The psychrophilic protease Apa1 consisting of a subtilisin-like region and a large insert (148 residues) with unknown structure has been crystallized. Collection of a data set to 1.78 Å resolution and molecular-replacement searches are reported.
Keywords: subtilisin-like protease, psychrophiles, Pseudoalteromonas
Abstract
The psychrophilic alkaline serine protease Apa1 secreted by the Antarctic psychrotroph Pseudoalteromonas sp. strain AS-11 consists of a subtilisin-like region (293 residues) and an additional insert region (148 residues) that does not show a sequence homology to any proteins in the RCSB Protein Data Bank. Apa1 inhibited with phenylmethanesulfonyl fluoride has been crystallized and X-ray diffraction data have been collected to 1.78 Å resolution. The crystals belong to space group C2, with unit-cell parameters a = 122.94, b = 138.48, c = 64.77 Å, α = γ = 90, β = 97.5°. A molecular-replacement solution has been found using the structure of the mesophilic counterpart subtilisin DY with 38% sequence identity to the catalytic domain of Apa1 as a search model. This is the first crystallographic study of a cold-adapted subtilisin-like protease.
1. Introduction
Psychrophilic or cold-adapted bacteria live at temperatures close to and often below 273 K, encountered in, for example, polar and alpine regions or the deep oceans. Enzymes from these organisms generally exhibit a higher specific activity at low temperatures and a lower thermal stability than their mesophilic counterparts. The high catalytic efficiency has been thought to originate from a highly flexible structure that enables the conformational changes required for catalysis to take place at low temperatures (Hochachka & Somero, 1984 ▶; Sheridan et al., 2000 ▶; Feller, 2003 ▶). There has recently been growing interest in psychrophilic enzymes as models in basic studies both to investigate the thermal stability of proteins and to understand the relationship between their stability, flexibility and catalytic efficiency and as potential candidates for industrial and biotechnological applications, e.g. washing, food processing, biosynthetic processes and environmental bioremediation (Gerday et al., 2000 ▶).
The crystal structures of six psychrophilic enzymes from cold-adapted microorganisms have been reported (Aghajari et al., 1998 ▶, 2003 ▶; Russell et al., 1998 ▶; Alvarez et al., 1998 ▶; Kim et al., 1999 ▶; Van Petegem et al., 2003 ▶) and various mechanisms for cold-adaptation have been proposed based on structural comparisons with mesophilic and thermophilic counterparts. The lower stability of cold-adapted enzymes may be attributed to many kinds of structural factors, including decreased proline content and increased glycine content as well as a general weakening of the intramolecular interactions, weakening of the intersubunit interactions, increased interactions with the solvent water and decreased interactions with solvent ions. All these factors may cause an increase in the overall or local flexibility of the enzyme structure. However, there appears to be difficulty in identifying the true structural determinants of cold activity, as permanently cold environments put selective pressure on enzymes to have high catalytic efficiency but also eliminates the pressure to maintain thermal stability. Indeed, directed evolution studies of temperature adaptation have recently indicated that there is not a strict correlation between increased activity at low temperatures and decreased thermostability (Miyazaki et al., 2000 ▶; Wintrode et al., 2000 ▶). To improve understanding of the structural basis of cold-adaptation, more crystal structures of psychrophilic enzymes need to be compared with the structures of closely related mesophilic and thermophilic counterparts, together with rationalized mutational analyses on the basis of knowledge from the comparative studies.
Subtilisin-like serine proteases (subtilases; EC 3.4.21.–) have been found in numerous bacteria, archaea and eukarya and can be divided into six main families A to F based on their sequence homologies (Siezen & Leunissen, 1997 ▶). The three catalytic residues (Asp, His, Ser) are perfectly conserved in all subtilases. The subtilisin members of family A, secreted by a large variety of Bacillus species, have been extensively studied and engineered to improve their stability and catalytic properties owing to their considerable industrial importance as the protein-degrading components of detergent formulations. The crystal structures of some representative subtilases have been solved at high resolution [e.g. BPN′ (McPhalen & James, 1988 ▶), Carlsberg (Bode et al., 1987 ▶), savinase (Kuhn et al., 1998 ▶), thermitase (Gros et al., 1989 ▶) and proteinase K (Betzel et al., 1988 ▶)]. Although several cold-adapted subtilases from psychrophiles have been reported (e.g. Davail et al., 1994 ▶; Arnórsdóttir et al., 2002 ▶), no crystal structure of a psychrophilic subtilase is so far available.
