The first crystal structure of a Mimosoideae lectin, Parkia platycephala has been solved by MAD phasing using 5-bromo-4-chloro-3-indolyl-α-d-mannose as an anomalous X-ray scatterer. This strategy may be useful for structure elucidation of novel lectins or when molecular replacement methods fail.
Keywords: protein–carbohydrate interactions, Parkia platycephala lectin, isothermal titration calorimetry, surface plasmon resonance, β-prism domain, MAD phasing
Abstract
Parkia platycephala belongs to the most primitive group of Leguminosae plants. Its seed lectin is made up of three homologous β-prism repeats and exhibits binding specificity for mannose/glucose. The properties of the association between the lectin from P. platycephala seeds and monosaccharide ligands were analysed by isothermal titration calorimetry and surface plasmon resonance. The results are consistent with the lectin bearing three thermodynamically identical binding sites for mannose/glucose per monomer with dissociation constants in the millimolar range. Binding of each ligand by the lectin is enthalpically driven. Crystals have been obtained of the lectin in complex with a brominated derivative of mannose (5-bromo-4-chloro-3-indolyl-α-d-mannose), which were suitable for deriving an electron-density map by MAD phasing. In agreement with the thermodynamic data, six Br atoms were found in the asymmetric unit of the monoclinic P21 crystals, which contained two P. platycephala lectin molecules. The availability of other Br derivatives of monosaccharides (glucose, galactose, fucose) may make this strategy widely useful for structure elucidation of novel lectins or when (as in the case of the P. platycephala lectin) molecular-replacement methods fail.
1. Introduction
Lectins are a structurally heterogeneous group of carbohydrate-binding proteins of nonimmune origin comprising distinct families of evolutionarily related proteins (Van Damme et al., 1998 ▶). Sugar-recognition mechanisms have evolved independently in diverse protein frameworks and lectins are ubiquitous in animals, plants and microorganisms. Lectins play biological roles in many cellular processes, such as fertilization, cell communication, differentiation and development, host defence, parasitic infection, tumour metastasis etc., by deciphering the glycocodes encoded in the structure of glycans attached to soluble and integral cell-membrane glycoconjugates (Gabius & Gabius, 1997 ▶).
The seed lectins of leguminous plants constitute the largest and most thoroughly studied lectin family (see the 3D Lectin Database at http://www.cermav.cnrs.fr/lectines). These lectins have represented paradigms for establishing the structural basis (Rini, 1995 ▶; Weis & Drickamer, 1996 ▶; Elgavish & Shaanan, 1997 ▶; Loris et al., 1998 ▶; Bouckaert et al., 1999 ▶; Vijayan & Chandra, 1999 ▶) and thermodynamics (Chervenak & Toone, 1995 ▶; Dam et al., 1998 ▶; Dam, Cavada et al., 2000 ▶; Dam, Roy et al., 2000 ▶) of selective sugar recognition. Most studies on Leguminoseae lectins involve members of the Papilionoideae subfamily, while investigations on lectins of the other two subfamilies, Caesalpinoideae and Mimosoideae, are scarce. The Mimosoideae subfamily of leguminous plants comprises six tribes (Adenanthereae, Mimoseae, Mimozygantheae, Parkiae, Acacieae and Ingeae) which contain 56 genera. Parkia platycephala is an important forage tree growing in parts of north-eastern Brazil. Of the Mimosoideae, the tribe Parkiae, comprising two genera (Penthalcletha and Parkia) together containing about 30 species and regarded as the most primitive group within the Leguminosae plants (Heywood, 1979 ▶), is the only taxon from which lectins have been biochemically characterized. These include the seed lectins from Parkia speciosa (Suvachittanont & Peutpaiboon, 1992 ▶), P. javanica (Utarabhand & Akkayanont, 1995 ▶), P. platycephala (Cavada et al., 1997 ▶; Ramos et al., 1999 ▶; Mann et al., 2001 ▶) and P. discolor (Cavada et al., 2000 ▶).
The lectin from seeds of P. platycephala (PPL) exhibits binding specificity for mannose/glucose, its best inhibitors being mannose and oligomannosides with α1–3 and α1–6 branch linkages (Ramos et al., 1999 ▶). The sugar-binding specificity of this lectin towards mannose, an abundant building block of surface-exposed glycoconjugates of viruses, bacteria and fungi, suggests a role for the P. platycephala lectin in the defence against plant pathogens or predators (Van Damme et al., 1998 ▶).
