). Bem3 and Rga1 contain cyclin-dependent kinase (CDK) consensus phosphorylation motifs, associate with Cln2 and can be phosphorylated by Cdc28 (Ubersax et al, 2003; Archambault et al, 2004). McCusker et al (2007) recently demonstrated that Cdk1/G1 cyclin complexes phosphorylate two Cdc24-associated proteins Boi1 and Rga2 in the control of cell surface growth. Ste20, a member of the p21-activated kinase family, is phosphorylated by Cdc28/Cln1, 2, which is thought to repress pheromone signaling and/or promote morphogenetic functions (Oehlen and Cross, 1998; Wu et al, 1998). Bem1, an interacting partner of Cdc24 and Ste20, can be phosphorylated by Cdc28/Cln3, but the phosphorylation mainly affects vacuolar morphogenesis (Han et al, 2005). Systematic studies in S. cerevisiae have revealed many polarity proteins as potential CDK substrates (Ubersax et al, 2003), indicating high regulatory complexity linking the cell-cycle engine with the polarity machinery.
Similar to S. cerevisiae, genetic data also suggest that CDK/cyclins control C. albicans morphogenesis. Two G1 cyclins, Cln1 and Cln3, are crucial for bud emergence and G1–S transition (Loeb et al, 1999; Bachewich and Whiteway, 2005; Chapa y Lazo et al, 2005), while two B-cyclins, Clb2 and Clb4, are important for S and M phase progression and negatively regulate polarized growth (Bensen et al, 2005). When G1 cells are induced for hyphal growth, the cellular level of Cln1 persists longer and the appearance of the B-type cyclins is delayed in comparison with the same cells released for yeast growth (Bensen et al, 2005). These data suggest that cyclins other than the hypha-specific Hgc1 may also contribute to constructing the highly elongated hyphal cells.
The Cdc42 polarity regulatory module also plays a key role in C. albicans hyphal development. Depletion of Cdc42 blocks both yeast and hyphal growth (Ushinsky et al, 2002). Bassilana et al (2003) observed that hyphal growth is more sensitive than yeast growth to decreases in cellular levels of Cdc42 and Cdc24, because reducing CDC42 and CDC24 expression can support normal yeast growth but results in defects in hyphal growth. Cdc42 and Cdc24 persistently localize to germ tube and hyphal tips (Hazan and Liu, 2002; Bassilana et al, 2005; Crampin et al, 2005). Consistently, the polarisome components, such as Spa2 and the formin Bni1, also polarize to hyphal tips in a similar manner, whereas they localize to different growth sites in different cell-cycle phases during yeast growth (Zheng et al, 2003; Crampin et al, 2005; Li et al, 2005; Martin et al, 2005).
In this study, we have investigated whether Cdc28/Hgc1 directly controls the polarity regulatory module Cdc42/Cdc24/GAP during hyphal morphogenesis. Here, we report the identification of Rga2, a Cdc42 GAP as a key regulatory target of Cdc28/Hgc1 based on biochemical, genetic and cell biology data. Our results establish a link between two central regulators of polarized growth.
Results
Characterization of Cdc42 GAPs in C. albicans
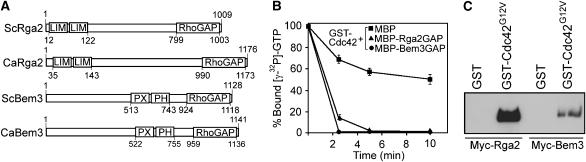
To understand how Cdc28/Hgc1 controls C. albicans hyphal morphogenesis, we set out to identify potential Cdc28/Hgc1 substrates. We first focused on Cdc42 GAPs, because previous studies had provided strong evidence that ScBem3 and ScRga1 may be substrates of ScCdc28 (Ubersax et al, 2003; Archambault et al, 2004). C. albicans genome contains two Cdc42 GAP homologues, one having the highest sequence identity of 23% with ScBem3 and the other ∼22% with ScRga2, which were thus named CaBem3 and CaRga2, respectively. Both ScBem3 and CaBem3 contain a C-terminal RhoGAP domain and a central PX/PH domain (Figure 1A), whereas ScRga2 and CaRga2 have two tandem LIM domains at the N-terminus and a RhoGAP domain at the C-terminus. To confirm that Rga2 and Bem3 are Cdc42 GAPs, we first demonstrated that the purified GAP domain of each protein expressed in Escherichia coli could enhance the hydrolysis of Cdc42-bound GTP (Figure 1B). To demonstrate association of the GAPs with Cdc42, we tagged Rga2 or Bem3 with a 6 × Myc epitope at the N-terminus and expressed them from the MET3 promoter. Extracts from cells expressing Myc-Rga2 or Myc-Bem3 were pulled down with beads bearing E. coli-expressed GST-Cdc42G12V (a constitutive active version of Cdc42; Davis et al, 1998) or with control beads carrying GST alone. Anti-Myc (αMyc) Western blotting detected Rga2 and Bem3 only in the pull down using the GST-Cdc42G12V beads, but not in pull downs using the control beads (Figure 1C), indicating association of both GAPs with Cdc42. These data support the idea that Rga2 and Bem3 are Cdc42 GAPs in C. albicans. Recently, Court and Sudbery (2007) made similar conclusions.
Figure 1.
Bem3 and Rga2 are Cdc42 GAPs. (A) CaRga2 and CaBem3 domain organizations. (B) The GAP domain of Rga2 and Bem3 were expressed as MBP fusions, purified and assayed for activities in promoting the hydrolysis of [γ-P32]-GTP prebound to Cdc42. In a control experiment, [γ-32P]-GTP was replaced with [α-32P]-GTP, showing that the decrease in Cdc42-bound γ-P32 is not due to dissociation of [γ-P32]-GTP (Supplementary Figure S1). The assays were performed in triplicates and standard errors are shown. (C) Myc-Rga2 and Myc-Bem3 associated with GST-Cdc42G12V. Myc-Rga2 in ZR5 (Table I lists all the strains used in this study) and Myc-Bem3 (ZR6) were purified by beads carrying GST-Cdc42G12V or GST before anti-Myc Western blotting (WB).
