The major SH2-binding site of p130Cas has been crystallized in complex with the SH3-SH2 regulatory domains of the Src-family kinase Lck.
Keywords: p130Cas, SH2-binding sites, Src-family kinases, Lck
Abstract
Cas-family proteins serve as docking proteins in integrin-mediated signal transduction. The founding member of this family, p130Cas, becomes tyrosine-phosphorylated in response to extracellular stimuli such as integrin-mediated cell adhesion and ligand engagement of receptor tyrosine kinases. Cas proteins are large multidomain molecules that transmit signals as intermediaries through interactions with signaling molecules such as FAK and other tyrosine kinases, as well as tyrosine phosphatases. After Cas is tyrosine-phosphorylated, it acts as a docking protein for binding SH2 domains of Src-family kinases. In order to examine the structural basis for a key step in propagation of signals by Cas, one of the major SH2-binding sites of Cas has been crystallized in complex with the SH3-SH2 regulatory domains of the Src-family kinase Lck. Crystallization conditions were identified by high-throughput screening and optimized with multiple rounds of seeding. The crystals formed at 295 K in space group P212121, with unit-cell parameters a = 77.4, b = 107.3, c = 166.4 Å, and diffract to 2.7 Å resolution.
1. Introduction
Stimuli from the extracellular matrix are transmitted across the plasma membrane by cell-surface receptors called integrins and result in different intracellular signals that regulate processes such as cell migration and proliferation, as well as growth and differentiation (Giancotti & Ruoslahti, 1999 ▶; O’Neill et al., 2000 ▶). Some of the major intracellular signals involve tyrosine phosphorylation. One group of proteins that serve as docking proteins in integrin-mediated signal transduction is the Cas family, which includes p130Cas (Crk-associated substrate; Sakai et al., 1994 ▶), Efs/Sin (embryonal Fyn-associated substrate/Src-interacting protein; Ishino et al., 1995 ▶; Alexandropoulos & Baltimore, 1996 ▶) and HEF1 (human enhancer of filamentation 1; Law et al., 1996 ▶; Minegishi et al., 1996 ▶). Cas, the founding member of this family, was originally identified as a major tyrosine-phosphorylated molecule in fibroblast cells transformed by v-Src and v-Crk oncoproteins (Kanner et al., 1991 ▶; Sakai et al., 1994 ▶). Cas becomes phosphorylated in response to extracellular stimuli, including integrin-mediated cell adhesion and ligand engagement of receptors such as G-protein-coupled receptors and receptor tyrosine kinases (Vuori & Ruoslahti, 1995 ▶; reviewed in O’Neill et al., 2000 ▶).
Cas-family proteins share several conserved domains that are key to their role as intermediaries in integrin-mediated signaling. The proteins contain an SH3-domain (Src-homology 3 domain) at their N-termini. This domain of Cas directly interacts with a proline-rich region in the FAK tyrosine kinase (Polte & Hanks, 1995 ▶) and also with tyrosine phosphatases such as PTP1B and PTP-PEST (Liu et al., 1996 ▶; Garton et al., 1997 ▶). Following interaction with FAK, Cas becomes tyrosine-phosphorylated by FAK or by Src-family tyrosine kinases that are bound to FAK. The main region in Cas that is the target for these tyrosine kinases is termed the substrate domain. This region bears many putative SH2 (Src-homology 2) binding sites for adaptor proteins such as Crk and Nck (Vuori et al., 1996 ▶; Schlaepfer et al., 1997 ▶). Another SH2-binding site is located in the C-terminal region of Cas (Nakamoto et al., 1996 ▶). Phosphorylation of the C-terminal SH2-binding site in Cas results in the formation of a high-affinity binding site for the SH2 domain of Src-family kinases (Kanner et al., 1991 ▶; Sakai et al., 1994 ▶). Upon binding to this high-affinity site, Src kinases further phosphorylate Cas in the substrate domain in a progressive manner. The three-dimensional configuration of these protein interaction domains of Cas is therefore critical for its functional role as a docking protein.