The P. platycephala seed lectin is a non-glycosylated single-chain polypeptide with a molecular weight of 47.9 kDa. Its structure is made up of three homologous β-prism repeats (Mann et al., 2001 ▶), each of which exhibits sequence similarity with jacalin-related lectin monomers from the angiosperm (Asteraceae, Convolvulaceae, Moraceae, Musaceae, Gramineae and Fagaceae; Mann et al., 2001 ▶) and gymnosperm (Cicadaceae; Yagi et al., 2002 ▶) plant families. Moreover, the P. platycephala lectin shows also sequence similarity with stress- and pathogen-upregulated defence and salt-stress-induced genes of a number of different plants (Zhang et al., 2000 ▶; Mann et al., 2001 ▶), suggesting a common ancestry for jacalin-related lectins and inducible defence proteins.
Oligomerization and the multivalency that results are of considerable importance for the function of lectins. In particular, the variability in the quaternary structure of legume lectins that are built up by subunits of essentially the same tertiary fold has attracted much attention (Srinivas et al., 2001 ▶). The apparent molecular weight of the P. platycephala seed lectin in buffered solutions of pH in the range 4.5–8.5 determined by analytical ultracentrifugation equilibrium sedimentation was 94 ± 3 kDa, indicating that the native lectin structure is a non-pH-dependent homodimer (Mann et al., 2001 ▶). A native dimeric PPL molecule may thus harbour up to six carbohydrate-binding sites. However, no structural data of lectins made up by tandem repeats of β-prism domains have been reported and the amino-acid side chains that interact with mannose residues in the crystal structure of the single β-prism lectins from Helianthus tuberosus (Bourne et al., 1999 ▶), Artocarpus integrifolia (Pratap et al., 2002 ▶) and Calystegia sepium (Bourne et al., 2004 ▶) are not absolutely conserved in the three P. platycephala lectin repeats. With the aim of determining the stoichiometry of sugar recognition sites per PPL monomer and of exploring the possibility of using a 5-bromo-derivative of mannose for MAD phasing, we have studied the energetics of the association of monosaccharides with the P. platycephala lectin.
2. Materials and methods
2.1. Isolation of P. platycephala lectin
Mature seeds from P. platycephala were collected in the state of Ceará (north-east Brazil). The P. platycephala seed lectin (PPL) was purified by ammonium sulfate precipitation followed by affinity chromatography on Sephadex G100, as described by Cavada et al. (1997 ▶). The apparent molecular weight and homogeneity of the purified P. platycephala seed lectin were estimated by SDS–PAGE and by MALDI–TOF mass spectrometry (Mann et al., 2001 ▶).
2.2. Isothermal titration calorimetry
Isothermal titration calorimetry experiments were carried out using an Omega instrument (MicroCal Inc., Nothampton, MA, USA) coupled to an external nanovoltimeter, which was used to improve the signal-to-noise ratio (Wiseman et al., 1989 ▶). Microlitre amounts of ligand solution (197.2 µM in 50 mM HEPES pH 7.5) were added sequentially to the calorimetric cell (1.37 ml) containing protein solution (7.9 µM in the same buffer). The amount of power required to maintain the reaction cell at constant temperature (298 K) after each injection was monitored as a function of time. To correct for the heats not directly related to the binding reaction (mainly dilution of the concentrated ligand solution into the calorimetric cell), control experiments were performed by making identical injections of the ligand solution into the titration cell containing only buffer. The heat arising from the binding reaction between the lectin and the ligand was obtained as the difference between the heat of reaction and the corresponding heat of dilution. The thermodynamic parameters of binding were obtained by analyzing the data with software provided by MicroCal.