Rga2 undergoes Cdc28/Hgc1-dependent hyperphosphorylation upon hyphal induction
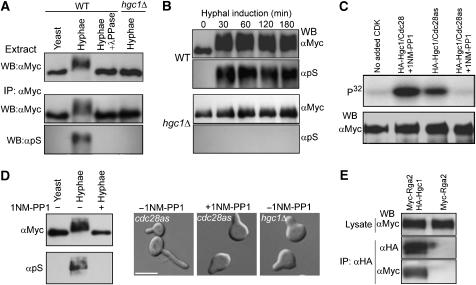
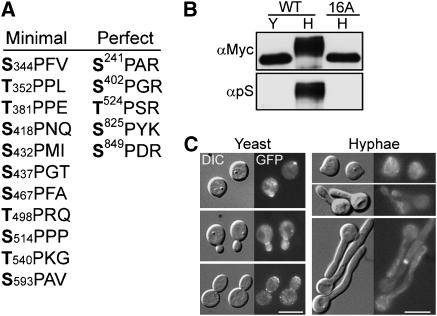
To determine whether Cdc28/Hgc1 controls Rga2 and Bem3 phosphorylation, we compared the electrophoretic mobility of the two proteins extracted from wild-type (WT) and hgc1Δ cells grown under yeast or hyphal growth conditions. Western-blot analyses (Figure 2A, upper panel) revealed that Myc-Rga2, but not Myc-Bem3 (data not shown), from WT hyphae migrated much slower than that from yeast cells, and this slow migration was completely abolished by lambda-phosphatase (λPPase) treatment of the lysate, confirming that it is the result of phosphorylation. Moreover, the mobility shift was not observed in hgc1Δ cells grown under the same induction condition. The results demonstrate that Rga2 undergoes hyperphosphorylation in an Hgc1-dependent manner in response to hyphal induction. Recently, Court and Sudbery (2007) also observed Rga2 hyperphosphorylation during hyphal growth. Rga2 contains multiple perfect CDK phosphorylation motifs (S/TPXR/K), including S241PAR, S402PGR, T524PSR, S825PYK and S849PDR, making it a likely substrate of Cdc28/Hgc1. To test this, we immunoprecipitated Rga2 and performed Western blotting by using an antibody that preferentially recognizes the phospho-serine residues in perfect CDK phosphorylation motifs. This antibody strongly reacted with Rga2 from WT hyphae, but not with that from yeast cells or that from hgc1Δ hyphal cells (Figure 2A, lower panel). Analyses at different time points of induction detected Rga2 hyperphosphorylation at as early as 30 min and throughout hyphal growth in WT cells, but not in hgc1Δ cells at any time of induction (Figure 2B).
Figure 2.
Cdc28/Hgc1 controls Rga2 phosphorylation. (A) Hgc1-dependent Rga2 hyperphosphorylation in hyphae. ZR5 (WT) and ZR9 (hgc1Δ) cells were grown as yeast or hyphae for 2 h. Myc-Rga2 in lysates or immunoprecipitates (IP) were probed with αMyc or an anti-phosphoserine-CDK-site antibody (αpS). (B) Rga2 hyperphosphorylation kinetics. ZR5 and ZR9 cells were collected at intervals of induction. Myc-Rga2 was immunoprecipitated with αMyc before Western blotting with αMyc or αpS. (C) Purified Cdc28/Hgc1 phosphorylated Rga2 in vitro. Anti-HA-purified Cdc28/HA-Hgc1 (ZR11) or Cdc28as/HA-Hgc1 (ZR12) was used to phosphorylate anti-Myc-purified Myc-Rga2 (ZR5), in the presence or absence of 1NM-PP1. (D) 1NM-PP1 inhibited Rga2 hyperphosphorylation and hyphal morphogenesis. ZR13 (cdc28as) and WYZ12 (hgc1Δ) G1 cells were induced in the presence or absence of 1NM-PP1 for 40 min for Western blotting, and 1 h for morphology. More than 90% of the hgc1Δ cells and the cdc28as cells treated with 1NP-PP1 exhibited similar morphology at the time of examination. (E) Rga2 was co-immunoprecipitated with Hgc1. Anti-HA immunoprecipitation was performed on lysates from cells coexpressing Myc-Rga2 and HA-Hgc1 (ZR15) or cells expressing Myc-Rga2 (ZR5) alone. The cells were grown as hyphae for 2 h before immunoprecipitation.
To gain further evidence that Cdc28/Hgc1 may directly phosphorylate Rga2, we constructed a strain expressing HA-tagged Hgc1 together with a mutant version of Cdc28 (Cdc28as), which is sensitive to the ATP analog 1NM-PP1 (Bishop et al, 2000). Cdc28as/HA-Hgc1 complexes were purified by affinity chromatography and used to phosphorylate Myc-Rga2 immunoprecipitated from yeast cells in the presence of γ-32P-ATP. Radioautography following SDS–PAGE detected Myc-Rga2 phosphorylation, which was significantly inhibited by 1NM-PP1 (Figure 2C), suggesting that Cdc28/Hgc1 may directly phosphorylate Rga2. Moreover, adding 1NM-PP1 to the hyphal induction medium eliminated the mobility retardation of Rga2 in the cells expressing Cdc28as (Figure 2D, left panel) and caused hyphal defects reminiscent of hgc1Δ cells (Figure 2D, right panel). Thus, we conclude that Rga2 hyperphosphorylation is dependent on Cdc28 in vivo. We also examined whether Hgc1 could associate with Rga2. Equal amounts of cell extracts prepared from hyphae coexpressing Myc-Rga2 and HA-Hgc1 or hyphae expressing Myc-Rga2 alone were subjected to anti-HA immunoprecipitation, followed by anti-Myc Western blotting. With amounts of the input Myc-Rga2 being equal, we detected Myc-Rga2 only from the cells coexpressing Myc-Rga2 and HA-Hgc1, but not from those expressing Myc-Rga2 alone (Figure 2E), indicating that Rga2 associates with Hgc1 in vivo. As an additional negative control, the interaction between Myc-Rga2 and HA-Hgc1 was not detected during yeast growth (Supplementary Figure S2).
Deletion of RGA2 suppresses hyphal defects of the hgc1Δ mutant
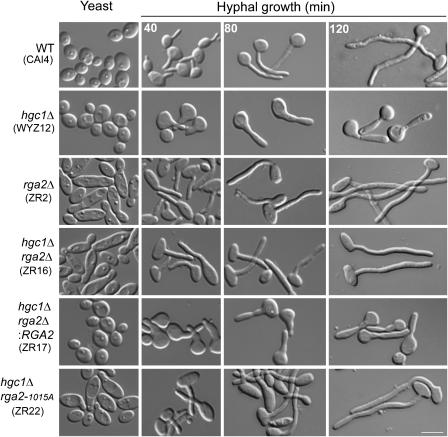
The Hgc1-dependent Rga2 hyperphosphorylation observed above suggests that Rga2 phosphorylation may play a role in regulating hyphal morphogenesis. To demonstrate this, we first constructed an rga2Δ mutant to determine whether Rga2 is required for hyphal growth. Figure 3 shows that under yeast growth conditions, nearly all rga2Δ cells exhibited an elongated morphology compared with WT, and many mutant cells also had markedly wide bud necks, consistent with defects in bud morphogenesis. Reintegrating a copy of WT RGA2 driven by its own promoter fully corrected the morphological defects of the mutant (strain ZR2.1, data not shown). However, rga2Δ cells could grow normal-looking hyphae. Since Rga2 is a negative regulator of Cdc42, one possible explanation of the apparent normal hyphal growth of rga2Δ cells is that Rga2 might function as a repressor of hyphal growth and Cdc28/Hgc1 inactivates it by phosphorylation upon hyphal induction. This hypothesis predicted that deleting RGA2 from the hgc1Δ mutant should restore or improve hyphal morphogenesis. Indeed, the double mutant hgc1Δ rga2Δ produced normal-looking hyphae (Figure 3). Unlike WT hyphae which emerge from a small surface area, hgc1Δ cells produced a broad surface protrusion that later grew into short and thick elongated cells (Zheng et al, 2004; and Figure 3). The WT-like hyphae of the hgc1Δ rga2Δ mutant indicate that deleting RGA2 corrected the morphological defects caused by deleting HGC1 from the beginning throughout hyphal development. Reintroducing a copy of WT RGA2 into the double mutant (hgc1Δ rga2Δ:RGA2) resulted in cells with the same morphological defects as the hgc1Δ mutant, further confirming that it was the loss of RGA2 in the hgc1Δ mutant that restored hyphal growth. Taken together, the results reveal that an important role of Cdc28/Hgc1 in hyphal growth is to repress Rga2 function. Deleting BEM3 had no effect on hgc1Δ hyphal growth (Supplementary Figure S3), indicating that Rga2 has a specific role in hyphal development.
