The crystallization of a hypothetical acetyltransferase from the archaeon P. furiosus by hanging-drop vapour diffusion is reported in space group P622.
Keywords: GCN5-related N-acetyltransferase, archaea, Pyrococcus furiosus
Abstract
The GCN5-related N-acetyltransferase (GNAT) superfamily has a primordial role in cellular processes such as transcription initiation and regulation by histone acetylation, aminoglycoside resistance and melatonin metabolism. To date, no acetyltransferase from the archaeal domain of life has been studied. This paper describes the cloning, expression, purification and crystallization of a Pyrococcus furiosus hypothetical acetyltransferase PfGNAT (MW = 22 007 Da). The crystals belong to space group P622, with one molecule in the asymmetric unit and unit-cell parameters a = b = 82.6, c = 105.92 Å, α = β = 90, γ = 120°. Crystals diffract X-rays to 3.0 Å resolution on a synchrotron-radiation source. Determination of this structure will provide new insights into the substrate-specificity of this acetyltransferase and the thermal stability of the N-acetyltransferase domain.
1. Introduction
The GCN5-related N-acetyltransferase (GNAT) superfamily catalyzes the transfer of the acetyl group from acetyl coenzyme A to the primary amine of a wide variety of substrates such as histones (Grunstein, 1997 ▶), aminoglycosides (Wright, 1999 ▶) and arylalkylamines (Zheng & Cole, 2002 ▶). This family is present in all kingdoms of life and the structural characterization of more than two dozen members of the family has revealed a common fold for the acetyltransferase domain despite low sequence similarities (Neuwald & Landsman, 1997 ▶; Dyda et al., 2000 ▶). The N-acetyltransferase domain is composed of four structurally conserved motifs, named A, B, C and D, which consist of a six-stranded mixed-polarity β-sheet with two parallel strands and four α-helices. This conserved domain can be duplicated and can also be specific of other acyl-group donors, as has been shown from the structure determination of several proteins including the FemABX family, the N-myristoyltransferases and the mycothiol synthase from Mycobacterium tuberculosis (Biarrotte-Sorin et al., 2004 ▶; Weston et al., 1998 ▶; Vetting et al., 2003 ▶).
To date, no biological and structural studies have been performed on an archaeal acetyltransferase. The Pfam database version 16.0 (http://www.sanger.ac.uk/Software/Pfam/) contains roughly 5000 members of the GNAT superfamily from the three domains of life, with 200 from archaeal species (Robb et al., 2001 ▶). Of the 2053 proteins of Pyrococcus furiosus genome, nine belong to this superfamily. P. furiosus is a hyperthermophilic archaeon isolated from thermally active regions with an optimal growth temperature of 373 K.
Here, we describe the cloning, expression, purification and crystallization of the 176-residue hypothetical acetyltransferase of P. furiosus (PfGNAT) encoded by the gene pf1456. The structure determination of PfGNAT will be an approach to determine the substrate-specificity of this protein and to understand the stabilizing interactions that govern the intrinsic thermal stability of its N-acetyltransferase domain.
2. Materials and methods
2.1. Cloning, expression and purification
The predicted open reading frame coding for the hypothetical acetyltransferase PfGNAT (PF1456, accession No. NC_003413.1) was amplified by PCR from a sample of P. furiosus DSM3638 generously donated by Professor Robert Kelly. The PCR-amplified fragment was cloned into the pPCR-ScriptTM Cam SK(+) vector (Stratagene). The DNA was then subcloned into pET28a(+) (Novagen) in order to give a hexa-His tag at the C-terminal part of the expressed protein. The PF1456-encoding plasmid was transformed into Escherichia coli strain BL21-CodonPlus (DE3)-RIL (Stratagene) and the cells were grown at 310 K in LB medium supplemented with 30 µg ml−1 kanamycin and 40 µg ml−1 chloramphenicol. Protein overproduction was induced at an OD600 of 0.6–0.8 with 1 mM IPTG and growth continued for an additional 6 h. Following harvesting, the cells were resuspended in buffer A (20 mM Tris pH 8.0, 10 mM imidazole pH 8.0, 150 mM NaCl) and lysed by sonication. The cell lysate obtained by centrifugation at 38 000g for 45 min was heated at 353 K for 30 min. After an additional centrifugation for 45 min at 3 000g, the supernatant was loaded onto an Ni2+-charged Hi-trap column (Qiagen) and washed first with buffer A supplemented with 1 M NaCl and then with buffer A supplemented with 50 mM imidazole. The recombinant protein was finally eluted with 20 mM Tris pH 8.0, 330 mM imidazole pH 8.0 and 150 mM NaCl. Fractions containing PfGNAT were concentrated, loaded onto a Superdex-75 gel-filtration column (Amersham Biosciences) and eluted with 20 mM Tris pH 8.0 and 150 mM NaCl as a species with an apparent molecular weight of approximately 19 kDa (the calculated molecular weight of the His-tagged recombinant protein is 22 kDa and a tag of sequence LEHHHHHH remains on the C-terminus). These observations indicate that PfGNAT is a monomer.
2.2. Crystallization
The protein was concentrated to 26 mg ml−1 in 150 mM NaCl and 20 mM Tris pH 8.0 using an Amicon Ultra-15 (Millipore) and preincubated overnight with 2 mM acetyl coenzyme A (Sigma). Crystallization attempts took place using commercially available crystallization screens from Hampton Research and Molecular Dimensions Ltd with the sitting-drop vapour-diffusion method at 291 K. Thin stick-like crystals were observed with 0.2 M lithium sulfate monohydrate, 0.1 M Tris–HCl pH 8.5 and 30%(w/v) polyethylene glycol (PEG) 4K.
