Mimivirus, the largest known double-stranded DNA virus, unexpectedly exhibits components of the protein-translation apparatus. The crystallization and functional assay of the viral tyrosyl tRNA synthetase are reported.
Keywords: nucleocytoplasmic large DNA virus, tyrosyl tRNA synthetase, translation apparatus, structural genomics
Abstract
The amoeba-infecting Mimivirus is the largest known double-stranded DNA virus, with a 400 nm particle size, comparable to that of mycoplasma. The complete sequence of its 1.2 Mbp genome has recently been determined [Raoult et al. (2004 ▶), Science, 306, 1344–1350] and revealed numerous genes that were not expected to be found in a virus, such as genes encoding translation components, including 4-amino-acyl tRNA synthetases and homologues to various translation initiation, elongation and termination factors. A comprehensive structural and functional study of these Mimivirus gene products was initiated, as they may hold important clues about the origin of DNA viruses. Here, the first preliminary crystallographic and functional results obtained on one of these targets, Mimivirus TyrRS, are reported. Preliminary phasing was obtained using an original combination of homology modelling and normal mode analysis. Experimental evidence that Mimivirus tyrosyl tRNA synthetase recombinant gene product does indeed activate tyrosine is also presented.
1. Introduction
In its particle (400 nm) and genome size (1.2 Mbp), the newly discovered Mimivirus (La Scola et al., 2003 ▶) blurs the established frontier between viruses and parasitic cellular organisms such as mycoplasma, chlamydia, treponema or rickettsia. However, the presence versus absence of protein-synthesis genes remained a key property that distinguished minimal cellular organisms from large DNA viruses. In addition to six tRNA-like genes, the discovery of numerous genes encoding protein-translation components in the genome of Mimivirus now further challenges our vision of viruses and suggests that they may be the result of a more ancient and more gradual reductive evolution than had previously been envisioned (Raoult et al., 2004 ▶). In this context, a detailed study of these newly found viral proteins becomes of fundamental interest. Mimivirus exhibits homologues to ten proteins with functions central to protein translation: 4-aminoacyl-tRNA synthetases (aaRS), translation-initiation factor 4E (e.g. mRNA cap-binding), translation factor eF-TU (GTP-binding translocation factor), translation-initiation factor SUI1, translation-initiation factor IF-4A (helicase) and peptide-chain release factor eRF1. The Mimivirus genome also encodes the first identified viral homologue of a tRNA-modifying enzyme [tRNA (uracil-5-)-methyltransferase]. Despite exhibiting significant sequence similarity to their eukaryotic counterparts, these genes do not appear to have been (recently) acquired from a host and thus might be important in investigating the evolutionary origin of the Mimivirus lineage. We thus initiated a comprehensive structural/functional genomics study of all unique Mimivirus genes, prioritizing those potentially involved in protein translation. Here, we describe the expression, crystallization and preliminary functional characterization of the first of them, the 346-amino-acid tyrosyl tRNA synthetase (TyrRS) Mimivirus protein, one of four Mimivirus tRNA synthetases.
2. Results and discussion
2.1. Expression of the TyrRS gene product
The 346-residue TyrRS-encoding gene was PCR amplified from Mimivirus genomic DNA and cloned into a Gateway system (Invitrogen; Hartley et al., 2000 ▶). Oligonucleotide primers were designed with AttB1 and AttB2 overhangs added for recombination cloning. The corresponding PCR products were inserted by homologous recombination using the ‘one-tube reaction’ in the pDEST17 expression plasmid in frame with an N-terminal His6 tag under the control of a T7 promoter. The purified plasmids were used for the overexpression of the recombinant proteins using our standard soluble expression screening protocol (Abergel et al., 2003 ▶). The best result is obtained when transforming BL21(DE3) pLyS. After initial growth at 310 K, the temperature was set to 298 K in 2YT medium containing 0.05% glucose and 2% lactose for autoinduction. Cells were harvested at an A 600 of around 2–2.5, resuspended in buffer A (20 mM sodium phosphate pH 8.0, 300 mM NaCl) containing 1.5% Triton X-100, 1.5% glycerol and total proteins were extracted by sonication. Selenomethionine derivatives were obtained by culturing transformed BL21(DE3) pLyS in M9 minimal medium at 303 K supplemented with 100 mg l−1 lysine, phenylalanine and threonine and 50 mg l−1 leucine, isoleucine, valine and selenomethionine. Induction was performed with 0.5 mM IPTG when A 600nm reached 0.4–0.8. After 24 h incubation, total proteins were extracted by sonication.