Figure 3.
Deletion of RGA2 or inactivating its GAP activity restored hyphal growth in the hgc1Δ mutant. Yeast cells of all strains were induced for hyphal growth for the indicated times. Scale bar, 5 μm.
Rga2 GAP activity is required for repressing hyphal growth
Some functions of GAPs do not require the GAP activity; an example is ScBem2's function in the morphogenesis checkpoint (Marquitz et al, 2002). Therefore, it was necessary to test whether the Rga2 GAP activity is required for repressing hyphal growth. We inactivated the GAP activity by replacing the conserved Arg-1015 residue with alanine in the ‘arginine finger' of the GAP domain. This mutation blocks GAP activity without compromising the binding of the domain to Cdc42 (Leonard et al, 1998; Supplementary Figures S1 and S4). When introduced into the rga2Δ mutant, Rga2R1015A exhibited the same localization pattern as its WT counterpart during yeast growth (data not shown, and see Figure 4 for Rga2 localization), and the morphology of the cells was similar to that of rga2Δ mutants (Figure 3). When we deleted one copy of RGA2 and replaced the second with the rga2R1015A allele in hgc1Δ cells, it restored hyphal growth similarly to deleting RGA2 (Figure 3, hgc1Δ rga2-1015A), indicating that Rga2 GAP activity is required for repressing hyphal growth.
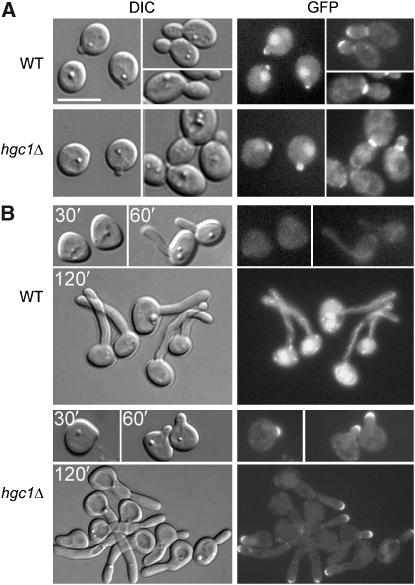
Figure 4.
Rga2 cellular localization. GFP-Rga2 was expressed in WT (ZR7) and hgc1Δ (ZR8) cells and grown under yeast (A) or hyphal induction conditions (B). Times of induction are indicated. Scale bar, 5 μm.
Rga2 localizes to polarized growth sites in hgc1Δ cells, but not in WT cells under hyphal induction
One mechanism by which phosphorylation may regulate the function of a GAP is to change its cellular localization (Bernards and Settleman, 2004). Next, we addressed the possibility that Cdc28/Hgc1 may control Rga2 localization for hyphal growth. To do this, we tagged Rga2 with the green fluorescence protein (GFP) at the N-terminus in WT and hgc1Δ cells. In both strains, GFP-Rga2 was the sole source of Rga2 expressed from the endogenous RGA2 promoter. The GFP fusion protein was functional because it allowed normal yeast morphology. Under yeast growth conditions, GFP-Rga2 exhibited similar localization patterns in WT and hgc1Δ cells: it was detected clearly at the incipient bud sites, tips of small buds and the bud necks of large-budded cells (Figure 4A). Under hyphal induction conditions, GFP-Rga2 was observed largely as cytoplasmic fluorescence in >90% of the WT cells throughout hyphal induction (Figure 4B), whereas in the rest of the cells (<10%), weak polarization of fluorescence was detected at germ tube or hyphal tips. In stark contrast, in hgc1Δ cells, GFP-Rga2 exhibited much stronger and persistent localization at every tip of the misshapen germ tubes and elongated cells (Figure 4B). The data suggest that in WT hyphae, Rga2 may be prevented from localizing to hyphal tips where Cdc42 accumulates. The lack of Rga2 localization at polarized growth sites during hyphal growth was also reported by Court and Sudbery (2007).
Mass spectrometry mapping of phosphorylation sites on Rga2
We next used mass spectrometry (MS) to map the phosphorylation sites on Rga2. Myc-Rga2 in WT hyphae was pulled down first by immunoprecipitation and then separated by SDS–PAGE. MS analyses revealed 35 phosphorylation sites (Supplementary Table S2; Supplementary Figure S5), among which five matched the perfect CDK phosphorylation motif (S/T)PX(R/K) and 11 matched the minimum motif (S/T)P (Figure 5A); 13 of the CDK phosphorylation sites are clustered in the region from amino acid (aa) 344–593, and two are at residues 825 and 849. To determine whether Cdc28/Hgc1 can phosphorylate these sites, the region from aa 344–596 was expressed as a GST-fusion protein in E. coli, purified and used as substrate for Cdc28/Hgc1 phosphorylation in vitro. Perhaps due to poor Cdc28/Hgc1 activity in in vitro kinase assay condition, MS analyses of the phosphorylation product identified only three phospho-serine/threonine residues including Ser-418, Thr-524 and Thr-540 (Supplementary Table S2), which nevertheless are a subset of the ones identified on hyphal Rga2 (Figure 5A).
Figure 5.
Evidence for Rga2 hyperphosphorylation by Cdc28. (A) MS mapping of phosphorylation sites on Rga2 purified from hyphae. The table lists all the perfect and minimal CDK phosphorylation sites identified by MS. (B) Rga2-16A did not undergo hyperphosphorylation under hyphal growth conditions. ZR5 (WT) and ZR19 (16A) were grown as yeast (Y) or hyphae (H) for 2 h before Western blotting with αMyc and αpS antibodies. (C) The rga2-16E allele significantly restored hyphal growth in the hgc1Δ mutant (compare with the morphology of rga2Δ and hgc1Δ cells shown in Figure 3). G1 cells of ZR23 (hgc1Δ rga2Δ GFP-rga2-16E) were grown as yeast or hyphae. Cells at different stages of budding or hyphal growth are shown. Scale bars, 5 μm.
