PAB0955 has been expressed, purified and crystallized, and it has been shown that this thermostable protein is dimeric in reductive conditions.
Keywords: PAB0955
Abstract
PAB0955 from Pyrococcus abyssi is a prototype of a new Walker-type ATPase/GTPase conserved in archaea and eukaryota but not found in bacteria. PAB0955 has been expressed, purified and crystallized, and it has been shown that this thermostable protein is dimeric in reductive conditions. Crystals have been obtained either without nucleotide or in the presence of GDP or GTPγS. Preliminary X-ray crystallographic data up to 2.08 Å resolution have been collected from these crystals.
1. Introduction
The sequencing of entire genomes from organisms from the three domains of life has allowed the definition of clusters of orthologous groups of proteins (COGs) of unknown biological function. Among them, 32 COGs, named PACE (proteins of archaea conserved in eukaryotes), are common to archaea and eukaryota but are not found in bacteria (Matte-Tailliez et al., 2000 ▶). Most of these proteins have been annotated as involved in translation, transcription, replication and DNA repair. We have started a systematic structural and functional analysis of these proteins in order to establish their biological function (Armengaud et al., 2003 ▶, 2004 ▶). PAB0955, a protein encoded by the Pyrococcus abyssi genome, was our first target for crystallographic study. This 29 kDa protein (248 amino acids) has been annotated as an ATP/GTP-binding protein and a putative Walker-type ATPase/GTPase. Its function should be crucial as all eukaryotes and archaea have at least one homologue encoded in their genomes. The genetic environment of the PAB0955 gene in the P. abyssi genome suggests a role for its product in the interplay between cell division and DNA replication (Matte-Tailliez et al., 2000 ▶). Its closest known eukaryotic homologue is XPA-binding protein 1 found in mice and humans, with 28% sequence identity over 248 amino acids. The closest bacterial homologue has only 13% sequence identity. Moreover, GTPase activity (and the absence of significant ATPase activity) has been demonstrated in vitro with the purified recombinant protein (J. Armengaud, unpublished results). Since no remotely related proteins were detected in the Protein Data Bank, we decided to determine the crystal structure of PAB0955. This archaeal three-dimensional template should greatly help in unveiling the biological function of the whole eukaryotic and archaeal family. Here, we describe the expression, purification and crystallization of PAB0955. Preliminary crystallographic data collected on either crystals of uncomplexed native protein or crystals of protein grown in the presence of guanosine diphosphate (GDP) and guanosine diphosphate monothiophosphate (GTPγS) are presented.
2. Expression and purification
2.1. Plasmid construction
The PAB0955 coding sequence was amplified from P. abyssi genomic DNA with two synthetic oligonucleotide primers: oAB18 (5′-GCTAGCATGATAGTCGTGTTT-3′) and oAB19 (5′-TCAAGTCAGGTCACCGCACGT-3′). The resulting 753 bp PCR product was cloned into a pCRT7/NT-topo vector (Invitrogen). One of the recombinant plasmids, named pSBTN-AB34, was selected and its Xpress epitope removed by digestion with NheI endonuclease and religation with T4-DNA ligase. The resulting plasmid, named pSBTN-AB32, was sequenced on both strands in order to ascertain the integrity of the nucleotide sequence. This construction resulted in the addition of 14 extra residues at the N-terminus (MRGSHHHHHHGMAS) of the recombinant protein. Prior to cloning, various prediction methods based on the amino-acid sequence (Odorico & Pellequer, 2003 ▶) were used to check that this N-terminal tag would not affect the secondary-structure prediction.
