An open reading frame from E. coli MG1655 has been cloned, expressed and purified. Crystals obtained from the purified recombinant protein have been obtained in a variety of different forms diffracting to 1.8 Å resolution.
Keywords: cell-division proteins, YgfE
Abstract
An open reading frame designated b2910 (ygfE) in the Escherichia coli K12-MG1655 genome sequence, identified as a possible homologue to the cell-division protein ZapA, was cloned into the high-expression plasmid pETDuet-1 and overexpressed in E. coli BL21 (DE3)-AI. The protein was purified in three steps to 99% purity. Crystals were obtained by the hanging-drop vapour-diffusion method at 291 K from a wide range of screened conditions, but principally from solutions containing 0.1 M HEPES pH 7.0, 18% PEG 6000, 5 mM CaCl2. Diffraction data to 1.8 Å were collected at the European Synchrotron Radiation Facility (ESRF). The crystals belong to space group P6122 or P6522, with unit-cell parameters a = 53.8, b = 53.8, c = 329.7 Å, α = β = 90, γ = 120°.
1. Introduction
The ancestral tubulin protein FtsZ has a crucial role in the process of bacterial cell division. FtsZ polymerizes into a ring (the ‘Z-ring’) around the middle of pre-divisional cells on the inner surface of the cytoplasmic membrane. The Z-ring then constricts, driving cell division (Addinall & Holland, 2002 ▶; Lutkenhaus & Addinall, 1997 ▶; Margolin, 2000 ▶). FtsZ is extensively conserved throughout bacterial species and plants (where it is required for plastid division). FtsZ has also been implicated in the division of some archaeal species and the mitochondria of some eukaryotes (Vaughan et al., 2004 ▶). As a cytoskeletal element with prokaryotic origins the Z-ring represents a fascinating topic of study and as a major player in a fundamental process for bacterial life FtsZ represents an exciting potential target for novel antibacterial compounds.
The mechanisms by which Z-rings form and constrict are poorly characterized and the detailed in vivo structure of the Z-ring is unknown. In vitro studies reveal that FtsZ is a GTPase which can polymerize into linear polymers in a GTP-dependent manner (Romberg & Levin, 2003 ▶), and that these polymers form lateral associations to a greater or lesser degree depending on polymerization conditions. Measurements indicate that cells contain sufficient FtsZ for the Z-ring to be composed of multiple protofilaments (Lu et al., 1998 ▶; Feucht et al., 2001 ▶). This is likely to be the case since cells containing GFP-FtsZ fusion protein incorporate approximately 30% of the GFP fluorescence into the Z-ring (Anderson et al., 2004 ▶). Therefore, lateral interactions between FtsZ polymers are likely to be important in Z-ring function.
Two FtsZ accessory proteins have been identified that promote lateral associations between FtsZ polymers. ZipA (from Escherichia coli) is a membrane-spanning protein with a cytoplasmic domain that interacts with the FtsZ C-terminus (Hale & de Boer, 1997 ▶). This domain of ZipA induces bundling of FtsZ polymers in vitro (Hale et al., 2000 ▶; RayChaudhuri, 1999 ▶) and its structure has been determined in complex with a small FtsZ peptide (Moy et al., 2000 ▶). ZapA from both Bacillus subtilis and Pseudomonas aeruginosa has been demonstrated to bind and to induce bundling between FtsZ polymers (Gueiros-Filho & Losick, 2002 ▶; Low et al., 2004 ▶), and the crystal structure of tetrameric P. aeruginosa ZapA has been solved (Low et al., 2004 ▶). We recently demonstrated that the E. coli YgfE protein binds to and bundles FtsZ polymers, inhibits the FtsZ GTPase and adopts a dimer–tetramer equilibrium in solution (Small et al., submitted work). YgfE is thus a functional homologue of ZapA as suggested by previous sequence and immunofluoresence data (Gueiros-Filho & Losick, 2002 ▶).