To evaluate the physiological function of the CDK phosphorylation sites and determine whether the Hgc1-dependent Rga2 band shift depends on them, we first constructed an RGA2 allele in which serine/threonine residues were replaced with alanine at all the 16 CDK motifs, and transformed it into the rga2Δ mutant under control of the native promoter. The cellular localization of GFP-Rga2-16A was indistinguishable from GFP-Rga2 during both yeast and hyphal growth (data not shown). We found that the mutations abolished Rga2 hyperphosphorylation upon hyphal induction (Figure 5B, upper panel), consistent with Rga2 phosphorylation at these sites. The rga2-16A cells appeared largely normal in both yeast and hyphal growth except that the yeast cells were moderately larger than WT (data not shown), indicating that lacking phosphorylation at the 16 CDK sites only weakly affects its role in morphogenesis. This might be because CDK phosphorylation can still occur at some of the identified non-CDK consensus sites, possibly compensating for the loss of phosphorylation at the consensus sites. Since phosphorylation at the 16 consensus sites may be required to inactivate Rga2 for hyphal growth, we reasoned that introducing phosphomimic mutations may reveal the physiological significance of phosphorylation at these sites. Hence, we created an Rga2-16E mutant in which the serine/threonine residues at all the 16 sites were replaced with glutamic acid. We then introduced the rga2-16E allele into the hgc1Δ rga2Δ mutant under control of the native promoter (see Supplementary Figure S6 for expression level) and made the following observations (Figure 5C). First, this mutant allele fully rescued the elongated yeast morphology associated with RGA2 deletion, indicating that it can provide the functions required for normal yeast morphogenesis. Second, the allele significantly restored hyphal growth, although the hyphae were thicker and shorter than WT (diameter, 2.0±0.1 versus 1.8±0.1 μm; hyphal length after 2 h induction, 19±1.5 versus 22±0.9 μm; n=100) and contained shallow septal constrictions. Third, although GFP-Rga2-16E localized strongly to the incipient bud sites and bud tips in yeast cells, it was not detected at the growth sites in >75% of the cells during hyphal growth; although Rga2 polarization at tips was observed in ∼25% of hyphal cells, the intensity of fluorescence was markedly weaker than in yeast cells. rga2-16E is recessive to the WT allele because the RGA2/rga2-16E strain is indistinguishable from WT during both yeast and hyphal growth (data not shown). Together, the results support our hypothesis that phosphorylation of Rga2 at the 16 consensus CDK phosphorylation sites is important for hyphal morphogenesis, and that the phosphorylation may prevent Rga2 from tip localization. The data also suggest that Rga2 cellular localization is regulated by different mechanisms during yeast and hyphal growth.
Rga2 undergoes Cdc28-dependent hyperphosphorylation around bud emergence
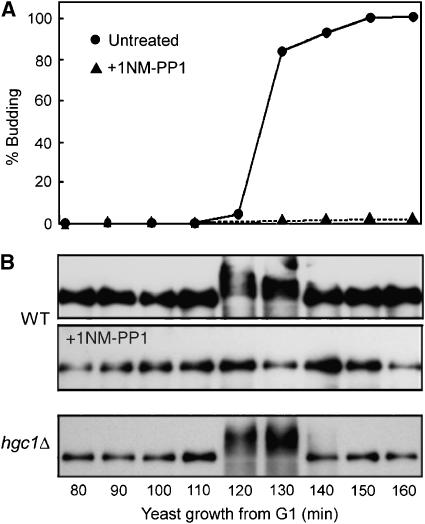
Based on the findings above of the roles of Cdc28/Hgc1 and Rga2 in hyphal growth, we speculated that Rga2 might also undergo hyperphosphorylation around bud emergence, which we may have failed to detect in earlier experiments due to the use of asynchronous cultures. To test this, we released synchronous G1 cells expressing Myc-Rga2 and Cdc28as for yeast growth and collected samples every 10 min for Western blotting. Indeed, we detected Rga2 hyperphosphorylation around the time of bud emergence, which was abolished by adding 1NM-PP1 to the medium (Figure 6). In addition, this event occurred normally in hgc1Δ cells, indicating that other CDK/cyclins are responsible for it during yeast growth. Together, our data suggest that Rga2 phosphorylation by CDK/cyclins might be part of a conserved mechanism that controls polarized growth in fungi. Consistent with this hypothesis, ScRga2 has recently been shown to be phosphorylated by G1 cyclin/Cdk1 complexes (McCusker et al, 2007).
Figure 6.
Rga2 underwent transient Cdc28-dependent hyperphosphorylation around bud emergence during yeast growth. Elutriated G1 cells of ZR13 (WT) and ZR9 (hgc1Δ) were released into yeast growth conditions in the presence or absence of 1NM-PP1. Aliquots were harvested at the indicated times for generating budding indices (A) and for αMyc Western blotting (B).
Discussion
CDKs drive an orderly progression of several key cellular processes through the cell cycle, such as DNA replication, chromosome segregation, cytokinesis and cell morphogenesis. However, the molecular pathways linking CDKs to the various effector machineries remain unclear in most cases (Nurse, 2000; Murray, 2004). In 1993, Lew and Reed proposed that G1 CDKs promotes bud emergence and apical growth. Despite extensive studies of the last 15 years, it remains unknown how CDKs regulate the central components of the polarity machinery, such as Rho GTPases, polarisomes and cytoskeleton. In this study, we have discovered that Cdc28/Hgc1, the key controller of hyphal morphogenesis in C. albicans, regulates Rga2, a GAP of the polarity GTPase Cdc42. This discovery establishes a key link in the signaling pathways that control C. albicans hyphal development. We also obtained evidence that other Cdc28/cyclin complexes may control bud formation through Rga2 as well.
Rga2 is a repressor of hyphal growth
Our genetic data reveal that while deleting RGA2 had little effect on hyphal morphogenesis, deleting it in the hgc1Δ mutant can nearly fully restore hyphal growth. The results are consistent with a model that Rga2 is a repressor of hyphal development, and a key role of Cdc28/Hgc1 is to inactivate Rga2 upon hyphal induction. Cdc28/Hgc1 may regulate Rga2 functions by several possible mechanisms, such as inhibiting the GAP activity, preventing its interaction with certain partners, sequestering Rga2 by direct binding, and changing its cellular localization. We observed that Rga2 strongly localized to polarized growth sites in hgc1Δ cells, whereas it was absent or weakly present at germ tube and hyphal tips in WT cells. Together with the persistent localization of Cdc42 and Cdc24 at hyphal tips (Hazan and Liu, 2002; Bassilana et al, 2005), our observation favors the following mechanism: Cdc28/Hgc1 might promote hyphal growth by keeping Rga2, a negative regulator of Cdc42, away from hyphal tips. Court and Sudbery (2007) reported persistent localization at hyphal tips of Bem3, another Cdc42 GAP, that appears to contradict our hypothesis. However, we found that deleting BEM3 from the hgc1Δ mutant did not improve hyphal growth, suggesting that the two GAPs may be involved in two different Cdc42 modules that regulate distinct cellular processes. Recently, Strickfaden et al (2007) reported the regulation of Ste5 localization by CDK in S. cerevisiae. Ste5, a scaffold protein in a MAP kinase cascade, is phosphorylated by G1 CDK at sites flanking a short polybasic membrane-binding motif, thereby abolishing Ste5 membrane localization. Similar polybasic amino-acid motifs are also present in Rga2 homologs in S. cerevisiae, Ashbya gossypii and Candida glabrata, raising the possibility that phosphorylation may regulate Rga2 localization by a similar mechanism. We are now investigating the role of this motif in determining Rga2 localization. Although our data support the idea that Cdc28/Hgc1 may function to prevent Rga2 from localizing to hyphal tips, we cannot exclude the other possible mechanisms described above. To determine whether Rga2 hyperphosphorylation may affect its interaction with Cdc42, we used GST-Cdc42G12V to pull down Rga2 from yeast and hyphal lysates. We found that Rga2 hyperphosphorylation does not significantly change its affinity for Cdc42 (Supplementary Figure S7).