2.2. Expression and purification of His6-tagged PAB0955
Overexpression of the PAB0955 construct was carried out essentially as previously described (Armengaud et al., 2003 ▶) using Escherichia coli Rosetta(DE3)pLysS strain (Novagen) transformed with pSBTN-AB32. This strain carries extra tRNA genes for arginine, isoleucine, proline and leucine corresponding to codons commonly found in many archaea including the Pyrococcus genus but rarely used by E. coli. Indeed, the presence of 33 rare codons and six tandems of such rare codons in the PAB0955 nucleotidic sequence could have prevented expression in a regular strain. Bacterial growth was performed at 303 K to an OD600 of 0.6. Expression was induced with 1 mM isopropyl-γ-d-thiogalactopyranoside (IPTG). The soluble cell extract was prepared as previously described (Armengaud et al., 2004 ▶), subjected to a 20 min heat treatment at 353 K and immediately centrifuged at 20 000g for 20 min at 277 K. Subsequent steps were performed at room temperature. NaCl was added to the resulting 300 ml clear supernatant to reach a final concentration of 250 mM. The sample was applied onto a Chelating Sepharose Fast Flow (50 ml, Amersham Biosciences) saturated with Ni2+. The major peak, which eluted from the column at about 200 mM imidazole, was concentrated and dialyzed overnight at 277 K against 50 mM KH2PO4/K2HPO4 buffer pH 7.2 containing 10 mM NaCl. The sample was then applied onto a Resource-Q ion-exchange column (6 ml, Amersham Biosciences). Recombinant PAB0955 was eluted at approximately 400 mM NaCl and desalted by dialysis in Spectra/Por1 tubing (Spectrum Laboratories, molecular-weight cutoff 6–8 kDa) against 20 mM KH2PO4/K2HPO4 buffer pH 7.2 containing 10 mM NaCl. A yield of approximately 6.6 mg of pure protein per litre of culture was obtained. Purified protein samples were flash-frozen in liquid nitrogen and stored at 193 K.
2.3. Gel-filtration analysis
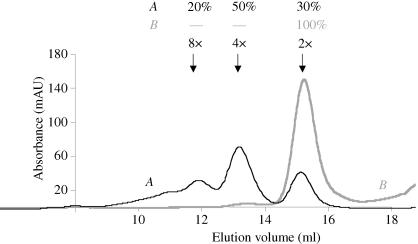
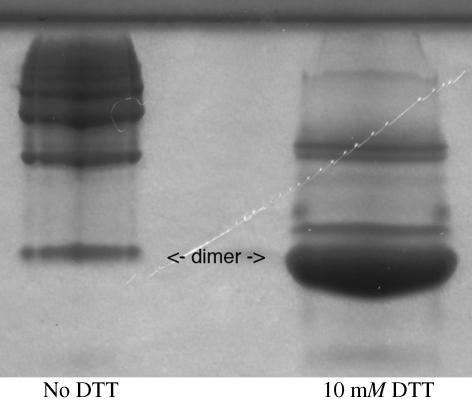
Purified PAB0955 protein was analyzed by size-exclusion chromatography. The elution profile obtained using a Superdex 200 calibrated column (curve A in Fig. 1 ▶) clearly indicated the presence of at least three species corresponding to different oligomerization states (30% dimer, 50% tetramer, 20% octamer). The presence of four cysteines in the polypeptide sequence prompted us to test whether this heterogeneity could result from the formation of disulfide bonds. Indeed, after incubation with 10–20 mM dithiotreitol (DTT), the protein fraction appeared to be mainly dimeric as analysed by Superdex200 gel filtration (curve B in Fig. 1 ▶) and native electrophoresis gel (Fig. 2 ▶). Accordingly, crystallization assays were carried out in the presence of DTT.
Figure 1.
Elution profiles of PAB0955 samples on a Superdex200 gel-filtration column equilibrated with 50 mM Tris–HCl buffer pH 8.0 containing 400 mM potassium glutamate. The absorbance at 280 nm was recorded using an Äkta Purifier 10 FPLC system (Amersham Biosciences). Curve A corresponds to the elution of 240 µg of PAB0955 without DTT. Curve B corresponds to the elution of 635 µg of PAB0955 after incubation with 20 mM DDT. The different quaternary states indicated on the figure (2×, 4× and 8×) correspond to dimers, tetramers and octamers, respectively. They were estimated from the elution volume of calibrators resolved under the same conditions.
Figure 2.