2. Methods and materials
2.1. Cloning, overexpression and purification
The gene encoding E. coli YgfE (b2910; 109 amino acids, 12 594 Da) was amplified by PCR from E. coli K12-MG1655 chromosomal DNA with primers incorporating an EcoRI site at the 5′ end of the gene and a HindIII site at the 3′ end. The PCR fragment was digested with these enzymes and ligated into a pETDuet-1 vector (Novagen), also digested with EcoRI and HindIII. This strategy provides YgfE with an amino-terminal six-histidine tag. After selection of recombinants containing the cloned gene by restriction digest, E. coli BL21 (DE3) AI (Invitrogen) was transformed with the resulting recombinant plasmid pETDuet-ygfE. Cells were grown at 310 K in 650 ml LB medium containing 50 µg l−1 ampicillin. Protein expression was induced by the addition of arabinose to a final concentration of 0.2% and growth was continued at 298 K for 16 h. Cells were harvested by centrifugation at 6400g and resuspended in 50 mM 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES) pH 7.5, 500 mM NaCl and 10 mM imidazole (buffer A) prior to lysis by sonication.
The sonicated extract was clarified by centrifugation at 50 000g and applied at 277 K onto a 25 ml chelating Sepharose (GE Healthcare) affinity column charged with three column volumes of 50 mM nickel chloride in 50 mM sodium phosphate buffer pH 4 prior to equilibration in buffer A. After loading of the crude extract, the column was washed with ten column volumes of A followed by a further ten column volumes of buffer A supplemented with 100 mM imidazole pH 8.0. YgfE was then eluted from the column in buffer A containing 500 mM imidazole pH 8.0. The protein was dialysed into 20 mM HEPES pH 7.5 and concentrated to 12 mg ml−1, as determined by BioRad protein assay.
3. Crystallization and DLS assay
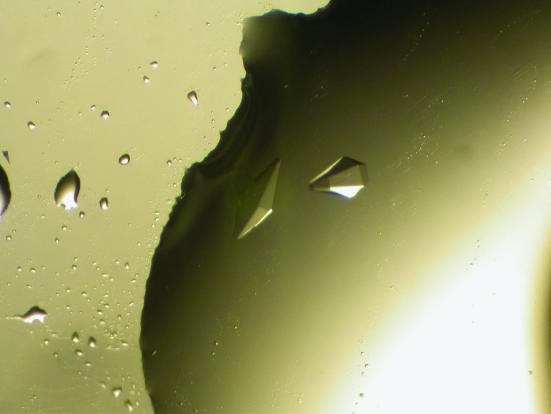
Prior to crystallization, samples of protein were filtered through a Whatman SpinX-centrifuge spin filter and subjected to dynamic light-scattering analysis using a DynaPro MS/X device to check for monodispersity. All crystallization experiments were performed using the hanging-drop vapour-diffusion method in a 24-well tissue-culture Linbro plate at 291 K. Initial crystallization trials were carried out using 0.5 ml reservoir solutions taken from the Hampton Research Crystal Screens I and II (Jancarik & Kim, 1991 ▶) or using 0.5 ml solutions taken from the Clear Strategy Screen buffered with 0.1 M MES pH 6.5 (Brzozowski & Walton, 2001 ▶). Drops consisting of 1 µl protein solution and 1 µl reservoir solution were used throughout. Crystals were obtained after several days using several conditions from the Clear Strategy Screen (CSS; Brzozowski & Walton, 2001 ▶) including CSS1 No. 1 (25% PEG 2K monomethylether, 0.3 M sodium acetate) and CSS1 No. 6 (0.1 M HEPES pH 7.0, 25% PEG 2K monomethylether, 0.8 M sodium formate). Additionally, crystals were obtained from a in-house screen developed for small proteins and peptides using conditions consisting of 0.1 M HEPES pH 7.0, 18% PEG 6000, 5 mM CaCl2. These latter crystals were of a quality suitable for X-ray data collection, with dimensions in excess of 0.1 × 0.5 × 0.2 mm (Fig. 1 ▶).
Figure 1.
Crystals of E. coli YgfE obtained from 0.1 M HEPES pH 7.0, 18% PEG 6000, 5 mM CaCl2. These crystals appear after within 2–3 d after incubation of the plate at 291 K and have a maximum size of 0.5 mm in any one dimension.
3.1. X-ray crystallographic studies
Preliminary diffraction data were collected at 100 K in-house using an Enraf–Nonius Cu Kα X-ray generator operating at 45 kV and 115 mA equipped with Osmic focusing mirrors and a MAR 345dtb image-plate detector. Complete data sets to the resolution quoted in this paper were collected at ESRF ID14.3. The crystals were vitrified in liquid nitrogen using 30%(v/v) glycerol in the mother liquor as cryoprotectant and were maintained at 100 K throughout data collection using an Oxford Cryostream system. Intensity data were indexed, integrated and scaled using the HKL programs DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶). Data-collection and processing statistics are given in Table 1 ▶.
