Human cyclophilin J is a member of the cyclophilin family and recent studies have indicated that it plays a role in the formation of liver cancers as a regulation factor or messenger. The preparation, crystallization and preliminary X-ray crystallographic studies of human cyclophilin J are reported.
Keywords: immunophilins, cyclophilin J
Abstract
Human cyclophilin J, a new member of the cyclophilin family, has been expressed and crystallized. Diffraction data have been collected to 2.0 Å resolution and preliminary crystallographic studies have been completed. The space group of the crystals is P3121, with unit-cell parameters a = b = 40.597, c = 170.732 Å, α = β = 90, γ = 120°.
1. Introduction
The cyclophilin (CyP) family is a group of intracellular receptors for the immunosuppressive drug cyclosporin A. These receptors have been identified in all cells of organisms examined to date and are highly conserved in sequence (Trandinh et al., 1992 ▶). It has been suggested that the CyP family plays a conserved role in signal transduction processes in diverse organisms (Chou & Gasser, 1997 ▶). Recently, they have been suggested to be messengers in intercellular communications (Bukrinsky, 2002 ▶), but their in vivo function remains a mystery. Some members of the CyP family are thought to participate in apoptosis (Montague et al., 1994 ▶, 1997 ▶; Capano et al., 2002 ▶; Cande et al., 2004 ▶). The CyP family can regulate cell growth; for example, a member of the CyP family, CyP A, has been reported to play an important role in neuronal growth and differentiation (Nahreini et al., 2001 ▶; Chiu et al., 2003 ▶). The structures of several CyP members from different sources have been determined. The structures of CyP in complex with proline-containing peptides have provided insight into the mechanism of peptidyl-prolyl cis–trans isomerization (Ke & Huai, 2004 ▶).
Human CyP J was first reported in 2001 (Zhou et al., 2001 ▶). A low-molecular-weight member of the CyP family, human CyP J has a single chain with a molecular weight of 18 kDa. Its sequence identity with CyP A, the structure of which is known, is only 46%. As a member of the CyP family, human CyP J also possesses intrinsic cis–trans peptidyl-prolyl isomerase activity and can bind cyclosporin A, although the binding is not as tight as that of CyP A. Research results have demonstrated the up-regulation of CyP J in liver cancer cells and a specific CyP J inhibitor at a suitable concentration can kill up to 90% of tumour cells in vitro (L. Yu, J. Chen & X. M. Zhao, unpublished work).
In this article, we report the expression, purification, crystallization and preliminary X-ray crystallographic studies of human CyP J. Determination of the high-resolution structure of human CyP J will help to understand how the CyP family exert their abundant functions.
2. Expression and purification
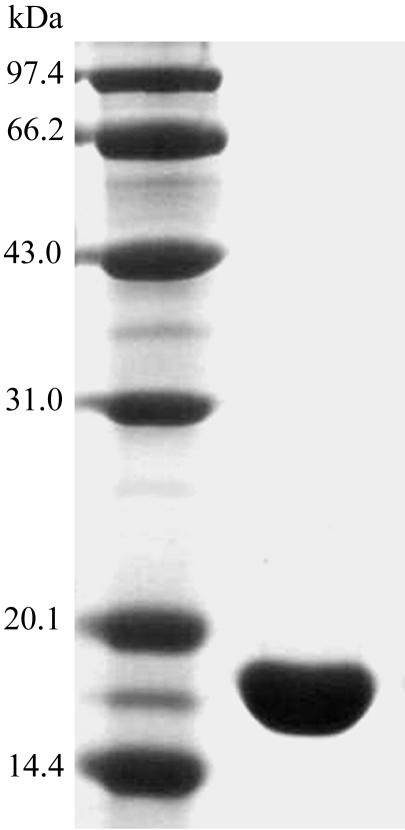
The human CyP J gene (GenBank accession No. NM_130906) was amplified from human cDNA. A PCR fragment encoding full-length human CyP J was inserted into the PGEXB1 vector (Amersham Bioscience) and then sequenced with an ABI 377 DNA Sequencer to ensure that there were no mistakes. The constructed plasmid was transformed into Escherichia coli BL21(DE3) cells and the bacteria were cultured for 4 h at 310 K in LB medium containing 100 µg ml−1 ampicillin. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.3 mM when A 550 reached 0.6; the culture was then allowed to grow at 301 K for 6 h before the cells were collected by centrifugation. The cell pellet was resuspended in lysis buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 pH 7.3) and lysed by sonication. The cell lysate was centrifuged at 12 000g for 30 min at 277 K. The recombinant protein in the supernatant was bound to a Glutathione Sepharose 4B (Amersham Bioscience) column, after which the column was washed with lysis buffer; 10 µl thrombin (Amersham Bioscience) was then added and the column was incubated at 298 K for 16 h before the recombinant protein was eluted with lysis buffer. The eluted recombinant protein was purified again by passage through another Glutathione Sepharose 4B column. The purified CyP J appeared to run as a single band on SDS–PAGE (Fig. 1 ▶). The yield of purified protein is approximately 4 mg per litre of culture. The buffer was then changed to 0.02 M bis-Tris pH 6.9 with a dialysis tube (Millipore) by centrifugation at 4000g; the purified protein was concentrated to the desired concentration in the same way.