Native 12.5% acrylamide gel electrophoresis of PAB0955. The concentration of PAB0955 sample was 4 mg ml−1 in a buffer containing 20 mM NaH2PO4 pH 7.2 and 50 mM NaCl. The migration was performed at 200 V. The migration of the protein without DTT and after incubation in 10 mM DTT are shown in the left and right column, respectively. The dimeric state clearly prevails in the presence of DTT.
3. Crystallization
3.1. Crystals of native PAB0955
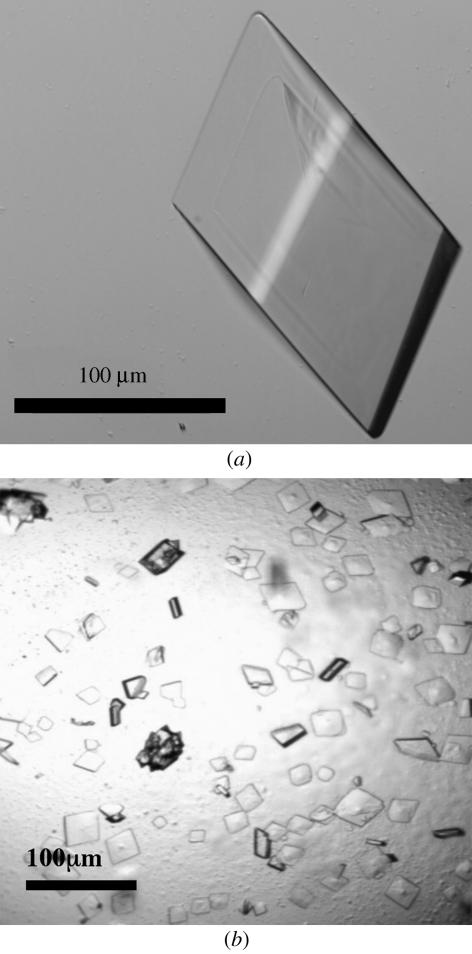
The first native protein crystals of about 20 µm in size were obtained by the sitting-drop vapour-diffusion method using a Tecan robot and Hampton Research Crystal Screens I and II, Ammonium Sulfate, Malonate, Formate and Quick Screen crystallization kits. The initial crystallization condition [7.8 mg ml−1 protein, 0.1 M trisodium citrate dihydrate pH 5.6, 20%(v/v) 2-propanol, 20%(w/v) PEG 4000 and 20 mM DTT] was optimized by substituting 2-propanol with different solvents provided by Additive Screen 1 (Hampton Research). Larger crystals (300 µm) were obtained in 15%(w/v) PEG 4000, 3%(v/v) dioxane, 0.1 M trisodium citrate dihydrate pH 5.6 and 20 mM DTT. Hanging drops were prepared by mixing 1 µl protein solution at a concentration of 7.8 mg ml−1 with 1 µl well solution at 291 K. Prismatic crystals (0.3 × 0.1 × 0.1 mm) were observed after 6 d (Fig. 3 ▶ a).
Figure 3.
(a) Native crystals of PAB0955 obtained in 15%(w/v) PEG 4000, 3%(v/v) dioxane, 0.1 M trisodium citrate dihydrate pH 5.6 and 20 mM DTT. (b) Crytals of PAB0955 in complex with GDP obtained in 13%(w/v) PEG 4000, 0.2 M ammonium acetate, 0.1 M trisodium citrate dihydrate pH 5.6 and 20 mM DTT.