In order to provide an understanding at the molecular level of the first step in the propagation of signals by Cas, we initiated biochemical and parallel structural studies of one of the major SH2-binding sites in Cas bound in complex with the SH3-SH2 domain of Lck, a Src-family member that is known to bind to Cas proteins (Kanda et al., 1997 ▶; Ohashi et al., 1998 ▶). Here, we report the crystallization of this complex and the preliminary X-ray analysis of the crystals.
2. Experimental
2.1. Expression and purification
Lck tyrosine kinase was selected as the representative of the Src family of kinases for this study because the regulatory domains of this kinase had previously been expressed in bacterial cells for structural studies (Eck et al., 1994 ▶) rather than mammalian or insect cell systems. Moreover, the sequence homology in this protein family is high and crystal structures are available for numerous members of the family, so direct correlation with Src kinases is ensured. The DNA sequence encoding the SH3-SH2 domains (residues 64–226 plus GSHM as the linker between a thrombin-cleavage site and Lck residue 64) of human Lck was cloned into a pET-28a plasmid (NdeI and BamHI) to express the protein as a His6-fusion protein with the hexahistidine sequence at the N-terminus of the protein. The primers used were N-terminal, 5′-GGCCCATATGAACCTGGTTATCGCTCTGCACAGCTATG-3′, and C-terminal, 5′-TTATGAATTCGGTCTGGCAGGGGCGGCTCAACCGTGTG-3′. The plasmid was transformed into Escherichia coli BL21 DE3 competent cells (Stratagene). For expression, overnight cultures of a single colony per 100 ml of LB medium containing 50 µg ml−1 kanamycin were diluted in a 1:10 ratio with the same medium and grown at 310 K to a cell density of A 550 = 0.5. Protein expression was induced by the addition of 0.1 mM isopropyl-1-thio-d-galactopyranoside for 4 h at room temperature. Cells were collected by centrifugation at 6000 rev min−1 and stored overnight at 193 K.
For the purification of Lck, frozen cells were thawed for 30 min on ice, resuspended in 10 ml extraction solution No. 1 (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol, 0.005% NaN3, 0.04 mg ml−1 lysozyme, 1 mM phenylmethylsulfonyl fluoride) per gram of cells and disrupted using a Dounce tissue grinder. After 30 min, 1 ml of 10× extraction solution No. 2 [1.5 M NaCl, 100 mM CaCl2, 100 mM MgSO4, 20 µg ml−1 DNase, 50 µg ovomucoid (trypsin inhibitor)] per 10 ml cell suspension was added to the solution with thorough mixing. The lysate was cleared of cell debris by centrifugation at 17 000 rev min−1 and then was mixed with 10 ml fresh Ni2+–nitrilotriacetic acid (NTA) beads (Qiagen) which had been equilibrated with loading buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol) and the suspension was mixed with constant gentle rotation at 277 K for 30 min. The suspension was loaded onto a column support and the beads were washed at 277 K for 4 h with wash buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 20 mM imidazole, 5 mM β-mercaptoethanol). His6-Lck-SH3-SH2 was eluted using an imidazole gradient from 20 to 200 mM in wash buffer. After elution, the protein solution was brought to 1 mM in EDTA in order to chelate extraneous metal ions and was then dialyzed against 10 mM Tris pH 8.0, 5 mM β-mercaptoethanol to remove imidazole and EDTA.
The hexahistidine tag was removed by digestion with human thrombin (1 unit per milligram of protein; Sigma) for 1 h at 295 K. After removal of the fusion tag by adsorption with fresh Ni2+–NTA beads, Lck protein was dialyzed against 10 mM Tris pH 8.5, 20 mM NaCl, 5 mM β-mercaptoethanol. The protein was then applied to tandem columns containing benzamidine-agarose (Sigma) and Q-Sepharose (Pharmacia) ion-exchange resin to remove thrombin and other cryptic contaminating proteases. The ion-exchange chromatography was developed using a linear 20–500 mM NaCl gradient in 10 mM Tris pH 8.5, 5 mM β-mercaptoethanol. The purified protein was homogeneous as judged by SDS–PAGE, with a single band and electrophoretic mobility corresponding to the expected molecular weight of 19 kDa. Typical yields were 7 mg purified protein per litre of culture. This sample was finally dialyzed against 10 mM Tris pH 8.5, 20 mM NaCl, 5 mM β-mercaptoethanol and concentrated by volume reduction to 12 mg ml−1.