Table 1. Data-collection and processing statistics.
Values in parentheses correspond to the outer resolution shell.
| Wavelength (Å) | 0.933 |
| Space group | P6122 |
| Unit-cell parameters | |
| a (Å) | 53.8 |
| b (Å) | 53.8 |
| c (Å) | 329.7 |
| α (°) | 90 |
| β (°) | 90 |
| γ (°) | 120 |
| Matthews coefficient (Å3 Da−1) | 2.7 |
| Molecules per AU | 2 |
| Solvent content (%) | 55 |
| Resolution range (Å) | 30–1.8 (1.83–1.80) |
| Total observations | 380518 |
| Unique reflections | 27357 |
| Average I/σ(I) | 47.9 (4.4) |
| Rmerge | 0.052 (0.242) |
| Completeness (%) | 98.3 (78.2) |
During the course of this study, the structure of P. aeruginosa ZapA (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1w2e) became available. Although ZapA is a functional homologue of YgfE, there is a low level of sequence similarity between the two proteins (Fig. 2 ▶) and thus far we have been unable to obtain a molecular-replacement solution using ZapA as search model. We are therefore investigating MIR and MAD strategies to obtain a solution to the structure of YgfE as part of a coordinated study investigating the interaction of YgfE, FtsZ and other cell-division proteins.
Figure 2.
CLUSTALW sequence alignment (http://www.ebi.ac.uk/clustalw/index.html) of E. coli K12-MG1655 YgfE (TIGR reference b2901) with P. aeruginosa PAO1 ZapA (TIGR reference PA5227).
Acknowledgments
We are grateful for access and user support at the synchrotron facilities of the ESRF, Grenoble and SRS, Daresbury. We would like to thank Klaus Fütterer (University of Birmingham) for assistance with data collection. AB is funded by a BBSRC studentship; RPG is supported via the EPSRC-supported Molecular Organisation and Assembly in Cells (MOAC) doctoral training centre at Warwick, KAJ is supported by the Karl Trygger Foundation, SA and TD are supported by the Wellcome Trust and MRC Career Development fellowships, respectively.
References
- Addinall, S. G. & Holland, B. (2002). J. Mol. Biol.318, 219–236. [DOI] [PubMed] [Google Scholar]
- Anderson, D. E., Gueiros-Filho, F. J. & Erickson, H. P. (2004). J. Bacteriol.186, 5775–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski, A. M. & Walton, J. (2001). J. Appl. Cryst.34, 97–101. [Google Scholar]
- Feucht, A., Lucet, I., Yudkin, M. D. & Errington, J. (2001). Mol. Microbiol.40, 115–125. [DOI] [PubMed] [Google Scholar]
- Gueiros-Filho, F. J. & Losick, R. (2002). Genes Dev.16, 2544–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale, C. A. & de Boer, P. A. (1997). Cell, 88, 175–185. [DOI] [PubMed] [Google Scholar]
- Hale, C. A., Rhee, A. C. & de Boer, P. A. (2000). J. Bacteriol.182, 5153–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Low, H. H., Moncrieffe, M. C. & Löwe, J. (2004). J. Mol. Biol.341, 839–852. [DOI] [PubMed] [Google Scholar]
- Lu, C., Stricker, J. & Erickson, H. P. (1998). Cell Motil. Cytoskeleton, 40, 71–86. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus, J. & Addinall, S. G. (1997). Annu. Rev. Biochem.66, 93–116. [DOI] [PubMed] [Google Scholar]
- Margolin, W. (2000). FEMS Microbiol. Rev.24, 531–548. [DOI] [PubMed] [Google Scholar]
- Moy, F. J., Glasfeld, E., Mosyak, L. & Powers, R. (2000). Biochemistry, 39, 9146–9156. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- RayChaudhuri, D. (1999). EMBO J.18, 2372–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg, L. & Levin, P. A. (2003). Annu. Rev. Microbiol.57, 125–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, S., Wickstead, B., Gull, K. & Addinall, S. G. (2004). J. Mol. Evol.58, 19–29. [DOI] [PubMed] [Google Scholar]