2.3. Surface plasmon resonance
All experiments were performed at 298 K in HBS buffer (10 mM HEPES pH 7.4 containing 150 mM NaCl, 3 mM EDTA and 0.005% surfactant P20) on a Biacore 2000 instrument. PPL was covalently immobilized on the carboxymethylated surface of two flowcells of a CM5 sensorchip, using the Amine Coupling Kit (Biacore) following the manufacturer’s instructions, to a level of 4037 resonance units (RU; low density = R1 immo) or 9269 RU (high density = R2 immo). Two reference flowcells were prepared, one with an immobilized non-relevant protein (BSA, 7411 RU); the second remained empty. The surface plasmon resonance (SPR) analyses were performed at a flow rate of 100 µl min−1 in HBS. Glycan solutions were prepared in 4% DMF at 50 mM and then diluted in HBS buffer to the desired concentration. 14 different concentrations (0.0196–10 mM) of X-mannopyranoside or X-galactopyranoside (X = 5-bromo-4-chloro-3-indolyl-α-d-) (Sigma) and 12 concentrations of d-glucose, d-mannose and d-galactose (Sigma) (0.12–10 mM) were injected across the four flowcells of the sensorchip for 1 min and the dissociation of the complexes was followed for another 60 s. Control experiments were performed by injecting the corresponding dilution of 4% DMF in HBS onto the lectin surface. The signals obtained on the low-density and on the high-density PPL surfaces were double-substracted from the signals obtained by injecting the glycans on the reference flowcells and from the signals obtained by injecting the DMF buffer on the PPL surfaces. The steady-state reponse (R eq) for each concentration was measured, and plotted against the glycan concentration (C). Fitting the plots with the equation
yielded R max (the maximum binding capacity of the PPL surface) and K d (the equilibrium dissociation constant) for each glycan. The stoichiometry of binding was determined using (2), where M lectin and M glycan are the molecular weights of PPL (47 950 Da) and the glycan used (X-Man/Gal, 408.6 Da; Man/Gal, 180.2 Da),
2.4. Crystallization and MAD phasing
The P. platycephala seed lectin was dissolved at a concentration of 5 mg ml−1 in 50 mM HEPES pH 7.5. X-Man was dissolved at a concentration of 50 mM in pure DMSO. For crystallization trials, X-Man was added to both the protein solution and the reservoir buffer at a final concentration of 3 mM. Crystals did not grow using the previously reported crystallization conditions for the isolated lectin (Gallego del Sol et al., 2002 ▶), and thus the sparse-matrix method (Jancarik & Kim, 1991 ▶) was employed with the Crystal Screen I and II formulations supplied by Hampton Research. Crystals suitable for diffraction experiments (maximal dimensions of 0.6 × 0.4 × 0.4 mm) grew within five weeks at 295 K by the vapour-diffusion method using hanging drops composed of equal volumes of protein solution and reservoir buffer (2.0 M ammonium dihydrogen phosphate, 0.1 M Tris–HCl pH 8.5 containing 3 mM X-Man). The crystals diffracted to 2.5 Å resolution and a complete MAD experiment was performed on beamline BM16 at ESRF (Grenoble) at three different wavelenths (edge, 0.91995 Å; peak, 0.91932 Å; remote, 0.92191 Å) using the bromide of X-Man as an anomalous scatterer. The data were indexed, integrated and scaled using MOSFLM (Leslie, 1997 ▶) and SCALA (Collborative Computational Project, Number 4, 1994 ▶). Br atoms were located with SOLVE (Terwilliger & Berendzen, 1999 ▶). After density modification using RESOLVE (Terwilliger, 2000 ▶), a density map suitable for polypeptide tracing was obtained and was skeletonized with the program MAPMAN (Kleywegt & Jones, 1996 ▶).
3. Results and discussion
Mannose-binding lectins are widely distributed in higher plants and may play a role in defence against plant pathogens or predators (Van Damme et al., 1998 ▶). Mannose-binding activity has evolved in distinct structural scaffolds (i.e. the β-sandwich of legume lectins, the β-barrel of monocot lectins and the β-prism of jacalin-related lectins; Barre et al., 2001 ▶, 2004 ▶; Jeyaprakash et al., 2004 ▶) and there is also large variation within each family in the quaternary association of lectins built up of essentially identical subunits. The number and spatial arrangement of carbohydrate-binding sites confers distinct biological properties to different oligomers with the same nominal carbohydrate-binding specificity. In the case of the mannose-specific jacalin-related lectins, calsepa from the rhizome of hedge bindweed Calystegia sepium is a dimer (Bourne et al., 2004 ▶), artocarpin from seeds of the jack fruit Artocarpus integrifolia behaves as a tetramer (Pratap et al., 2002 ▶) and heltuba from the Jerusalem artichoke (Helianthus tuberosus) tuber displays a toroid-shaped structure composed of eight subunits arranged into a dimer of tetramers with the eight carbohydrate-binding sites located at the periphery and occupied by Manα1–3Man dimannosides (Bourne et al., 1999 ▶).