We found that Rga2 also undergoes Cdc28-dependent hyperphosphorylation during bud emergence. However, unlike its absence at hyphal tips, Rga2 localized to the incipient bud site and the tips of small buds, suggesting that the mechanisms for Rga2 localizations may be different between yeast and hyphae. The differential effects might be due to some intrinsic differences between these two processes. During bud growth, a septin ring forms simultaneously with actin polarization, whereas during hyphal growth, septin ring formation is delayed and separated temporally from actin polarization (Sudbery, 2001; Warenda and Konopka, 2002). As the function of Rga2 is important for septin ring assembly (Court and Sudbery, 2007), it is possible that the process of septin ring construction may provide another localization platform, which is not affected by G1 CDK phosphorylation. It remains to be determined why the rga2-16E allele in hgc1Δ cells has little effect on the morphogenesis of yeast cells. In S. cerevisiae, four GAPs including Rga1, Rga2, Bem2 and Bem3 are involved in the regulation of bud morphogenesis and exhibit considerable functional redundancy (Smith et al, 2002; Park and Bi, 2007); C. albicans Bem3 and Rga2 also exhibit partial functional redundancy in bud morphogenesis during yeast growth (Court and Sudbery, 2007). This may partially account for the lack of discernible effect of the rga2-16E allele in hgc1Δ cells during yeast growth.
GAP in the regulation of Cdc42 in polarity control
Cdc42 is the master regulator of polarity control in both S. cerevisiae and C. albicans. It acts as a molecular switch toggling between an active GTP-bound form and an inactive GDP-bound form. GAPs promote GTP hydrolysis and thus inactivate Cdc42, while the GEF Cdc24 functions to regenerate active Cdc42. GTP-bound Cdc42 is thought to directly interact with several effectors, such as the PAK family kinases Ste20 and Cla4, the formins Bni1 and Bnr1, and the exocyst subunit Sec3 (Park and Bi, 2007). Through these effectors, Cdc42 directly or indirectly controls actin assembly, septin ring formation and exocytosis, all of which are crucial for polarity establishment and maintenance. Despite the understanding of the general biochemical roles of GEFs and GAPs in regulating GTPase activity, how these Cdc42 regulators differentially control Cdc42 activities in different cellular processes is poorly understood. GEFs have long been thought to play a major role in regulating GTPase activities in response to signal inputs, while GAP's role is secondary. However, recent findings indicate that GAP activity can be regulated by a number of mechanisms as well, such as protein–protein interactions, phospholipid interactions, phosphorylation, subcellular localization and proteolytic degradation (Bernards and Settleman, 2004). Our finding that the deletion of RGA2, but not BEM3, can rescue the hyphal growth defects of the hgc1Δ mutant, provides a convincing example that the role of Cdc42 in different cellular processes may be dictated by which GAP it interacts with. The functional specificity of individual GAP may also be determined by each one's unique binding partners including proteins and lipids. Identification of these partners is crucial for understanding the specific functions of each GAP in polarity control.
Is Rga2 a direct substrate of Cdc28/Hgc1?
Several lines of evidence we have obtained in this study support the view that Cdc28/Hgc1 regulates Rga2 function by direct phosphorylation. First, Rga2 undergoes Cdc28/Hgc1-dependent phosphorylation. Second, immunoprecipitated Cdc28as/Hgc1 complexes could phosphorylate Rga2, which was inhibited by 1NM-PP1. Third, when the mutant expressing Cdc28as as the sole source of Cdc28 was induced in media containing 1NM-PP1, both Rga2 hyperphosphorylation and hyphal growth were impaired. With the development of advanced MS, theoretically, all posttranslational modifications of any protein become accessible. Unfortunately, identification of a bona fide CDK substrate remains difficult. Previous studies have shown that CDKs can phosphorylate proteins at sites lacking the proline +1 of consensus CDK target sites (Garrett et al, 2001; Murray, 2004; Harvey et al, 2005), and mutating all or most of the consensus CDK phosphorylation sites on a single substrate, such as Swe1 (Harvey et al, 2005) and Slk19 (Ubersax et al, 2003) and Cdc24 (Gulli et al, 2000) had no significant consequences. In Swe1, over half of the CDK phosphorylation sites are non-consensus sites (Harvey et al, 2005). Thus, it poses a great challenge to validate phosphorylation sites in relation to the consequence of phosphorylation of a substrate. Using MS, we identified 35 phosphorylation sites on Rga2, among which 16 match either the perfect or the minimal consensus CDK target motifs. However, mutating the target serine/threonine residues to alanine in all the 16 sites had little effect on morphogenesis, except that the yeast cells exhibited a moderate increase in size. A possible explanation of the weak phenotype is that the other 19 non-consensus sites may also be phosphorylated by Cdc28/Hgc1 and important for polarized morphogenesis. Therefore, the physiological consequence of Rga2 phosphorylation may not be fully revealed by mutating only the consensus CDK phosphorylation sites. Nevertheless, the significant restoration of hyphal growth in hgc1Δ mutants by expressing the phosphomimic rga2-16E allele is consistent with a role for Rga2 phosphorylation in regulating cell morphogenesis.
Being a large protein with multiple functional domains, Rga2 is likely to have multiple interaction partners important for cell morphogenesis. Is it possible that Cdc28/Hgc1 also control the phosphorylation of some of Rga2 interacting partners for hyphal morphogenesis? In support of this, studies in S. cerevisiae have found at least six proteins that associate with Rga2 (Aguilar et al, 2006; Krogan et al, 2006). McCusker et al (2007) identified Rga2, Boi1 and several other proteins in a protein complex containing Cdc24 in S. cerevisiae; they demonstrated that Boi1 is phosphorylated by Cdc28/G1 cyclin complexes, which is thought to be important in regulating cell-surface growth. It would be interesting to identify Rga2 interacting partners in C. albicans in order to gain a better understanding of how Cdc42 is regulated during hyphal morphogenesis.
Materials and methods
Strains and culture conditions
Table I lists the strains used in this study. C. albicans was routinely grown at 30°C in YPD medium (2% yeast extract, 1% bactopeptone and 2% glucose) or in GMM (2% glucose and 6.79 g/l yeast nitrogen base without amino acids). For hyphal induction, 20% serum was added to liquid YPD or GMM medium, and cells were incubated at 37°C. To prepare G1 cells by centrifugal elutriation, cells were grown overnight at 30°C in a medium containing 2% galactose and 6.79 g/l yeast nitrogen base supplemented with amino acids. G1 cells were obtained using a Beckman (Fullerton, CA) J6-MC elutriator.
Table 1.