2.2. Peptide synthesis
A peptide representing residues 758MEDpYDYVHL767 from the carboxyl region of Cas, with a phosphotyrosine modification at residue 762, was synthesized by the University of Utah DNA/Peptide core facility (http://www.cores.utah.edu/peptide.htm). The peptide was acetylated at the N-terminus and amidated at the C-terminus. The final product was purified by HPLC and the peptide sequence confirmed by automated sequencing and mass spectrometry (not shown).
2.3. Formation of the Cas–Lck complex
The peptide was dissolved in 10 mM Tris buffer pH 8.5 at a concentration of 50 mM and mixed with Lck protein at a molar ratio of 5:1 peptide:protein to ensure stability of the complex. This ratio was optimal since subsequent crystallization trials with peptide:protein ratios of 3:1 and 7:1 produced only small poorly formed crystals. Peptide was added at 299 K to the concentrated protein solution (12 mg ml−1) directly prior to crystallization trials and mixed thoroughly.
2.4. Crystallization
Initial crystallization trials implementing the sitting-drop vapor-diffusion method were conducted using Hampton Research Crystal Screen Kits I and II (Aliso Viejo, CA, USA). Although crystals of native Lck were produced, priority was given to crystallization of the preformed Cas–Lck complex. In the initial screens, a systematic search using the full range of organic and inorganic precipitants was made with 2 µl drops (1:1 protein:reservoir solution) and the Crystal Clear Strips apparatus (Hampton Research, Inc.). Crystallization trials were performed at 295 K. No crystals were observed from these screens.
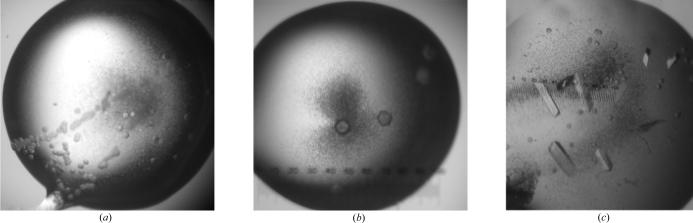
Samples of the Cas–Lck complex were also tested in trials at the high-throughput crystallization facility at the Hauptman–Woodward Institute (Luft et al., 2003 ▶). High-throughput experiments screened 1536 distinct mixtures in microassay plates in batch mode under paraffin oil. The ‘cocktails’ tested were designed to screen a broad range of conditions to induce supersaturation. Small well formed crystals were observed in one trial in solutions of 2.3 M K2 HPO4 in 0.1 M TAPS pH 9.0. These conditions were optimized in the home laboratory at 295 K using hanging drops (5 µl drops and 1:1 protein:reservoir solution) as well as batch (4 µl) format. While vapor diffusion produced only twinned or irregular crystals, the experiments in batch mode produced small single crystals (see Fig. 1 ▶).
Figure 1.
Crystals of the Cas–Lck complex. (a) shows twinned crystals that were crushed and used for seed stocks for subsequent streak-seeding. A fine hair was touched to the seed stock and transferred to freshly prepared drops containing Cas–Lck complex and crystallization buffer as described in the text. After the first seeding experiment, the crystals grew larger but were still twinned (b). Finally, single prismatic crystals were produced following repeated rounds of streak-seeding (c). These crystals diffracted to high resolution and were suitable for data collection.
The size and morphology of these crystals was improved by a combination of microseeding and streak-seeding (Stura & Wilson, 1990 ▶, 1991 ▶). During the seeding experiments, precipitant concentration, pH, protein:peptide ratio and equilibration time before seeding were varied. Seed stocks were produced by crushing a single crystal and were used for streak-seeding into drops with no equilibration prior to the introduction of seeds. Seeds were transferred to fresh solutions containing the same protein concentration as the mother liquor in the seed stock, but with different concentrations of K2HPO4 (1.1, 1.15, 1.2 and 1.25 M). Solutions containing 1.15 M K2HPO4 in 0.1 M TAPS pH 9.0 provided the best conditions for seeding. Single crystals were produced and the size was progressively increased through multiple (e.g. up to four repetitions) consecutive seeding rounds (see Fig. 1 ▶). Crystals suitable for X-ray diffraction (approximately 0.7 mm in length) formed in 5 d. The space group was identified as P212121 and confirmed by examination of systematic absences; the measured unit-cell parameters were a = 77.4, b = 107.3, c = 166.4 Å (see Table 1 ▶). The solvent content of these crystals was 54.6%. Subsequent refinement and structure solution with data collected from these crystals indicated that there are six complexes per asymmetric unit (to be published elsewhere).