3.2. Crystals of PAB0955 grown in the presence of nucleotides
A large-scale screening strategy was applied using a Cartesian automat and 15 crystallization screen kits (Crystal Screens I, II, Lite, PEG-Ion, MembFac, Natrix, Ammonium Sulfate, Screen Malonate, Formate, Quick Screen, PEG 6000, PEG LiCl, MPD, MME 5000 and Index from Hampton Research). Systematically, 2 µl of 1 M DTT solution was added to every 100 µl of well solution prior to setting up the crystallization drops. Sitting drops were then prepared using 100 nl protein solution (final concentration of 7.8 mg ml−1) mixed with GTPγS (final concentration of 0.13 mM) and 100 nl well solution. After 1 d, 97 of the 596 crystallization conditions tested provided crystals. Conditions that led to larger crystals were reproduced by the hanging-drop technique using 1 µl protein solution at 7.8 mg ml−1, 1 µl GTPγS at 0.65 mM and 1 µl well solution. The largest crystal (0.17 × 0.16 × 0.25 mm) grew from 13%(w/v) PEG 4000, 0.2 M ammonium acetate, 0.1 M trisodium citrate dihydrate pH 5.6 and 20 mM DTT after two weeks at 291 K. Attempts to crystallize PAB0955 in the presence of 0.65 mM GTPγS and under the same conditions as the native protein were unsuccessful. Crystals of PAB0955 grown in the presence of GDP were obtained under the same conditions after 2 d and had dimensions of 0.22 × 0.12 × 0.04 mm (Fig. 3 ▶ b).
3.3. Data collection
Prior to being flash-cooled in liquid nitrogen, crystals were soaked in a cryoprotectant solution containing mother liquor with the PEG concentration increased to 30%(w/v). All data were collected at 100 K. The data set for a native protein crystal was collected on beamline ID14-eh2 of the European Synchrotron Radiation Facility (ESRF) at a wavelength of 0.933 Å using an ADSC Q4 CCD detector. The data set of the PAB0955–GDP crystal was collected in-house with a Rigaku RU-200 rotating-anode X-ray generator at a wavelength of 1.542 Å (Cu Kα) using an R-AXIS IV++ imaging-plate detector. The PAB0955–GTPγS crystallographic data set was collected on beamline BM-30A of the ESRF at a wavelength of 0.979 Å using a MAR CCD detector. Data processing was performed using XDS (Kabsch, 1993 ▶).
4. Results
4.1. The tagged PAB0955 product is hyperthermostable
To design a PAB0955 overexpression system, the Met-ATG codon located on the Watson strand at 1392033 (numbering of the whole P. abyssi genome sequence; Cohen et al., 2003 ▶; http://www-archbac.u-psud.fr/genomes/newpab/newpab.html) was taken as the initiation codon. The upstream Met-TTG codon (at 1391946) that the genome annotators first mentioned in the database is rarely used as an initiator in archaea and the resulting N-terminal PAB0955 sequence aligns better with those of the other archaeal homologues. Rosetta (DE3)pLysS E. coli cells containing pSBTN-AB32 plasmid produced a large amount of the expected 30 kDa polypeptide after induction at 303 K. The PAB0955 protein remained soluble when the cell extract was heated at 358 K for 20 min, thus confirming the hyperthermostability of the tagged recombinant product.
4.2. Preliminary crystallographic data
Complete crystallographic data sets were collected from crystals of either native PAB0955 or PAB0955 co-crystallized with GTPγS or GDP. The crystals of native PAB0955 and PAB0955–GTPγS belong to the trigonal space group P3121 or P3221. The unit-cell parameters are very similar (Table 1 ▶) and according to the Matthews coefficient (V
M = 2.35 Å3 Da−1) there is one molecule per asymmetric unit. The structure factors of the native and GTPγS co-crystallized PAB0955 were compared with the program SCALEIT from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). The overall ΔF/F (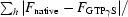
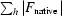 ) value of 0.32 indicates a significant difference in the unit-cell content and may arise from the binding of the nucleotide. The crystals of the PAB0955–GDP complex belong to the monoclinic space group P21. The Matthews coefficient indicates the presence of two molecules per asymmetric unit (V
M = 2.51 Å3 Da−1). Crystals belonging to the P21 space group were only obtained in the presence of GDP in the crystallization solution. Experimental phases will be needed in order to check the specific binding of the nucleotide. The statistics of the different data sets are detailed in Table 1 ▶.