Strains used in this study
| Straina | Genotype | Source |
|---|---|---|
| CAI4 | Ura3∷imm434/ura3∷imm434 | Fonzi and Irwin (1993) |
| BWP17 | Ura3∷imm434/ura3∷imm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG | Wilson et al (1999) |
| WYZ12 | Same as BWP17 except hgc1Δ∷ARG4/hgc1Δ∷HIS1 | Zheng et al (2004) |
| WYZ13.1 | Same as BWP17 except hgc1Δ∷ARG4/hgc1Δ∷hisG | Zheng et al (2004) |
| ZR1 | RGA2/rga2Δ∷ARG4 | This study |
| ZR2 | Rga2Δ∷ARG4/rga2Δ∷HIS1 | This study |
| ZR2.1 | Rga2Δ∷ARG4/rga2Δ∷HIS1, RGA2-URA3 | This study |
| ZR3 | BEM3/bem3Δ∷ARG4 | This study |
| ZR4 | Bem3Δ∷ARG4/bem3Δ∷HIS1 | This study |
| ZR5 | Rga2Δ∷ARG4/PMET3-6Myc-RGA2-URA3 | This study |
| ZR6 | Bem3Δ∷ARG4/PMET3-6Myc-BEM3-URA3 | This study |
| ZR7 | Rga2Δ∷ARG4/GFP-RGA2-URA3 | This study |
| ZR8 | Hgc1Δ∷ARG4/hgc1Δ∷HisG rga2Δ∷HIS1/GFP-RGA2-URA3 | This study |
| ZR9 | Hgc1Δ∷ARG4/hgc1Δ∷HIS1 RGA2/PMET3-6Myc-RGA2-URA3 | This study |
| ZR10 | Hgc1Δ∷ARG4/hgc1Δ∷HIS1 BEM3/PMET3-6Myc-BEM3-URA3 | This study |
| ZR11 | Same as CAI4, except HGC1/6HA-HGC1-URA3 | This study |
| ZR12 | Cdc28Δ∷ARG4/PMET3-cdc28as-URA3 HGC1/6HA-HGC1-HIS1 | This study |
| ZR13 | Cdc28Δ∷ARG4/PMET3-cdc28as-URA3 RGA2/6Myc-RGA2-HIS1 | This study |
| ZR14 | Rga2Δ∷ARG4/PMET3-GFP-RGA2-URA3 | This study |
| ZR15 | Rga2Δ∷ARG4/PMET3-6Myc-RGA2-URA3 HGC1/6HA-HGC1-HIS1 | This study |
| ZR16 | Hgc1Δ∷ARG4/hgc1Δ∷hisG rga2Δ∷HIS1/rga2Δ∷URA3 | This study |
| ZR17 | Hgc1Δ∷ARG4/hgc1Δ∷hisG rga2Δ∷HIS1/rga2Δ∷hisG, RGA2-URA3 | This study |
| ZR18 | Hgc1Δ∷ARG4/hgc1Δ∷hisG bem3Δ∷HIS1/bem3Δ∷URA3 | This study |
| ZR19 | Rga2Δ∷HIS1/rga2Δ∷ARG4 own-promoter-6Myc-rga216A-URA3 | This study |
| ZR20 | Rga2Δ∷HIS1/rga2Δ∷ARG4 PMET3-6Myc-rga2R1015A-URA3 | This study |
| ZR21 | Rga2Δ∷ARG4/rga2R1015A-URA3 | This study |
| ZR22 | Hgc1Δ∷ARG4/hgc1Δ∷hisG rga2Δ∷HIS1/rga2R1015A-URA3 | This study |
| ZR23 |
Hgc1Δ∷ARG4/hgc1Δ∷hisG rga2Δ∷HIS1/ rga2Δ∷hisG, own-promoter-GFP-rga216E-URA3 |
This study |
aThe strains constructed in this study are all derivatives of BWP17, unless otherwise indicated.
Plasmid and strain constructions
All plasmids and their construction are described in Supplementary Table S1. rga2Δ and bem3Δ mutants were constructed by sequentially deleting the two copies of a gene from BWP17 (Wilson et al, 1999). Gene deletion cassettes were constructed by flanking a selectable marker gene (ARG4, HIS1 or URA3) with the AB and CD fragments, which correspond to the 5′- and 3′-untranslated regions (UTRs) of the target gene, respectively. Strain genotypes were verified by Southern blotting or PCR. hgc1Δ rga2Δ and hgc1Δ bem3Δ mutants were constructed from strain WYZ13.1 using HIS1 and URA3 cassettes to delete RGA2 or BEM3 gene, thereby yielding ZR16 and ZR18.
Fluorescence microscopy
A Leica DMR fluorescence microscope and a Hamamatsu digital camera interfaced with METAMORPH software (Molecular Devises Corporation, Downingtown, PA) were used for imaging. Cells were examined without prefixing.
Western blotting
After SDS–PAGE, proteins were transferred to the PVDF membrane (Millipore, Billerica, MA) before being probed with the anti-Myc antibody (9E10, Roche, Indianapolis, IN) or the anti-phosphoserine-CDK-site antibody (#2324, Cell Signaling technology Inc., Danvers, MA) at 1:500 and 1:1000 dilutions, respectively. For anti-Myc Western blotting, the membranes were incubated for 1 h at room temperature and then washed three times with phosphate-buffered saline (PBS) containing 0.1% Tween-20. This was followed by incubation with the horseradish peroxidase-conjugated anti-mouse antibody (Amersham, Buckinghamshire, UK) at 1:5000 dilutions for 1 h, followed by three more washes in PBS containing 0.1% Tween-20. Proteins were detected by using the chemiluminescence kit of Pierce Biotechnology (Rockford, IL). Similar conditions were used for Western blotting using the anti-phospho-CDK-site antibody, with the following modifications: membranes were incubated at 4°C overnight, all washes were performed in TBS (50 mM Tris–HCl, pH 7.6, 0.9% NaCl) containing 0.1% Tween-20, and the secondary antibody used was horseradish peroxidase-conjugated anti-rabbit antibody at 1:10 000 dilutions (Amersham). For anti-GFP Western blotting, the anti-GFP antibody from Clontech (California, USA) was used at 1:500 dilutions.
GST and MBP fusion protein purification
Expression in E. coli of GST and MBP fusion proteins and their purification were carried out by using commercial vectors and protocols (Amersham; and New England Biolabs, Ipswich, MA). Briefly, recombinant proteins were expressed by growing E. coli cells to mid-log phase and inducing them with 1 mM IPTG for 2 h at 37°C. To purify GST-Cdc42, the cells were lysed by sonication in lysis buffer containing 50 mM Tris–HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT and EDTA-free protease inhibitor mix (Roche). The lysate was cleared by centrifugation at 30 000 g for 15 min, and incubated with glutathione beads (Amersham) on a roller-mixer at 4°C for 1 h. The beads with bound GST-fusion proteins were washed three times with and stored as 50% slurry in the lysis buffer. To purify MBP-GAP fusion proteins, the cells were lysed by sonication in lysis buffer containing 20 mM Tris–HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA and EDTA-free protease inhibitor mix (Roche). The cell lysate was centrifuged at 30 000 g and the supernatant was incubated with MBP beads (New England Biolabs) at 4°C for 1 h. The beads were washed three times with the lysis buffer and then eluted with the same buffer supplemented with 10 mM maltose. For phosphorylation site determination of Rga2 (aa 344–596)-GST fusion, the fusion protein was purified using lysis buffer containing 50 mM Tris–HCl, pH 7.5, 150 mM NaCl and the complete protease inhibitor mix (Roche).