Table 1. Data-collection statistics.
Values in parentheses are for the outer resolution shell (2.85–2.70 Å).
| Source | ESRF ID14-1 |
| Temperature (K) | 108 |
| Wavelength (Å) | 0.934 |
| Resolution range (Å) | 30–2.7 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 77.4, b = 107.3, c = 166.4 |
| Total No. of reflections | 128725 |
| No. of unique reflections | 34665 |
| Completeness (%) | 96.8 (96.0) |
| Redundancy | 3.5 (3.5) |
| Rmerge (%) | 7.3 (48.2) |
| Average I/σ(I) | 7.2 (1.5) |
2.5. Data collection
For data collection, a crystal was transferred through a series of cryoprotectant solutions of increasing glycerol concentration [0.5, 10, 15, 20, 25, 30%(v/v) glycerol in 1.2 M K2HPO4, 0.1 M TAPS pH 9.0, with 1.5 min soaking time in each solution] prior to flash-cooling. This was necessary because the crystals were sensitive to direct transfer to cryoprotectant containing 20–30% glycerol. After cryocooling, the crystals diffracted to 2.7 Å resolution. X-ray diffraction data were collected at 108 K at beamline ID14-1 at the ESRF synchrotron facility in Grenoble, France using a MAR CCD detector. Data were indexed and processed using MOSFLM and SCALA (Evans, 1997 ▶) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶). Data-collection statistics are provided in Table 1 ▶.
3. Results and discussion
The crystals obtained by high-throughput screening were tiny and required scaling up to produce larger crystals. When the same conditions were tested in batch mode in larger volumes, the crystal size did not improve. In hanging drops in the vapor-diffusion mode, somewhat larger crystals formed but were of very poor quality and exhibited serious twinning (Fig. 1 ▶). The morphology of these crystals could not be improved by changing conditions, including altering the protein and precipitant concentrations, pH or temperature or introducing additives known to minimize twinning. However, these crystals did serve well as seed stock. After multiple consecutive rounds of streak-seeding, large single untwinned crystals were produced.
The SH2 domain of Src-family kinases mediates a high-affinity interaction with Cas by binding to a tyrosine-phosphorylated region near the carboxyl-terminal domain of Cas (Petch et al., 1995 ▶; Vuori & Ruoslahti, 1995 ▶; Nojima et al., 1995 ▶; Nakamoto et al., 1996 ▶; Minegishi et al., 1996 ▶). Our studies demonstrate that this tyrosine-phosphorylation site within Cas is predicted to be extended and accessible without ordered secondary-structural features. Within this region, there are three tyrosines that represent potential candidates for phosphorylation: Tyr751, Tyr762 and Tyr764. Tyr751 is present in Cas but not the other Cas-family members. Tyr762 and Tyr764 are close within the motif 762YDYVHL767 that is strictly conserved in all the known human and rodent members of the Cas family, i.e. HEF1 and Efs/Sin. It has been proposed that Tyr762 is the primary SH2-binding site for Src-family kinases, but this has not been unequivocably established to date. Biochemical studies are under way to characterize this binding interaction. The crystals produced in the present study are being used to determine the crystal structure of the Cas–Lck complex and to directly examine the interaction of this tyrosine, in the phosphorylated state, with the Src-family kinase Lck.
Acknowledgments
This work was supported by Grant No. CA071560 from the National Cancer Institute, Grant No. 00-00512V-10030 from the California Research Program, Grant No. 81B-0187 from the California Breast Cancer Research Program, NINDS Grant No. NS35181 and pre-doctoral fellowship No. DAMD17-01-1-0169 and an award from the Swedish–American Foundation (to FN). The authors are grateful to the staff at the Hauptman–Woodward high-throughput screening facility and the ESRF synchrotron for experimental support and consultation, to Arnold C. Satterthwait for helpful discussions on peptide synthesis and to Lisa O’Brien for preparing the manuscript for publication.