The subtilisin-like alkaline serine protease Apa1 is secreted from the Antarctic psychrotroph Pseudoalteromonas sp. strain AS-11. It has the characteristics of cold-adapted enzymes, with a higher specific activity on protein substrate at low temperatures and a lower thermal stability than the mesophilic subtilisin BPN′. The nucleotide sequence of the apa1 gene encodes a polypeptide of 817 amino acids including signal peptide, N-terminal propeptide, mature peptide and C-terminal propeptide (accession No. AB120714 in the DDBJ database). The mature enzyme has a molecular weight of 45 kDa and carries a large insert consisting of 148 amino acids that does not show sequence homology to any proteins deposited in the RCSB Protein Data Bank. The amino-acid sequence of the subtilisin-like region reveals that Apa1 belongs to the family A subtilases and the whole sequence of mature Apa1 has 91% identity to the psychrophilic protease SapSh from the psychrotroph Shewanella sp. (Kulakova et al., 1999 ▶) and 60% identity to the mesophilic VapT from Vibrio metschnikovii (Kwon et al., 1995 ▶). Determination of the three-dimensional structure of Apa1 will allow detailed structural analysis and comparative studies with the mesophilic enzyme in order to gain insight into the cold-adaptation of the protein molecule.
2. Materials and methods
2.1. Purification of Apa1
The psychrotroph Pseudoalteromonas sp. strain AS-11 was isolated from the shellfish Neobuccinum eatoni living in the Antarctic ice-covered sea around the Japanese Syowa Station (39° 35′ E, 69° 00′ S). The psychrotroph was continuously cultured using a 10 l fermentor (Eyela MBF-1000M, Japan) at 277 K with stirring at 200 rev min−1 and with vigorous airflow in a medium consisting of artificial marine water (Aquamarine from Yashima Pure Chemicals, Japan), 2.5 g l−1 yeast extract, 10 g l−1 glucose and 10 g l−1 Hammerstein casein pH 7.0. The culture was centrifuged at 10 000g in a continuous rotor (Kubota RC-800) at a flow rate of 1 l h−1 to remove bacteria. The supernatant obtained was applied onto a bacitracin-Sepharose 4B affinity column (2.5 × 13 cm; Stepanov & Rudenskaya, 1983 ▶) equilibrated with 10 mM MES pH 6.0, 1 mM CaCl2 (buffer A) at a flow rate of 4 ml min−1. After the column had been washed with buffer A and then with the same buffer containing 1 M NaCl, the protease Apa1 was eluted with 50% ethylene glycol and 1 M NaCl in buffer A at a flow rate of 1 ml min−1. To measure the protease activity of each fraction, 5 µl diluted sample was added to 295 µl 40 mM Tris–HCl pH 8.0, 10 mM CaCl2 containing 0.1 mM N-succinyl-Ala-Ala-Pro-Phe-p-nitroanilide as substrate and the production of p-nitroaniline was monitored at 405 nm at 298 K using a thermostated spectrophotometer (model Lambda 11, Perkin–Elmer). The eluted protease was collected, inhibited by adding phenylmethanesulfonyl fluoride (PMSF) to 5 mM and stored at 253 K.
The sample of Apa1 inhibited with PMSF (PMS-Apa1) was dialyzed against 10 mM Tris–HCl pH 7.0, 1 mM CaCl2 (buffer B), loaded onto a HiPrep 16/10 Q XL column (1.6 × 10 cm, Amersham Biosciences) and then eluted with 200 ml of a linear gradient of NaCl from 0 to 2 M in buffer B at a flow rate of 2 ml min−1. The eluted PMS-Apa1 was collected and concentrated to 24 mg ml−1 with a centrifugal concentrator (Amicon Centrifugal Filter Devices, 10 kDa molecular-weight cutoff, Millipore, USA). The solvent was exchanged for Milli-Q water because of the high solubility of PMS-Apa1 in water and 100 µl aliquots of the sample were stored at 193 K. A total of 40 mg PMS-Apa1 was purified from 40 l culture supernatant.