2.2. Purification
The cleared lysate was applied onto a 5 ml HiTrap Chelating Column (Pharmacia) charged with Ni2+ and equilibrated with buffer A (50 mM sodium phosphate pH 8.0, 300 mM NaCl). The column was washed with ten column volumes of buffer A, ten column volumes of buffer A containing 25 mM imidazole and five column volumes of buffer A containing 50 mM imidazole at a flow rate of 1 ml min−1. Elution was performed with a linear gradient over eight column volumes from 50 to 500 mM imidazole. The eluates were collected and the presence and purity of the recombinant TyrRS in the different fractions were assessed by SDS–PAGE. The recombinant TyrRS protein thus corresponds to native protein in which the N-terminus methionine has been replaced by an extended His tag inherent to the use of the Gateway system (21-residue tag: SYYHHHHHHLESTSLYKKAGL). The fractions corresponding to the elution of TyrRS with 150–200 mM imidazole were run on a desalting column (Fast Desalting Column HR 10/10, Pharmacia) and analysed by N-terminal sequencing. The fractions contained at least 98% pure protein in 20 mM Tris buffer pH 7.4. Isoelectric focusing revealed a band around 5.5. The same protocol was applied to the selenomethionine-substituted TyrRS. The produced protein desalted in 20 mM MOPS buffer pH 8.0 was analyzed by mass spectroscopy to confirm the selenomethionine incorporation. Isoelectric focusing of the selenomethionine protein revealed a band around 5.8.
2.3. Crystallization
The TyrRS protein was concentrated to 6.9 mg ml−1 in 20 mM Tris pH 7.4 using a centrifugal filter device (Ultrafree Biomax 30K, Millipore, Bedford MA, USA). Precipitation experiments were carried out on the TyrRS protein using various precipitating agents [i.e. (NH4)2SO4, PEG, NaCl, MPD] at various pH values (i.e. 5, 6, 7, 8, 9). The screening for crystallization conditions was performed on 3 × 96-well crystallization plates (Greiner) loaded by an eight-needle dispensing robot (Tecan WS 100/8 workstation modified for our needs) using one 1 µl sitting drop per condition. The Mimivirus TyrRS was initially tested at 293 K against 480 different conditions at a concentration determined by phase-diagram analysis (precipitation experiments). The tested crystallization conditions include in-house-designed (Audic et al., 1997 ▶) and commercially available solution sets (Crystal Screen from Hampton Research and Wizards Screens from Emerald BioStructures). After analyzing the results, the most promising conditions were obtained in PEG 4000 pH 5–7. Conditions were refined using the incomplete factorial design approach (Audic et al., 1997 ▶). Crystals were obtained at 293 K by hanging-drop vapour diffusion using 24-well culture plates. Each hanging drop was prepared by mixing 1 µl of 6.9 mg ml−1 TyrRS in the presence and absence of its various cofactors (Mg, K, ATP) and ligand (tyrosine) with 0.5 µl reservoir solution. The hanging drop on the cover glass was vapour-equilibrated against 500 µl reservoir solution in each well of the tissue-culture plate. The best crystals were obtained using 0.1 M sodium citrate pH 5.5, 6%(w/v) PEG 4000 when the TyrRS protein had been incubated in 20 mM Tris buffer pH 7.4, 1 mM MgCl2, 0.1 M KCl, 0.2 mM tyrosine.
2.4. Data collection and processing
One crystal was collected in a Hampton Research 0.2 × 0.2 mm loop, flash-frozen to 105 K in a cold nitrogen-gas stream and subjected to X-ray diffraction. This data set was collected on a MAR CCD camera at the ESRF synchrotron-radiation facility (ID14-EH4) at a wavelength of 0.9798 Å. Data collection was carried out with an oscillation angle of 0.75° and a crystal-to-detector distance of 200 mm. The total oscillation range collected was 180°. MOSFLM and SCALA from the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶) were used for the processing, scaling and data reduction of the native data set. The crystals belong to the monoclinic space group P21, with unit-cell parameters a = 49.2, b = 156.5, c = 55.2 Å, β = 94.2°. The crystal mosaicity estimate is 0.8. The packing density for two monomers of TyrRS (42.25 kDa) in the asymmetric unit of these crystals (volume = 425 018.6 Å3) is 2.51 Å3 Da−1, a reasonable value for globular proteins, indicating an approximate solvent content of 51% (Matthews, 1968 ▶). The crystal diffracted to 2.6 Å and 260 601 reflections were measured in the resolution range 2.56–37.8 Å. This reduced to a data set of 19 642 unique reflections with an R sym value of 12.9. This represents a completeness of 87.4% with a multiplicity of 3.3 and an average I/σ(I) of 3.9. For the highest resolution shell 1555 reflections were measured in the resolution range 2.64–2.6 Å, corresponding to 669 unique hkl, an R sym value of 23.5 and an average I/σ(I) of 3.2, a completeness of 72.1% and a multiplicity of 2.0.