Immunoprecipitation
Myc-Rga2 was immunoaffinity-purified as follows. Briefly, lysates were prepared by bead beating in RIPA lysis buffer (50 mM Tris–HCl, pH 7.2, 1% Triton-X 100, 1% sodium deoxycholate, 0.1% SDS, EDTA-free protease inhibitor mix (Roche) and phosphatase inhibitor cocktail (2 mM NaF, 4 mM sodium orthovanadate, 0.2 mM Na4P2O7, 2.3 mM sodium molybdate and 0.2 mM β-glycerol phosphate) and incubated with anti-Myc antibody-coupled beads (Sc40, Santa Cruz Biotechnology) at 4°C for 1 h. The beads were washed three times with RIPA wash buffer (50 mM Tris–HCl, pH 7.2, 1% Triton-X 100, 1% sodium deoxycholate, 0.1% SDS and 150 mM NaCl). For the experiment shown in Figure 2A, Myc-Rga2 was pulled down from yeast and hyphal lysates. For the in vitro kinase assay (Figure 2C), Myc-Rga2 was pulled down from yeast lysates in the presence of 500 mM NaCl during incubation with anti-Myc antibody-coupled beads (Sc40, Santa Cruz Biotechnology) and during wash to remove associated proteins. For phosphorylation site mapping, hyphae were lysed in RIPA lysis buffer containing 150 mM NaF and 150 mM β-glycerol phosphate, and beads with bound Myc-Rga2 proteins were washed with the RIPA wash buffer.
Co-immunoprecipitation of Rga2 and Hgc1
For the experiment described in Figure 2E, co-immunoprecipitation of Rga2 and Hgc1 was performed using conditions described by Archambault et al (2004). HA-Hgc1 was precipitated by using anti-HA antibody-coupled beads (Sc-805, Santa Cruz) from the hyphal extract of strain ZR5 or ZR15. The presence of Rga2-Myc in the precipitates was determined by anti-Myc Western blotting. The extraction buffer contained 25 mM Na-HEPES (pH 7.4), 150 mM KCl, 10% glycerol, 0.1% Tween-20, 1 mM DTT and EDTA-free protease inhibitor mix (Roche).
Co-purification of Cdc42 with Rga2 and Bem3
GST-Cdc42G12V and GST alone were expressed in E. coli and purified as described above. C. albicans cells expressing Myc-Bem3 (ZR6) or Myc-Rga2 (ZR5) were grown in GMM at 30°C to mid-log phase. Cell lysates were prepared by bead beating in NP40 buffer containing 25 mM HEPES (pH 7.5), 100 mM NaCl, 50 mM NaF, 1% IGEPAL, 10 mM Na2H2P2O7, 5 mM MgCl2 and 1 mM DTT. After centrifugation in a microfuge at 14 000 r.p.m. for 10 min at 4°C, the supernatant was incubated with the purified GST-Cdc42G12V for 2 h at 4°C with continuous gentle mixing. Then the beads were washed three times with the lysis buffer. The bound proteins were eluted with glutathione, resolved by SDS–PAGE and detected by Western blotting using the anti-Myc antibody (9E10, Roche).
MS mapping of phosphorylation sites on Rga2
To map phosphorylation sites on Rga2, 2.5 l of log-phase ZR5 yeast cells were induced for hyphal growth in GMM+20% serum at 37°C for 1 h. Hyphae were harvested by centrifugation and washed once with a solution containing 0.9% NaCl, 10 mM EDTA, 1 mM sodium azide and 50 mM NaF, followed by lysis of the cells by bead beating. Proteins were eluted from the beads by boiling in 1% SDS and concentrated by freeze-drying. The protein sample was resolved by SDS–PAGE and stained with Coomassie Brilliant Blue. The portion of the gel containing Myc-Rga2 was cut out. For MS, the gel pieces were washed, reduced, alkylated, followed by in-gel tryptic digestion for overnight. The peptide mixture was extracted, dried in speed-vac and dissolved in 20 μl of 5% formic acid+10% acetonitrile. The phosphopeptides were enriched using TiO2 TopTip (Glygen Inc., Columbia, NY). The bound peptides were eluted with 0.5% NH3OH. Both flow-through and eluant were analyzed by Micromass Q-TOF2 LC-MS/MS. MASCOT 2.1 from Matrix Science (London, UK) was used to search all of the tandem mass spectra against the target protein, with an MS and MS/MS mass tolerance of 1.2 and 0.6 Da, respectively. PKL files were created using the software Masslynx 3.5 from Waters, which has a processing macro that smoothes, centroids and assesses the quality of data. The parameters used for the searches were as follows: trypsin specificity restriction with two missing cleavage sites and variable modifications including oxidation (M), deamidation (NQ), alkylation (C) and phosphorylation (STY) with neutral losses of phosphoric acid. The identified phosphorylation sites on Rga2 are listed in Supplementary Table S2. An example of the MS result is shown in Supplementary Figure S1.
GAP activity assay
GAP activity was assayed as described by Smith et al (2002), with the following modifications: purified MBP-GAP proteins were added to the slurry of beads with bound GST-Cdc42 and incubated at room temperature. Aliquots of the beads were collected at intervals and washed with the wash buffer containing 20 mM Tris–HCl, pH 7.5, 1 mM DTT and 5 mM MgCl2. The amount of [γ-32P]-GTP bound to the beads was determined using a scintillation counter.
In vitro kinase assay
Immunoaffinity purification of HA-Cdc28/Hgc1 was carried out as described previously (Surana et al, 1993), with some modifications. For the kinase assay described in Figure 2C, Cdc28/HA-Hgc1 and Cdc28as/HA-Hgc1 complexes were pulled down by using anti-HA antibody-coupled beads (Santa Cruz) and then mixed with the Myc-Rga2 purified from yeast cells in kinase reaction buffer containing 5 μCi γ-P32-ATP. 1NM-PP1 (Calbiochem, San Diego, CA) was used at a final concentration of 25 μM and preincubated with the beads for 10 min before the addition of γ-P32-ATP. Phosphorylated proteins were resolved by SDS–PAGE and visualized by autoradiography. For mapping phosphorylation sites on Rga2344–596-GST fusion, ∼10 μg of the protein was used as substrate.
1NM-PP1 inhibition of Cdc28as
1NM-PP1 was dissolved in DMSO to prepare a 10 mM stock. For the experiment described in Figure 2D, yeast cells expressing the cdc28as allele were induced for hyphal growth in GMM+20% serum at 37°C for 1 h in the presence of 25 μM 1NM-PP1 or DMSO alone. Cell lysates were prepared from the induced cells as well as untreated yeast cells. For Western blotting of Rga2 phosphorylation, Myc-Rga2 was immunoprecipitated with anti-Myc antibody before SDS–PAGE and probed with the anti-Myc (9E10, Roche) and anti-phospho-CDK-site (#2324, Cell Signaling technology) antibodies.
Supplementary Material
Supplementary data
Acknowledgments
We thank members of YW laboratory for comments and suggestions. This work was supported by the Agency for Sciences, Technology and Research of Singapore.