2.2. Crystallization
The initial crystallization conditions were screened by the hanging-drop vapour-diffusion method in 24-well Falcon plates using Crystal Screen kits (Hampton Research, USA; Jancarik & Kim, 1991 ▶). Since calcium ions improved the stability of Apa1, all crystal screening experiments were performed with at least 1 mM CaCl2. After 1 µl of 100 mM CaCl2 had been added to 100 µl 24 mg ml−1 PMS-Apa1 in water, 1 µl aliquots of the protein sample were mixed with an equal volume of screening solution and equilibrated against 300 µl screening solution in the reservoir at 298 K.
2.3. X-ray data collection and processing
X-ray diffraction data were collected on beamline BL40B2 at SPring-8, Japan at a wavelength of 1.000 Å. A crystal was immersed in reservoir solution containing 40%(w/v) polyethylene glycol 5000 monomethyl ether (PEG 5K MME, Fluka) as a cryoprotectant, flash-frozen in a stream of supercooled nitrogen gas and maintained at 100 K during data collection. The crystal-to-detector distance was set to 150 mm and a total of 360° of data were recorded from a single crystal on an ADSC Quantum 4R CCD detector with an oscillation angle of 1.0° and 10 s exposure per frame. Raw diffraction data were integrated, scaled and reduced using the HKL2000 package (Otwinowski & Minor, 1997 ▶). Molecular-replacement and refinement calculations were performed using CNS v.1.1 (Brünger et al., 1998 ▶) running on VineLinux 2.6. Manual model building was performed with the program XtalView (McRee, 1999 ▶).
3. Results and discussions
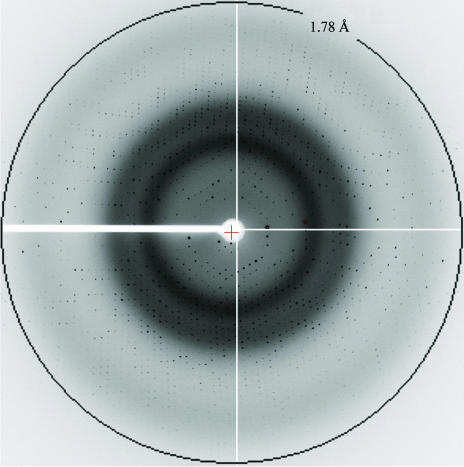
Condition 22 of Crystal Screen [30%(w/v) polyethylene glycol 4000, 0.2 M sodium acetate, 0.1 M Tris–HCl pH 8.5] yielded multiply twinned crystals of PMS-Apa1 after 10 d at 298 K. After optimization of the pH and the types and concentrations of polyethylene glycols and salts of the crystallization conditions, reproducible single crystals with a good diffraction quality were finally obtained using 30%(w/v) PEG 5K MME, 0.1 M Na2SO4, 1 mM CaCl2, 0.1 M Tris–HCl pH 8.5 using 22 mg ml−1 protein solution mixed with an equal volume of the reservoir solution. The plate-shaped crystals appeared after incubation at 298 K for 1 d and grew to dimensions of 0.5 × 0.2 × 0.03 mm within one week (Fig. 1 ▶). The crystal diffracted to a maximum resolution of around 1.78 Å (Fig. 2 ▶). The diffraction data were 98.2% complete to 1.78 Å resolution and displayed good statistics (Table 1 ▶). The space group of the crystal is C2, with unit-cell parameters a = 122.94, b = 138.48, c = 64.77 Å, α = γ = 90, β = 97.5°. Assuming two Apa1 molecules in the asymmetric unit and a monomeric molecular weight of 45.1 kDa, the volume per unit weight (V M) and the solvent content were calculated to be 3.03 Å3 Da−1 and 59%, respectively. These values are within the range usually found for protein crystals (Matthews, 1968 ▶).