The primary structure of the P. platycephala seed lectin consists of three tandemly arranged homologous domains, each of which shares the signature of the β-prism fold (Mann et al., 2001 ▶) observed in the crystal structures of the lectins heltuba (Asteraceae), artocarpin (Moraceae) and calsepa (Convolvulaceae). However, in contrast to these three jacalin-related lectins, which exhibit an exclusive specificity towards mannose or oligomannosides, the P. platycephala seed lectin (Mimosoideae) belongs to the glucose/mannose-recognition lectins of leguminous plants.
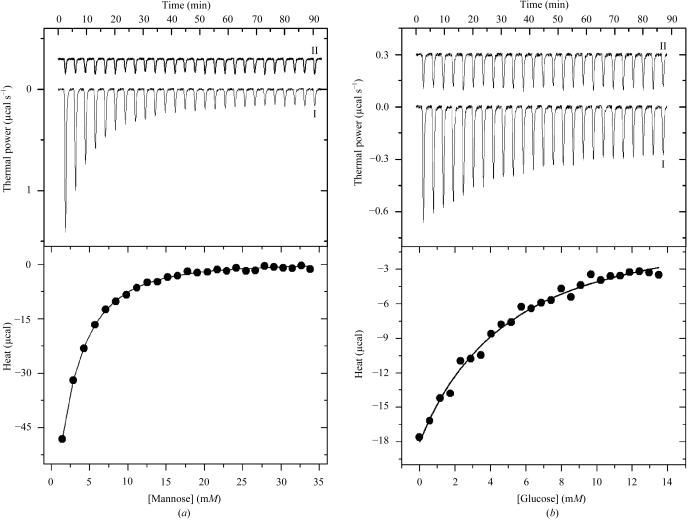
The energetics of association between P. platycephala lectin and both mannose and glucose was measured by isothermal titration calorimetry. Fig. 1 ▶(a) shows the titration of P. platycephala lectin with mannose in 50 mM HEPES buffer pH 7.5 at 298 K together with the corresponding dilution experiment. The binding reaction is characterized by a significant exothermic heat effect which decreases (in absolute value) as the concentration of ligand increases, reaching the dilution level when all the available binding sites become saturated. The binding isotherm (lower panel in Fig. 1 ▶ a) was fitted to the identical binding sites model. Since the experiments were performed at a protein concentration far below the dissociation constant of the lectin–sugar complex, analysis of the binding energetics was performed fixing the stoichiometry of the complex to three molecules of sugar per protein molecule as established by surface plasmon resonance experiments (see below). Leaving stoichiometry as a variable parameter in the fitting procedure could lead to an increase in the individual errors of both the stoichiometry and the enthalpy change upon binding. The data are consistent with the lectin having three thermodynamically identical binding sites for mannose with an affinity binding constant of 415 ± 20 M −1 (K d = 2.41 ± 0.12 mM; Table 1 ▶). Fitting of the data to a model considering the binding sites to be non-identical leads to virtually the same thermodynamical parameters. The situation is similar when the binding of glucose to the protein is considered (Fig. 1 ▶ b) though the ligand is recognized by the lectin with a much lower affinity. The binding constant for glucose was K a = 103 ± 28 M −1 (K d = 9.7 ± 2.6 mM; Table 1 ▶), which represents a fourfold decrease compared with mannose. Binding of both ligands by the lectin is governed primarily by enthalpic forces (ΔH = −7.6 kcal mol−1 for glucose and −6.2 kcal mol−1 for mannose; 1 kcal = 4.186 kJ; Table 1 ▶). Glucose binding to PPL has to overcome a larger entropic penalty (−TΔS = +4.86 kcal mol−1) than mannose (−TΔS = +2.6 kcal mol−1), which results in a more favourable Gibbs free energy upon binding for mannose (ΔG = −3.6 kcal mol−1) than for glucose (ΔG = −2.7 kcal mol−1; Table 1 ▶). The enthalpically driven reaction suggests that hydrogen bonds and van der Waals interactions are the main forces that stabilize the binding of the monosaccharide in the carbohydrate-combining site of PPL. These types of interactions are typically conserved in crystal structures of legume lectin–saccharide complexes (Rini, 1995 ▶; Elgavish & Shaanan, 1997 ▶; Loris et al., 1998 ▶) and have also been observed in the mannose-bound structures of β-prism lectins, i.e. heltuba (Bourne et al., 1999 ▶), calsepa (Bourne et al., 2004 ▶), artocarpin (Pratap et al., 2002 ▶; Jeyaprakash et al., 2004 ▶) and jacalin (Bourne et al., 2002 ▶). Enthalpy–entropy compensation has been reported for many other lectin–carbohydrate interactions and has been attributed to the reorganization of structural water molecules around the protein binding site and the carbohydrate ligand (Lemieux et al., 1991 ▶; Mandal et al., 1994 ▶; Chervenak & Toone, 1995 ▶; Dam, Cavada et al., 2000 ▶).