Figure 1.
SDS–PAGE analysis of purified human CyP J.
3. Crystallization
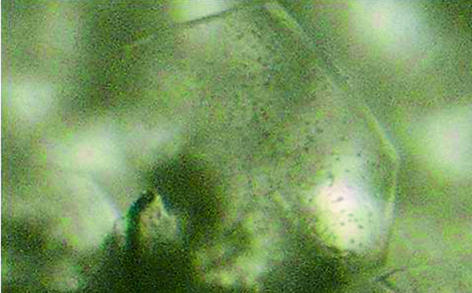
Crystallization was carried out by the hanging-drop vapour-diffusion method. Preliminary screenings were carried out at 277 and 291 K with Crystal Screen and Crystal Screen 2 kits (Hampton Research) and MD1-13 3D Structure Screen Kit (Molecular Dimensions). After the ‘first hit’ was obtained, intensive efforts were made to optimize the crystal quality by varying the pH, precipitant concentration, temperature and protein concentration. Better quality crystals were grown at 277 K according to the following method: a 5 µl drop of reservoir solution containing 0.1 M Tris pH 8.3, 0.01 M nickel(II) chloride hexahydrate, 17%(w/v) polyethylene glycol monomethyl ether 2000 was mixed with a 5 µl drop of protein solution at a concentration of 15 mg ml−1 dissolved in 0.02 M bis-Tris pH 6.9 and equilibrated against 500 µl reservoir solution. Flake-shaped crystals grew within 5 d to maximum dimensions of 270 × 530 × 45 µm. A typical crystal of human CyP J is shown in Fig. 2 ▶. To attempt to improve the crystal shape, various concentrations of additives such as MPD and Mg2+ were added during the optimization, but failed to produce any improvement.
Figure 2.
A crystal of human CyP J.
4. Data collection and processing
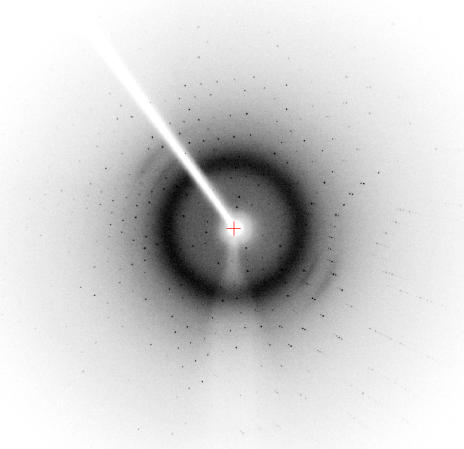
Diffraction data were collectedon an R-AXIS IV++ detector in 0.5° oscillation steps over a 123° range (Fig. 3 ▶) using a Rigaku FR-E SuperBright X-ray generator with a wavelength of 1.5418 Å. Data collection was performed at a temperature of 100 K and NVH oil (Hampton Research) was used as a cryoprotectant.
Figure 3.
Diffraction pattern of the human CyP J crystal.
Data were processed with the HKL2000 program package (Otwinowski & Minor, 1997 ▶). A summary of data collection and processing is given in Table 1 ▶.
Table 1. Data-collection statistics for the human CyP J crystal.