Figure 1.
A crystal of Apa1. The longest dimension of the crystal is 0.5 mm.
Figure 2.
A 1° oscillation image collected from an Apa1 crystal at beamline BL40B2 of SPring-8 at 100 K.
Table 1. X-ray diffraction data.
Values in parentheses are for the highest resolution shell.
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 122.94, b = 138.48, c = 64.77, α = γ = 90, β = 97.5 |
| Resolution (Å) | 45.75–1.78 (1.84–1.78) |
| Measured reflections | 752927 |
| Unique reflections | 101031 |
| Redundancy (%) | 7.5 (6.4) |
| Completeness (%) | 98.2 (94.1) |
| I/σ(I) | 14.3 (2.3) |
| Rmerge (%) | 7.7 (31.7) |
| Mosaicity (°) | 0.410 |
Although no homologous protein was found in the PDB for the insert region (148 amino-acid residues) of Apa1, its subtilisin-like region (293 residues) has 38% sequence identity to subtilisin DY (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1bh6; Betzel et al., 1993 ▶). It was therefore used as a search probe for molecular replacement. A cross-rotation search and a subsequent translation search for the top ten cross-rotation peaks using data in the resolution range 15–4 Å resulted in two individual solutions with correlation coefficients of 13.0 and 10.2%, which were relatively small but significantly larger than those of all other solutions. These top two peaks appeared to correspond to the two possible orientations of the molecules in the asymmetric unit, as predicted from the unit-cell parameters. A second translation search to complete the two molecules in the asymmetric unit, with the best solution fixed, gave a solution with a correlation coefficient of 21.8%. Subsequent rigid-body refinement for the resolution range 50–2.0 Å resulted in an R factor of 51.9% with a free R factor of 51.9% for the two http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1bh6 molecules in the asymmetric unit. In the initial stage of model building and refinement for the Apa1 molecule, strict non-crystallographic symmetry (NCS) was used to generate the two molecules in the asymmetric unit. In the first round of model building and refinement, about 200 amino acids in the catalytic domain of Apa1 could be manually included and their main-chain and side-chain geometry was optimized according to the 2F o − F c and F o − F c electron-density maps. Residues that did not show reasonable 2F o − F c electron density for their side chains were replaced by alanine. If a residue did not show reasonable 2F o − F c electron density for its main chain, it was removed from the model. With fixed B factors (15.0 Å2) for all atoms, simulated annealing using torsion-angle dynamics gave an R factor of 43.3% and free R factor of 44.6%. During the subsequent iterative rounds of manual model building, torsion-angle simulated annealing and B-factor refinement, all residues of the insert and subtilisin-like regions omitted from the initial model could be modelled as their respective amino acids according to the F o − F c maps, except for the C-terminal seven residues. All residues initially replaced by alanine were also substituted back to their respective amino acids. The current model of Apa1 without solvent molecules had R and free R factors of 28.8 and 28.9%, respectively. Further refinement of the model is currently in progress.
Acknowledgments
This work was supported by National Project on Protein Structural and Functional Analyses and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan. The organizers of the Japan Synchrotron Radiation Research Institute (JASRI) are acknowledged for supplying beamtime on the beamline BL40B2 at SPring-8.