Figure 1.
Upper panels: high-sensitivity isothermal titration calorimetry of P. platycephala seed lectin with mannose (a) and glucose (b) (trace I) together with the dilution experiment (trace II). The monosaccharide solution was injected in small increments of 5 µl into the reaction cell containing the protein solution. The power output, in units of µcal s−1, is shown as a function of time for successive injections of ligand. Lower panels: calorimetric binding isotherms corresponding to the experiments presented in the upper panels. The area under each peak (amount of heat released by the binding event at a given degree of saturation) is plotted against the concentration of ligand in the cell. The continuous line represents the result of the non-linear least-squares fitting of the data to the identical binding sites model. Thermodynamic parameters are shown in Table 1 ▶.
Table 1. Thermodynamic parameters governing the binding of monosaccharides to PPL derived by isothermal titration calorimetry.
| Ligand | Ka (M−1) | ΔG (kcal mol−1) | ΔH (kcal mol−1) | ΔS (cal K−1 mol−1) |
|---|---|---|---|---|
| Mannose | 415 ± 20 | −3.6 ± 0.2 | −6.2 ± 0.8 | −8 ± 3 |
| Glucose | 103 ± 28 | −2.7 ± 0.3 | −7.6 ± 0.9 | −16 ± 4 |
The interaction of the P. platycephala lectin with monosaccharides was also analyzed by surface plasmon resonance. In line with its reported specificity (Cavada et al., 1997 ▶), no interaction with d-galactose or with its X-derivative could be detected. The results also confirmed the low affinity for glucose, which could not however be quantified. On the other hand, the K a for d-mannose and X-mannose were (1.64 ± 0.25) × 10−3 M −1 (K d = 0.61 ± 0.09 mM) and (3.79 ± 1.05) × 10−3 M −1 (K d = 0.26 ± 0.07 mM), respectively, and the number of binding sites per PPL monomer was 3.1 ± 0.5. These values are in the same range as those measured by isothermal titration calorimetry and the slight differences can be attributed to the loss of degrees of freedom of rotation/diffusion of PPL upon immobilization.
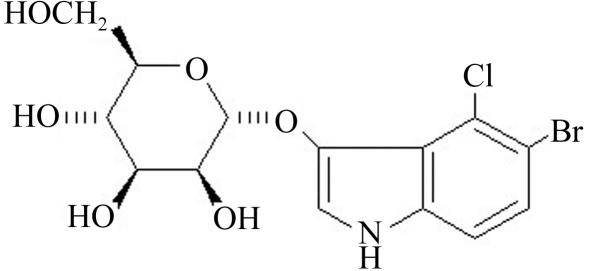
The rationale for characterizing the binding of the X-mannose (Fig. 2 ▶) was to evaluate the possibility of using this sugar as a carrier of Br (an anomalous X-ray scatterer) for MAD (multiple-wavelength anomalous dispersion) experiments in order to solve the X-ray crystal structure of the P. platycephala lectin, since the molecular-replacement approach using the available coordinates of single β-prism domains (PDB codes http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1c3k, http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1ouw, http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1j4s, http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1j4t and http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1j4u) failed (Gallego del Sol et al., 2002 ▶). Buts et al. (2003 ▶) reported a similar approach, using a selenium derivative of N-acetyl-d-glucosamine, to solve the structure of the bacterial F17-G lectin domain by MAD phasing. The 5-bromo-4-chloro-3-indolyl-α-d-mannose molecule shown in Fig. 2 ▶ was chosen as a Br-containing potential ligand of the P. platycephala lectin because according to the structures of other Man-specific β-prism lectins (Bourne et al., 1999 ▶, 2004 ▶; Pratap et al., 2002 ▶), the derivatized anomeric O1 atom is not expected to be involved in direct interactions with the protein. On the other hand, the O3, O4, O5 and O6 hydroxyl groups of mannose participate in a network of hydrogen bonds with residues of heltuba, artocarpin and calsepa.