Values in parentheses are for the highest resolution shell.
| Space group | P3121 or P3221 |
| Unit-cell parameters (Å) | a = b = 40.597, c = 170.732 |
| No. molecules in asymmetric unit | 1 |
| Observed reflections | 48415 |
| Unique reflections | 20135 |
| Resolution (Å) | 30.0–2.0 (2.07–2.00) |
| Completeness (%) | 94.6 (91.6) |
| I/σ(I) | 20.56 (6.78) |
| Rmerge† (%) | 3.9 (13.1) |
R
merge = 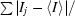
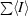 , where I
j is the intensity of reflection j and 〈I
j〉 is the average intensity of reflection j.
, where I
j is the intensity of reflection j and 〈I
j〉 is the average intensity of reflection j.
Molecular-replacement calculations were performed using the program MOLREP (Vagin & Teplyakov, 1997 ▶). The O chain of the CyP-like domain from Brugia malayi (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1c5f; Ellis et al., 2000 ▶) was used as search model because of its relatively high sequence identity (46%) with human CyP J, which gave better results than using the structure of CyP A as the search model. Using 27.0–3.0 Å resolution data, calculations in space groups P3121 and P3221 gave different results: the preliminary solution in P3121 has a better R factor and correlation coefficient than that in P3221. In the case of space group P3121, the highest peak has an R factor of 52.3 and a correlation coefficient of 40.6 with equivalent values of 57.6 and 29.1 for the second peak, but in the case of P3221 space group the R factor and correlation coefficient are 58.9 and 24.4, respectively, for the highest peak. The crystal packing in space group P3121 was also checked using O (Jones et al., 1991 ▶) and no unreasonable contacts were observed. This led us to conclude that the space group is P3121; the Matthews coefficient is 2.2 Å3 Da−1 and the solvent content is 43.2% (Matthews, 1968 ▶). The relatively high value of the R factor (52.3%) is likely to be a consequence of the structural differences between the search model and human CyP J, including the low sequence identity and lack of the last seven residues in the search model and possible conformational differences between the two proteins. This may increase the difficulties in solving the crystal structure, but the structure solution at 2.0 Å resolution may provide further new structural information contributing to understanding the complicated functions of this family of proteins.
Acknowledgments
This work was supported by the Knowledge Innovation Program of the Chinese Academy of Sciences.
References
- Bukrinsky, M. I. (2002). Trends Immunol.23, 323. [DOI] [PubMed] [Google Scholar]
- Cande, C., Vahsen, N., Kourant, I., Schmitt, E., Daugas, E., Spahr, C., Luban, J., Kroemer, R. T., Giordanetto, F., Garrido, C., Penninger, J. M. & Kroemer, G. (2004). Oncogene, 23, 1514–1521. [DOI] [PubMed] [Google Scholar]
- Capano, M., Virji, S. & Crompton, M. (2002). Biochem J.363, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu, R., Rey, O., Zheng, J. Q., Twiss, J. L., Song, J., Pang, S. & Yokoyama, K. K. (2003). Cell. Mol. Neurobiol.23, 929–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, I. T. & Gasser, C. S. (1997). Plant Mol. Biol.35, 873–892. [DOI] [PubMed] [Google Scholar]
- Ellis, P. J., Carlow, C. K., Ma, D. & Kuhn, P. (2000). Biochemistry, 39, 592–598. [DOI] [PubMed] [Google Scholar]
- Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Ke, H. & Huai, Q. (2004). Front. Biosci.9, 2285–2296. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Montague, J. W., Gaido, M. L., Fryen, C. & Cidlowski, J. A. (1994). J. Biol. Chem.269, 18877–18880. [PubMed] [Google Scholar]
- Montague, J. W., Hughes, F. M. & Cidlowski, J. A. (1997). J. Biol. Chem.272, 6677–6684. [DOI] [PubMed] [Google Scholar]
- Nahreini, P., Hovland, A. R., Kumar, B., Andreatta, C., Edwards-Prasad, J. & Prasad, K. N. (2001). Cell. Mol. Neurobiol.21, 65–79. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Trandinh, C. C., Pao, G. M. & Saier, M. H. (1992). FASEB J.6, 3410–3420. [DOI] [PubMed] [Google Scholar]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]
- Zhou, Z., Ying, K., Dai, J., Tang, R., Wang, W., Huang, Y., Zhao, W., Xie, Y. & Mao, Y. (2001). Cytogenet. Cell Genet.92, 231–236. [DOI] [PubMed] [Google Scholar]