) value of 0.32 indicates a significant difference in the unit-cell content and may arise from the binding of the nucleotide. The crystals of the PAB0955–GDP complex belong to the monoclinic space group P21. The Matthews coefficient indicates the presence of two molecules per asymmetric unit (V
M = 2.51 Å3 Da−1). Crystals belonging to the P21 space group were only obtained in the presence of GDP in the crystallization solution. Experimental phases will be needed in order to check the specific binding of the nucleotide. The statistics of the different data sets are detailed in Table 1 ▶.
Table 1. Crystallographic data statistics.
Values in parentheses are for the highest resolution shell.
| Native | GTPγS | GDP | |
|---|---|---|---|
| Space group | P3121 or P3221 | P3121 or P3221 | P21 |
| Unit-cell parameters (Å, °) | a = b = 60.3, c = 117.1 | a = b = 60.2, c = 115.9 | a = 59.1, b = 84.9, c = 60.2, β = 95 |
| Resolution (Å) | 50–2.15 (2.23–2.15) | 50–2.08 (2.15–2.08) | 50–2.30 (2.38–2.30) |
| No. observations | 140472 (10187) | 83727 (5701) | 88157 (8488) |
| No. unique reflections | 13998 (1345) | 12892 (1245) | 26182 (2560) |
| 〈I〉/〈σ(I)〉 | 8.2 (2.2) | 6.6 (2.3) | 5.2 (2.1) |
| Completeness (%) | 100 (100) | 87.6 (94.0) | 99.9 (100) |
| Rmerge (%) | 5.7 (34.8) | 6.7 (28.4) | 6.8 (36.1) |
5. Conclusions
The presence of a nucleotide appears to facilitate crystallization, since only one condition of the 192 present in the Hampton Research Crystal Screens I and II, Ammonium Sulfate, Malonate, Formate and Quick Screen crystallization kits led to crystals for the native protein. In the presence of GTPγS, crystals of PAB0955 were obtained from 12 conditions from the same screening kits. These preliminary data are promising and searches for heavy-atom derivatives are under way. If useful derivatives are not readily obtained, recombinant SeMet-substituted proteins will be used for phasing. The three-dimensional structure should confirm the GTP specificity of PAB0955 and allow us to identify the residues involved in GTP hydrolysis. It will be also very useful in identifying putative partners. Along with other biochemical and genetic approaches, it may thus help to unveil the biological role of PAB0955 and by extension that of its human homologue.
Acknowledgments
We thank J. Dupuy (beamline FIP-BM30A) and S. Monaco (beamline ID14-eh2) for help with synchrotron data collection at the ESRF (Grenoble). This work was supported in part by a grant from the Centre National de la Recherche Scientifique (Program Protéomique et Génie des Protéines, Appel à propositions 2001).
References
- Armengaud, J., Fernandez, B., Chaumont, V., Rollin-Genetet, F., Finet, S., Marchetti, C., Myllykallio, H., Vidaud, C., Pellequer, J. L., Gribaldo, S., Forterre, P. & Gans, P. (2003). J. Biol. Chem.278, 31078–31087. [DOI] [PubMed] [Google Scholar]
- Armengaud, J., Urbonavicius, J., Fernandez, B., Chaussinand, G., Bujnicki, J. M. & Grosjean, H. (2004). J. Biol. Chem.279, 37142–37152. [DOI] [PubMed] [Google Scholar]
- Cohen, G. N., Barbe, V., Flament, D., Galperin, M., Heilig, R., Lecompte, O., Poch, O., Prieur, D., Querellou, J., Ripp, R., Thierry, J. C., Van der Oost, J., Weissenbach, J., Zivanovic, Y. & Forterre, P. (2003). Mol. Microbiol.47, 1495–1512. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800. [Google Scholar]
- Matte-Tailliez, O., Zivanovic, Y. & Forterre, P. (2000). Trends Genet.16, 533–536. [DOI] [PubMed] [Google Scholar]
- Odorico, M. & Pellequer, J. L. (2003). J. Mol. Recognit.16, 20–22. [DOI] [PubMed] [Google Scholar]