Figure 2.
Schematic drawing of the structure of the 5-bromo-4-chloro-3-indolyl derivative of α-d-mannose used to for MAD phasing. The axial 3-OH and the equatorial 4-OH of mannose used by lectins as a major factor for discriminating between mannose/glucose and galactose (axial 4-OH) (Elgavish & Shaanan, 1997 ▶) are numbered.
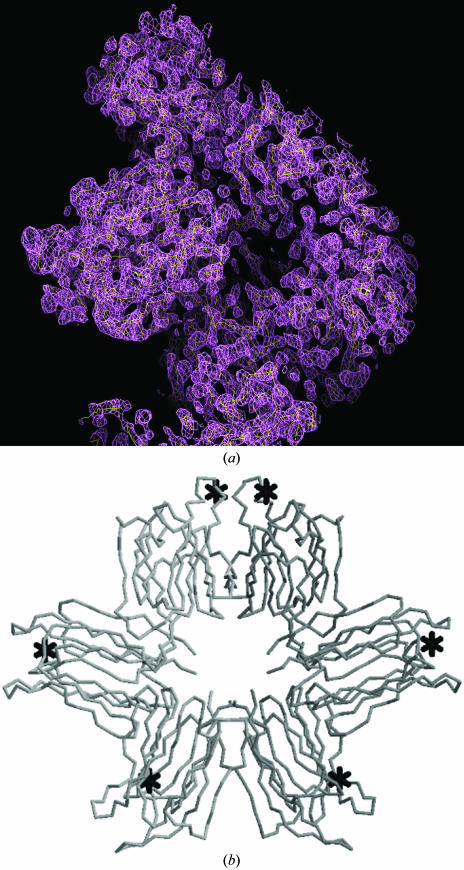
We set up crystallization trials of the P. platycephala lectin in the presence of 3 mM (10 × K d) X-mannose. Crystals suitable for X-ray diffraction experiments (maximal dimensions of 0.6 × 0.4 × 0.4 mm) grew in about five weeks at 295 K. The crystals belong to the monoclinic P21 space group (unit-cell parameters a = 80.2, b = 114.2, c = 80.3 Å, α = β = 90, γ = 119.9°) and diffracted to 2.5 Å resolution. A complete MAD data set, performed at the absorption edge (0.91995 Å), peak (0.91932 Å) and remote (0.92191 Å) wavelengths of Br as determined by a fluorescence scan for bromide, was collected at the BM16 beamline of ESRF (Grenoble). Two lectin molecules occupied the asymmetric unit, which had a solvent content of approximately 61% (corresponding to a Matthews coefficient, V M, of 3.20 Å3 Da−1); six Br atoms were found in the unit cell using the program SOLVE v.2.08 (Terwilliger & Berendzen, 1999 ▶). The overall figure of merit for phasing was 0.45. The data confirmed the presence of three X-mannose-binding sites per P. platycephala lectin molecule. After density modification using the program RESOLVE (Terwilliger, 2000 ▶), an electron-density map was obtained (Fig. 2 ▶). RESOLVE built 457 amino-acid residues in the experimental electron-density map of the P21 unit cell. The solution showed a circular arrangement of six β-prisms (Fig. 3 ▶), suggesting that the two subunits of the P. platycephala lectin homodimer adopt a toroid-like structure resembling the shape of the octameric assembly observed in crystals of the H. tuberosus lectin (Bourne et al., 1999 ▶). A detailed description of the quaternary structure of the P. platycephala lectin, including the relative orientation of and the interacting surface between the two subunits and the architecture of the six mannose-binding sites, awaits the refinement of the crystal structure, which is under way in our laboratory.
Figure 3.