Sequence comparison of mimivirus TyrRS reveals it to be most similar to eukaryotic TyrRS. Its closest homologues are the putative TyrRS from rice (Swiss-Prot Q84PX0, 43% identity over 342 residues) and Plasmodium yoelii (Q7RH02, 43% identity over 344 residues). However, no structures are available for these TyrRSs and we thus used the Methanococcus jannaschii TyrRS structure (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1j1u; Kobayashi et al., 2003 ▶; 29% identity over 274 residues) to perform standard molecular replacement using the AMoRe software (Navaza, 2001 ▶), which failed to produce a solution.
Meanwhile, a structural comparison of tyrosyl tRNA synthetase structures available in the PDB suggested a change in the conformation of the molecule upon binding of the cofactors (ATP, Mg, tyrosine) and of the tRNA ligand. Given this observation, we decided to perform a normal mode analysis (Suhre & Sanejouand, 2004 ▶; http://www.igs.cnrs-mrs.fr/elnemo/) on the available M. jannaschii TyrRS structure (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1j1u; Kobayashi et al., 2003 ▶) in order to describe this change in conformation. The various modes were tested using molecular replacement (AMoRe; Navaza, 2001 ▶) and the conformation producing the best solution was used through the CaspR server (http://www.igs.cnrs-mrs.fr/Caspr/index.cgi) to generate homology models of the mimivirus tRNA synthetase and screen for a molecular-replacement solution (Claude et al., 2004 ▶). This procedure produced a two-molecule solution which after preliminary refinement using CNS (one step of rigid-body refinement and minimization) led to R work = 41.8 and R free = 47.7. Refinement of this solution is currently in progress.
2.5. Activity measurements
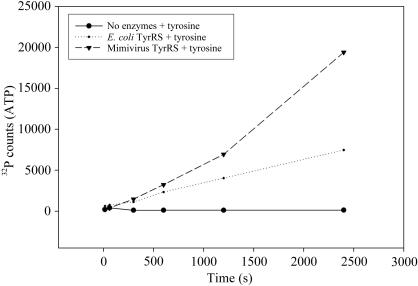
The tyrosine-dependent exchange of ATP with 32PPi was assayed as described by Retailleau et al. (2003 ▶). Assays were performed for a 320 µl reaction at 303 K in 100 mM HEPES pH 7.2, 10 mM MgCl2, 2 mM KF, 2 mM 32PPi (∼2000 counts min−1 nmol−1), 1 mM ATP, 2 mM tyrosine with purified recombinant tyrosyl-tRNA synthetases from mimivirus (142 nM) or a mixture of purified E. coli amino-acyl tRNA synthetases (Sigma, A3646; 148 nM). 50 µl aliquots were removed at intervals indicated in Fig. 1 ▶, quenched with 200 µl 15%(v/v) perchloric acid, 1%(w/v) activated charcoal and 0.4 M sodium pyrophosphate, filtered onto Whatman GF/C filters which were washed three times with 5 ml water and twice with 5 ml 95% ethanol and dried before scintillation counting of [32P]ATP in the presence of 8 ml scintillation cocktail (CytoScint, MP Biomedicals) in a scintillation counter (Beckman LS 1801). Results are shown in Fig. 1 ▶. Given that the exact proportions of the 20 E. coli tRNA synthetases within the commercial mixture are not available, a quantitative comparison of the mimivirus versus E. coli TyrRS specific activity is not possible. However, our results (Fig. 1 ▶) unambiguously establish that the purified mimivirus TyrRS protein exhibits the predicted enzymatic activity and is able to activate tyrosine at a rate comparable to that achieved by the E. coli enzyme.
Figure 1.
Enzymatic activity of mimivirus TyrRS versus E. coli TyrRS.