References
- Aguilar RC, Longhi SA, Shaw JD, Yeh LY, Kim S, Schon A, Freire E, Hsu A, McCormick WK, Watson HA, Wendland B (2006) Epsin N-terminal homology domains perform an essential function regulating Cdc42 through binding Cdc42 GTPase-activating proteins. Proc Natl Acad Sci USA 103: 4116–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault V, Chang EJ, Drapkin BJ, Cross FR, Chait BT, Rout MP (2004) Targeted proteomic study of the cyclin–Cdk module. Mol Cell 14: 699–711 [DOI] [PubMed] [Google Scholar]
- Bachewich C, Whiteway M (2005) Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot Cell 4: 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M, Blyth J, Arkowitz RA (2003) Cdc24, the GDP–GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot Cell 2: 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M, Hopkins J, Arkowitz RA (2005) Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot Cell 4: 588–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Clemente-Blanco A, Finley KR, Correa-Bordes J, Berman J (2005) The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol Biol Cell 16: 3387–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J, Sudbery PE (2002) Candida albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet 3: 918–930 [DOI] [PubMed] [Google Scholar]
- Bernards A, Settleman J (2004) GAP control: regulating the regulators of small GTPases. Trends Cell Biol 14: 377–385 [DOI] [PubMed] [Google Scholar]
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407: 395–401 [DOI] [PubMed] [Google Scholar]
- Chapa y Lazo B, Bates S, Sudbery P (2005) The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot Cell 4: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court H, Sudbery P (2007) Regulation of Cdc42 GTPase activity in the formation of hyphae in Candida albicans. Mol Biol Cell 18: 265–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampin H, Finley K, Gerami-Nejad M, Court H, Gale C, Berman J, Sudbery P (2005) Candida albicans hyphae have a Spitzenkorper that is distinct from the polarisome found in yeast and pseudohyphae. J Cell Sci 118: 2935–2947 [DOI] [PubMed] [Google Scholar]
- Davis CR, Richman TJ, eliduka SB, Blaisdell JO, Collins CC, Johnson DI (1998) Analysis of the mechanisms of action of the Saccharomyces cerevisiae dominant lethal cdc42G12V and dominant negative cdc42D118A mutations. J Biol Chem 273: 849–858 [DOI] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY (1993) Isogenic strain construction and gene mapping in Candida albicans. Genetics 134: 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett S, Barton WA, Knights R, Jin P, Morgan DO, Fisher RP (2001) Reciprocal activation by cyclin-dependent kinases 2 and 7 is directed by substrate specificity determinants outside the T loop. Mol Cell Biol 21: 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR (1992) Unipolar cell divisions in the yeast S cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090 [DOI] [PubMed] [Google Scholar]
- Gow NA, Brown AJ, Odds FC (2002) Fungal morphogenesis and host invasion. Curr Opin Microbiol 5: 366–371 [DOI] [PubMed] [Google Scholar]
- Gulli MP, Jaquenoud M, Shimada Y, Niederhauser G, Wiget P, Peter M (2000) Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol Cell 6: 1155–1167 [DOI] [PubMed] [Google Scholar]
- Han BK, Bogomolnaya LM, Totten JM, Blank HM, Dangott LJ, Polymenis M (2005) Bem1p, a scaffold signaling protein, mediates cyclin-dependent control of vacuolar homeostasis in Saccharomyces cerevisiae. Genes Dev 19: 2606–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SL, Charlet A, Haas W, Gygi SP, Kellogg DR (2005) Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell 122: 407–420 [DOI] [PubMed] [Google Scholar]
- Hazan I, Liu H (2002) Hyphal tip-associated localization of Cdc42 is F-actin dependent in Candida albicans. Eukaryot Cell 1: 856–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Kumamoto CA, Vinces MD (2005) Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol 7: 1546–1554 [DOI] [PubMed] [Google Scholar]
- Leonard DA, Lin R, Cerione RA, Manor D (1998) Biochemical studies of the mechanism of action of the Cdc42-GTPase-activating protein. J Biol Chem 273: 16210–16215 [DOI] [PubMed] [Google Scholar]
- Lew DJ, Reed SI (1993) Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol 120: 1305–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI (1995) Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev 5: 17–23 [DOI] [PubMed] [Google Scholar]
- Li CR, Wang YM, De Zheng X, Liang HY, Tang JC, Wang Y (2005) The formin family protein CaBni1 has a role in cell polarity control during both yeast and hyphal growth in Candida albicans. J Cell Sci 118: 2637–2648 [DOI] [PubMed] [Google Scholar]
- Liu H (2001) Transcriptional control of dimorphism in Candida albicans. Curr Opin Microbiol 4: 728–735 [DOI] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR (1994) Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266: 1723–1926 [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR (1997) Nonfilamentous C albicans mutants are avirulent. Cell 90: 939–949 [DOI] [PubMed] [Google Scholar]
- Loeb JD, Sepulveda-Becerra M, Hazan I, Liu H (1999) G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol Cell Biol 19: 4019–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquitz AR, Harrison JC, Bose I, Zyla TR, McMillan JN, Lew DJ (2002) The Rho-GAP Bem2p plays a GAP-independent role in the morphogenesis checkpoint. EMBO J 21: 4012–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R, Walther A, Wendland J (2005) Ras1-induced hyphal development in Candida albicans requires the formin Bni1. Eukaryot Cell 4: 1712–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker D, Denison C, Anderson S, Egelhofer TA, Yates JR III, Gygi SP, Kellogg DR (2007) Cdk1 coordinates cell-surface growth with the cell cycle. Nat Cell Biol 9: 506–515 [DOI] [PubMed] [Google Scholar]
- Moffat J, Andrews B (2004) Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat Cell Biol 6: 59–66 [DOI] [PubMed] [Google Scholar]
- Murray AW (2004) Recycling the cell cycle: cyclins revisited. Cell 116: 221–234 [DOI] [PubMed] [Google Scholar]
- Nurse P (2000) A long twentieth century of the cell cycle and beyond. Cell 100: 71–78 [DOI] [PubMed] [Google Scholar]
- Oehlen LJ, Cross FR (1998) Potential regulation of Ste20 function by the Cln1–Cdc28 and Cln2–Cdc28 cyclin-dependent protein kinases. J Biol Chem 273: 25089–25097 [DOI] [PubMed] [Google Scholar]
- Park HO, Bi E (2007) Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev 71: 48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GR, Givan SA, Cullen P, Sprague GF Jr (2002) GTPase-activating proteins for Cdc42. Eukaryot Cell 1: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt VR, Sonneborn A, Leuker CE, Ernst JF (1997) Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J 16: 1982–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickfaden SC, Winters MJ, Ben-Ari G, Lamson RE, Tyers M, Pryciak PM (2007) A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell 128: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery PE (2001) The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol Microbiol 41: 19–31 [DOI] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K (1993) Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J 12: 1969–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Ushinsky SC, Harcus D, Ash J, Dignard D, Marcil A, Morchhauser J, Thomas DY, Whiteway M, Leberer E (2002) CDC42 is required for polarized growth in human pathogen Candida albicans. Eukaryot Cell 1: 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warenda AJ, Konopka JB (2002) Septin function in Candida albicans morphogenesis. Mol Biol Cell 13: 2732–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP (1999) Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181: 1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Leeuw T, Leberer E, Thomas DY, Whiteway M (1998) Cell cycle- and Cln2–Cdc28-dependent phosphorylation of the yeast Ste20 protein kinase. J Biol Chem 273: 28107–28115 [DOI] [PubMed] [Google Scholar]
- Zheng XD, Wang Y, Wang Y (2004) Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J 23: 1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XD, Wang YM, Wang Y (2003) CaSPA2 is important for polarity establishment and maintenance inCandida albicans. Mol Microbiol 49: 1391–1405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data