(a) Electron-density map of a P. platycephala lectin dimer obtained by MAD using the Br of X-mannose as an anomalous scatterer. The map was calculated with the program FFT (CCP4 package) using the phases calculated after density modification and contouring at 1σ. The map was skeletonized using MAPMAN (base level for the electron density 1.5σ and step size for the skeletonization 0.5σ). (b) Cα-backbone tracing of the six β-prism domains building up a P. platycephala lectin dimer in the asymmetric cell unit of the monoclinic crystal. The position of the six Br atoms, presumably near the monosaccharide binding sites, are indicated by asterisks.
Acknowledgments
This work was supported in part by grants BMC2001/3337, BFU2004-01432/BMC (to JJC) and BIO2001/1081 (to JG) from the Ministerio de Ciencia y Tecnología, Madrid, Spain. The authors wish to thank Professor Pedro Alzari (Institut Pasteur, Paris) for providing laboratory facilities during the short-term stay of FGS in his Department. FGS is a recipient of a Beca de Formación de Personal Investigador from the Ministerio de Ciencia y Tecnología, Madrid, Spain. BSC is a senior investigator from CNPq (Conselho Nacional de Desenvovimento Científico e Tecnológico, Brazil).
References
- Barre, A., Bourne, Y., Van Damme, E. J. M., Peumans, W. J. & Rougé, P. (2001). Biochimie, 83, 645–651. [DOI] [PubMed] [Google Scholar]
- Barre, A., Peumans, W. J., Rossignol, M., Borderies, G., Culerrier, R., Van Damme, E. J. M. & Rougé, P. (2004). Biochimie, 86, 685–691. [DOI] [PubMed] [Google Scholar]
- Bouckaert, J., Hamelryck, T., Wyns, L. & Loris, R. (1999). Curr. Opin. Struct. Biol.9, 572–577. [DOI] [PubMed] [Google Scholar]
- Bourne, Y., Astoul, C. H., Zamboni, V., Peumans, W. J., Menu-Bouaouiche, L., Van Damme, E. J. M., Barre, A. & Rougé, P. (2002). Biochem. J.364, 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne, Y., Roig-Zamboni, V., Barre, A., Peumans, W. J., Astoul, C. H., Van Damme, E. J. M. & Rougé, P. (2004). J. Biol. Chem.279, 527–533. [DOI] [PubMed] [Google Scholar]
- Bourne, Y., Zamboni, V., Barre, A., Peumans, W. J., van Damme, E. J. M. & Rougé, P. (1999). Structure, 7, 1473–1482. [DOI] [PubMed] [Google Scholar]
- Buts, L., Loris, R., De Genst, E., Oscarson, S., Lahmann, M., Brosens, E., Wyns, L., De Greve, H. & Bouckaert, J. (2003). Acta Cryst. D59, 1012–1015. [DOI] [PubMed] [Google Scholar]
- Cavada, B. S., Madeira, S. V. F., Calvete, J. J., Sousa, L. A. G., Bomfim, L. R., Dantas, A. R., Lopes, M. C., Grangeiro, T. B., Freitas, B. T., Pinto, V. P. T., Leite, K. B. & Ramos, M. V. (2000). Prep. Biochem. Biotechnol.30, 271–280. [DOI] [PubMed] [Google Scholar]
- Cavada, B. S., Santos, C. F., Grangeiro, T. B., Moreira da Silva, L. I. M., Campos, M. J. O., de Sousa, F. A. M. & Calvete, J. J. (1997). Physiol. Mol. Biol. Plants, 3, 109–115.
- Chervenak, M. C. & Toone, E. J. (1995). Biochemistry, 34, 5685–5695. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Dam, T. K., Cavada, B. S., Grangeiro, T. B., Santos, C. F., Ceccatto, V. M., Sousa, F. A. M., Oscarson, S. & Brewer, C. F. (2000). J. Biol. Chem.275, 16119–16126. [DOI] [PubMed] [Google Scholar]
- Dam, T. K., Cavada, B. S., Grangeiro, T. B., Santos, C. F., Sousa, F. A. M., Oscarson, S. & Brewer, C. F. (1998). J. Biol. Chem.273, 12082–12088. [DOI] [PubMed] [Google Scholar]
- Dam, T. K., Roy, R., Das, S. K., Oscarson, S. & Brewer, C. F. (2000). J. Biol. Chem.275, 14223–14230. [DOI] [PubMed] [Google Scholar]
- Elgavish, S. & Shaanan, B. (1997). Trends Biochem. Sci.22, 462–467. [DOI] [PubMed] [Google Scholar]
- Gabius, H.-J. & Gabius, S. (1997). Editors. Glycosciences: Status and Perspectives. Weinheim: Chapman & Hall.