The multiple alignment of the mimivirus TyrRS sequence with archeal and eukaryotic TyrRS sequences (Fig. 2 ▶) suggest the enzyme is fully functional and specific for eukaryotic tRNA. Further experiments are currently in progress in order to quantify this activity, verify the amino-acid specificity and assess its ability to load cognate eukaryotic tRNAs.
Figure 2.
Multiple alignment of the mimivirus TyrRS sequence with structural homologues and related archeal and eukaryotic sequences. j1u and n3l correspond to the PDB structures http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1j1u (Kobayashi et al., 2003 ▶) and http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1n3l (Yang et al., 2002 ▶). The accession numbers of the homologous sequences are as follow: Ehisto, Entamoeba histolytica unfinished genome (gnl|TIGR_5759, ENTER63TR); Osati, Oryza sativa putative TyrRS (Q84PX0); Atha1, Arabidopsis thaliana putative TyrsRS (Q8S9J2); Atha2, Arabidopsis thaliana putative TyrsRS (P93018); Tobac, Nicotiana tabacum putative TyrRS (P93363); Pyoeli, Plasmodium yoelii putative TyrRS (Q7RH02); CParvum, Cryptosporidium parvum putative TyrRS(Q7YYA0). Black triangles correspond to residues known to be involved in interaction with the tyrosine, green triangles correspond to residues known to be involved in interaction with the acceptor and red stars correspond to residues known to be involved in interaction with the anticodon. The multiple alignment was generated using the T-COFFEE software (Poirot et al., 2004 ▶) in order to combine structural and sequence information and was used through the CaspR procedure to generate homology-based models of the mimivirus structure.
Acknowledgments
We thank Dr Joanne McCarthy for expert assistance on the ESRF ID14 beamlines, Professor Richard Giegé and Dr Joëlle Rudinger for helpful discussions on mimivirus TyrRS activity measurements and Dr Patricia Renesto for the PCR amplification of the mimivirus genes studied in our structural genomics project. We also thank the referees for helpful comments on the manuscript.
References
- Abergel, C., Coutard, B., Byrne, D., Chenivesse, S., Claude, J.-B., Deregnaucourt, C., Fricaux, T., Gianesini-Boutreux, C., Jeudy, S., Lebrun, R., Maza, C., Notredame, C., Poirot, O., Suhre, K., Varagnol, M. & Claverie, J.-M. (2003). J. Struct. Funct. Genomics, 4, 141–157. [DOI] [PubMed] [Google Scholar]
- Audic, S., Lopez, F., Claverie, J.-M., Poirot, O. & Abergel, C. (1997). Proteins, 29, 251–256. [DOI] [PubMed]
- Claude, J.-B., Suhre, K., Notredame, C., Claverie, J.-M. & Abergel, C. (2004). Nucleic Acids Res.32, W606–W609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Hartley, J. L., Temple, G. F. & Brasch, M. A. (2000). Genome Res.10, 1788–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., Nureki, O., Ishitani, R., Yaremchuk, A., Tukalo, M., Cusack, S., Sakamoto, K. & Yokoyama, S. (2003). Nature Struct. Biol.10, 425–432. [DOI] [PubMed] [Google Scholar]
- La Scola, B., Audic, S., Robert, C., Jungang, L., de Lamballerie, X., Drancourt, M., Birtles, R., Claverie, J.-M. & Raoult, D. (2003). Science, 299, 2033. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (2001) Acta Cryst. D57, 1367–1372. [DOI] [PubMed]
- Poirot, O., Suhre, K., Abergel, C., O’Toole, E. & Notredame, C. (2004). Nucleic Acids Res.32, W37–W40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult, D., Audic, S., Robert, C., Abergel, C., Renesto, P., Ogata, H., La Scola, B., Susan, M. & Claverie, J.-M. (2004). Science, 306, 1344–1350. [DOI] [PubMed] [Google Scholar]
- Retailleau, P., Huang, X., Yin, Y., Hu, M., Weinreb, V., Vachette, P., Vonrhein, C., Bricogne, G., Roversi, P., Ilyin, V. & Carter, C. W. Jr (2003). J. Mol. Biol.325, 39–63. [DOI] [PubMed] [Google Scholar]
- Suhre, K. & Sanejouand, Y. H. (2004). Acta Cryst. D60, 796–799. [DOI] [PubMed] [Google Scholar]
- Yang, X. L., Skene, R. J., McRee, D. E. & Schimmel, P. (2002). Proc. Natl Acad. Sci. USA, 99, 15369–15374. [DOI] [PMC free article] [PubMed] [Google Scholar]