References
- Alexandropoulos, K. & Baltimore, D. (1996). Genes Dev.10, 1341–1355. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Eck, M. J., Atwell, S. K., Shoelson, S. E. & Harrison, S. C. (1994). Nature (London), 368, 764–769. [DOI] [PubMed] [Google Scholar]
- Evans, P. R. (1997). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.33, 22–24.
- Garton, A. J., Burnham, M. R., Bouton, A. H. & Tonks, N. K. (1997). Oncogene, 15, 877–885. [DOI] [PubMed] [Google Scholar]
- Giancotti, F. G. & Ruoslahti, E. (1999). Science, 285, 1028–1032. [DOI] [PubMed] [Google Scholar]
- Ishino, M., Ohba, T., Sasaki, H. & Sasaki, T. (1995). Oncogene, 11, 2331–2338. [PubMed] [Google Scholar]
- Kanda, H., Mimura, T., Morino, N., Hamasaki, K., Nakamoto, T., Hirai, H., Morimoto, C., Yazaki, Y. & Nojima, Y. (1997). Eur. J. Immunol.27, 2113–2117. [DOI] [PubMed] [Google Scholar]
- Kanner, S. B., Reynolds, A. B., Wang, H. C., Vines, R. R. & Parsons, J. T. (1991). EMBO J.10, 1689–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law, S. F., Estojak, J., Wang, B., Mysliwiec, T., Kruh, G. & Golemis, E. A. (1996). Mol. Cell. Biol.16, 3327–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., Hill, D. E. & Chernoff, J. (1996). J. Biol. Chem.271, 31290–31295. [DOI] [PubMed] [Google Scholar]
- Luft, J. R., Collins, R. J., Fehrman, N. A., Lauricella, A. M., Veatch, C. K. & DeTitta, G. T. (2003). J. Struct. Biol.142, 170–179. [DOI] [PubMed] [Google Scholar]
- Minegishi, M., Tachibana, K., Sato, T., Iwata, S., Nojima, Y. & Morimoto, C. (1996). J. Exp. Med.184, 1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamoto, T., Sakai, R., Ozawa, K., Yazaki, Y. & Hirai, H. (1996). J. Biol. Chem.271, 8959–8965. [DOI] [PubMed] [Google Scholar]
- Nojima, Y., Morino, N., Mimura, T., Hamasaki, K., Furuya, H., Sakai, R., Sato, T., Tachibana, K., Morimoto, C. & Yazaki, Y. (1995). J. Biol. Chem.270, 15398–15402. [DOI] [PubMed] [Google Scholar]
- Ohashi, Y., Tachibana, K., Kamiguchi, K., Fujita, H. & Morimoto, C. (1998). J. Biol. Chem.273, 6446–6451. [DOI] [PubMed] [Google Scholar]
- O’Neill, G. M., Fashena, S. J. & Golemis, E. A. (2000). Trends Cell Biol.10, 111–119. [DOI] [PubMed] [Google Scholar]
- Petch, L. A., Bockholt, S. M., Bouton, A., Parsons, J. T. & Burridge, K. (1995). J. Cell Sci.108, 1371–1379. [DOI] [PubMed] [Google Scholar]
- Polte, T. R. & Hanks, S. K. (1995). Proc. Natl Acad. Sci. USA, 92, 10678–10682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, R., Iwamatsu, A., Hirano, N., Ogawa, S., Tanaka, T., Mano, H., Yazaki, Y. & Hirai, H. (1994). EMBO J.13, 3748–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer, D. D., Broome, M. A. & Hunter, T. (1997). Mol. Cell. Biol.17, 1702–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stura, E. A. & Wilson, I. A. (1990). Methods, 1, 38–49.
- Stura, E. A. & Wilson, I. A. (1991). J. Cryst. Growth, 11, 270–282.
- Vuori, K., Hirai, H., Aizawa, S. & Ruoslahti, E. (1996). Mol. Cell. Biol.16, 2606–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuori, K. & Ruoslahti, E. (1995). J. Biol. Chem.270, 22259–22262. [DOI] [PubMed] [Google Scholar]