- Gallego del Sol, F., Ramón-Maiques, S., Santos, C. F., Grangeiro, T. B., Nagano, C. S., Farias, C. M. S. A., Cavada, B. S. & Calvete, J. J. (2002). Acta Cryst. D58, 167–169. [DOI] [PubMed]
- Heywood, V. H. (1979). Chemotaxonomy of the Leguminosae, edited by J. B. Harborne & D. Boulter, pp. 1–29. London: Academic Press.
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Jeyaprakash, A. A., Srivastav, A., Surolia, A. & Vijayan, M. (2004). J. Mol. Biol.338, 757–770. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G. J. & Jones, T. A. (1996). Acta Cryst. D52, 826–828. [DOI] [PubMed] [Google Scholar]
- Lemieux, R. U., Delbaere, L. T. J., Beierbeck, H. & Spohr, U. (1991). Ciba Found. Symp.158, 231–248. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1997). MOSFLM User’s Guide: MOSFLM Version 5.50. Laboratory of Molecular Biology, Cambridge.
- Loris, R., Hamelryck, T., Bouckaert, J. & Wyns, L. (1998). Biochim. Biophys. Acta, 1383, 9–36. [DOI] [PubMed] [Google Scholar]
- Mandal, D. K., Kishore, N. & Brewer, C. F. (1994). Biochemistry, 33, 1149–1156. [DOI] [PubMed] [Google Scholar]
- Mann, K., Farias, C. M. S. A., Gallego Del Sol, F., Santos, C. F., Grangeiro, T. B., Nagano, C. S., Cavada, B. S. & Calvete, J. J. (2001). Eur. J. Biochem.218, 4414–4422. [DOI] [PubMed]
- Pratap, J. V., Jeyaprakash, A. A., Rani, P. G., Sekar, K., Surolia, A. & Vijayan, M. (2002). J. Mol. Biol.317, 237–247. [DOI] [PubMed] [Google Scholar]
- Ramos, M. V., Cavada, B. S., Bomfim, L. R., Debray, H., Mazard, A.-M., Calvete, J. J., Grangeiro, T. B. & Rougé, P. (1999). Protein Pept. Lett.6, 215–222.
- Rini, J. M. (1995). Annu. Rev. Biomol. Struct.24, 551–577. [DOI] [PubMed]
- Srinivas, V. R., Reddy, G. B., Ahmad, N., Swaminathan, C. P., Mitra, N. & Surolia, A. (2001). Biochim. Biophys. Acta, 1527, 102–111. [DOI] [PubMed] [Google Scholar]
- Suvachittanont, W. & Peutpaiboon, A. (1992). Phytochemistry, 31, 4065–4070.
- Terwilliger, T. C. (2000). Acta Cryst. D56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utarabhand, P. & Akkayanont, P. (1995). Phytochemistry, 38, 281–285.
- Van Damme, E. J. M., Peumans, W. J., Barre, A. & Rougé, P. (1998). Crit. Rev. Plant Sci.17, 575–692.
- Vijayan, M. & Chandra, N. (1999). Curr. Opin. Struct. Biol.9, 707–714. [DOI] [PubMed] [Google Scholar]
- Weis, W. I. & Drickamer, K. (1996). Annu. Rev. Biochem.65, 441–473. [DOI] [PubMed] [Google Scholar]
- Wiseman, T., Williston, S., Brandts, J. F. & Lin, L. N. (1989). Anal. Biochem.179, 131–137. [DOI] [PubMed] [Google Scholar]
- Yagi, F., Iwaya, T., Haraguchi, T. & Goldstein, I. J. (2002). Eur. J. Biochem.269, 4335–4341. [DOI] [PubMed] [Google Scholar]
- Zhang, W., Peumans, W. J., Barre, A., Astoul, C. H., Rovira, P., Rougé, P., Proost, P., Truffa-Bachi, P., Jalali, A. A. H. & Van Damme, E. J. M. (2000). Planta, 210, 970–978. [DOI] [PubMed] [Google Scholar]