References
- Aghajari, N., Feller, G., Gerday, C. & Haser, R. (1998). Protein Sci.7, 564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajari, N., Van Petegem, F., Villeret, V., Chessa, J.-P., Gerday, C., Haser, R. & Van Beeumen, J. (2003). Proteins, 50, 636–647. [DOI] [PubMed] [Google Scholar]
- Alvarez, M., Zeelen, J. P., Mainforid, V., Rentier-Delrue, F., Martial, J. A., Wyns, L., Wierenga, R. K. & Maes, D. (1998). J. Biol. Chem.273, 2199–2206. [DOI] [PubMed] [Google Scholar]
- Arnórsdóttir, J., Smáradóttir, R. B., Magnússon, Ó. T., Thorbjarnardóttir, S. H., Eggertsson, G. & Kristjánsson, M. M. (2002). Eur. J. Biochem.269, 5536–5546. [DOI] [PubMed] [Google Scholar]
- Betzel, C., Pal, G. P. & Saenger, W. (1988). Eur. J. Biochem.178, 155–171. [DOI] [PubMed] [Google Scholar]
- Betzel, C., Visanji, M., Eschenburg, S., Wilson, K. S., Peters, K., Fittkau, S., Singh, T. P. & Genov, N. (1993). Arch. Biochem. Biophys.302, 499–502. [DOI] [PubMed] [Google Scholar]
- Bode, W., Papamokos, E. & Musil, D. (1987). Eur. J. Biochem.166, 673–692. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Davail, S., Feller, G., Narinx, E. & Gerday, C. (1994). J. Biol. Chem.269, 17448–17453. [PubMed] [Google Scholar]
- Feller, G. (2003). Cell Mol. Life Sci.60, 648–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerday, C., Aittaleb, M., Bentahir, M., Chessa, J.-P., Claverie, P., Collins, T., D’Amico, S., Dumont, J., Garsoux, G., Georiette, D., Hoyoux, A., Lonhience, T., Meuwis, M.-A. & Feller, G. (2000). Trends Biotechnol.18, 103–107. [DOI] [PubMed] [Google Scholar]
- Gros, P., Betzel, C., Dauter, Z., Wilson, K. S. & Hol, W. G. J. (1989). J. Mol. Biol.210, 347–367. [DOI] [PubMed] [Google Scholar]
- Hochachka, P. W. & Somero, G. N. (1984). Biochemical Adaptation, pp. 355–449. Princeton: Princeton University Press.
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Kim, S. Y., Hwang, K. Y., Kim, S. H., Sung, H. C., Han, Y. S. & Cho, Y. (1999). J. Biol. Chem.274, 11761–11767. [DOI] [PubMed] [Google Scholar]
- Kuhn, P., Knapp, M., Soltis, S. M., Ganshaw, G., Thoene, M. & Bott, R. (1998). Biochemistry, 37, 13446–13452. [DOI] [PubMed] [Google Scholar]
- Kulakova, L., Galkin, A., Kurihara, T., Yoshimura, T. & Esaki, N. (1999). Appl. Environ. Microbiol.65, 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, Y. T., Kim, J. O., Moon, S. Y., Yoo, Y. D. & Rho, H. M. (1995). Gene, 152, 59–63. [DOI] [PubMed] [Google Scholar]
- McPhalen, C. A. & James, M. N. G. (1988). Biochemistry, 27, 6582–6598. [PubMed] [Google Scholar]
- McRee, D. E. (1999). J. Struct. Biol.125, 156–165. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Miyazaki, K., Wintrode, P. L., Grayling, R. A., Rubingh, D. N. & Arnold, F. H. (2000). J. Mol. Biol.297, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Russell, R. J., Gerike, U., Danson, M. J., Hough, D. W. & Taylor, G. L. (1998). Structure, 6, 351–361. [DOI] [PubMed] [Google Scholar]
- Sheridan, P. P., Panasik, N., Coombs, J. M. & Brenchley, J. E. (2000). Biochim. Biophys. Acta, 1543, 417–433. [DOI] [PubMed] [Google Scholar]
- Siezen, R. J. & Leunissen, J. A. (1997). Protein Sci.6, 501–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanov, V. M. & Rudenskaya, G. N. (1983). J. Appl. Biochem.5, 420–428. [PubMed] [Google Scholar]
- Van Petegem, F., Collins, T., Meuwis, M.-A., Gerday, C., Feller, G. & Van Beeumen, J. (2003). J. Biol. Chem.278, 7531–7539. [DOI] [PubMed] [Google Scholar]
- Wintrode, P. L., Miyazaki, K. & Arnold, F. H. (2000). J. Biol. Chem.275, 31635–31640. [DOI] [PubMed] [Google Scholar]