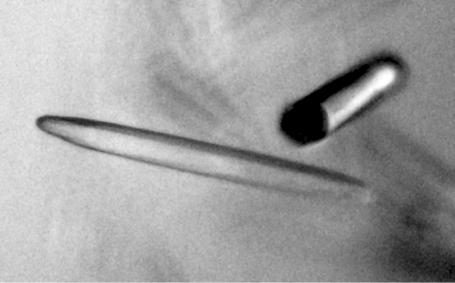
This initial condition was then optimized by variations of the concentration and the molecular weight of the PEG and of the concentration of lithium sulfate monohydrate. Crystals suitable for diffraction experiments were grown by mixing 2 µl protein solution with 2 µl 0.1 M Tris–HCl pH 8.5 and 25%(w/v) PEG 6K and reached maximum dimensions after 6 d (400 × 50 × 30 µm; Fig. 1 ▶).
Figure 1.
Typical stick-like crystals of P. furiosus putative N-acetyltransferase with dimensions of 0.4 × 0.05 × 0.03 mm.
2.3. Data collection
Crystals were flash-frozen with 85 mM Tris–HCl pH 8.5, 15.5%(w/v) PEG 4K and 15%(v/v) glycerol. Diffraction data were collected using a MAR CCD detector on beamline FIP-BM30A of the European Synchrotron Radiation Facility, Grenoble (Roth et al., 2002 ▶). The data set was collected at 3.0 Å resolution with a 120° sweep. Data were processed using XDS and XSCALE (Kabsch, 1988 ▶). Autoindexing and consideration of systematically absent reflections revealed the crystal to belong to the hexagonal space group P622, with one molecule in the asymmetric unit (V M = 2.37 Å3 Da−1) and 48.1% solvent content. The data-collection statistics are summarized in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell (3.2–3.0 Å).
| X-ray source | FIP-BM30A |
| Space group | P622 |
| Unit-cell parameters (Å) | a = b = 82.60, c = 105.92 |
| Wavelength (Å) | 0.9795 |
| Resolution range (Å) | 27-3 |
| No. of observations | 60747 |
| No. of unique reflections | 4610 |
| Completeness (%) | 98.6 (99.0) |
| Redundancy | 12.9 (13.1) |
| Rsym† (%) | 4.7 (16.8) |
| 〈I/σ(I)〉 | 21.7 (6.1) |
R
sym = 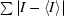 /
/ , where I is the intensity of an individual reflection and 〈I〉 is its mean value.
, where I is the intensity of an individual reflection and 〈I〉 is its mean value.
3. Conclusion
Pfam version 16.0 and an additional multiple sequence alignment predicted the hypothetical protein encoded by the gene pf1456 to be an acetyltransferase. The resulting thermostable protein was expressed and purified. Crystals were grown in the presence of acetyl coenzyme A in the mother liquor, which is presumed to be the acetyl group donor. As the N-acetyltransferase fold is highly conserved, we are attempting to solve the structure of PfGNAT by molecular replacement using AMoRe (Navaza, 1994 ▶) with the Bacillus subtilis putative phosphinothricin N-acetyltransferase (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1vhs; 33% sequence similarity) as a model.
Acknowledgments
We thank Dr Michel Pirrochi from the FIP-BM30A beamline at the ESRF for assistance with data collection and Sabrina Tachdjian and Professor Robert Kelly for the generous gift of P. furiosus genomic DNA.
References
- Biarrotte-Sorin, S., Maillard, A. P., Delettre, J., Sougakoff, W., Arthur, M. & Mayer, C. (2004). Structure, 12, 1–20. [DOI] [PubMed] [Google Scholar]
- Dyda, F., Klein, D. C. & Burgess-Hickmann, A. (2000). Annu. Rev. Biophys. Struct.29, 81–103. [DOI] [PMC free article] [PubMed]
- Grunstein, M. (1997). Nature (London), 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. (1988). J. Appl. Cryst.21, 67–71. [Google Scholar]
- Navaza, J. (1994). Acta Cryst A50, 157–163. [Google Scholar]
- Neuwald, A. F. & Landsman, D. (1997). Trends Biochem. Sci.22, 154–155. [DOI] [PubMed] [Google Scholar]
- Robb, F. T., Maeder, D. L., Brown, J. R, DiRuggiero, J., Stump, M. D., Yeh, R. K., Weiss, R. B. & Dunn, D. M. (2001). Methods Enzymol.330, 134–157. [DOI] [PubMed] [Google Scholar]
- Roth, M., Carpentier, P., Kaikati, O., Joly, J., Charrault, P., Pirocchi, M., Kahn, R., Fanchon, E., Jacquamet, L., Borel, F. & Ferrer, J. L. (2002). Acta Cryst. D58, 805–814. [DOI] [PubMed] [Google Scholar]
- Vetting, M. W., Roderick, S. L., Yu, M. & Blanchard, J. S. (2003). Protein Sci.12, 1954–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston, S. A., Camble, R., Colls, J., Rosenbrock, G., Taylor, I., Egerton, M., Tucker, A. D., Tunnicliffe, A., Mistry, A., Mancia, F., de La Fortelle, E., Irwin, J., Bricogne, G. & Pauptit, R. A. (1998). Nature Struct. Biol.5, 213–221. [DOI] [PubMed] [Google Scholar]
- Wright, G. D. (1999). Curr. Opin. Microbiol.2, 499–503. [DOI] [PubMed] [Google Scholar]
- Zheng, W. & Cole, P. A. (2002). Curr. Med. Chem.9, 1187–1199. [DOI] [PubMed] [Google Scholar]