Two outer membrane factor family proteins, OprM and OprN, from a tripartite efflux pump found in P. aeruginosa were crystallized. A diffraction data set was collected to 3.8 Å resolution in the space group C2 for OprM crystals.
Keywords: Efflux pumps, antibiotic resistance, outer membrane factor family
Abstract
OprM and OprN belong to the outer membrane factor family proteins. These ∼52 kDa proteins are part of the tripartite efflux pumps found in Pseudomonas aeruginosa and are responsible in part for the antibiotic resistance observed in these bacteria. Both proteins have been expressed in Escherichia coli as His-tag proteins and purified accordingly by affinity chromatography in the presence of n-octyl-β-d-glucopyranoside detergent. OprM and OprN were crystallized using PEG 20 000/ammonium citrate and ammonium sulfate as precipitating agents, respectively. Crystals belong to space group C2, with unit-cell parameters a = 152.6, b = 87.9, c = 355.9 Å, β = 98.9° and a = 151.3, b = 87.6, c = 356.5 Å, β = 98.1° for OprM and OprN, respectively. Using the ESRF synchrotron-radiation source, OprM diffraction data extended to 3.4 Å.
1. Introduction
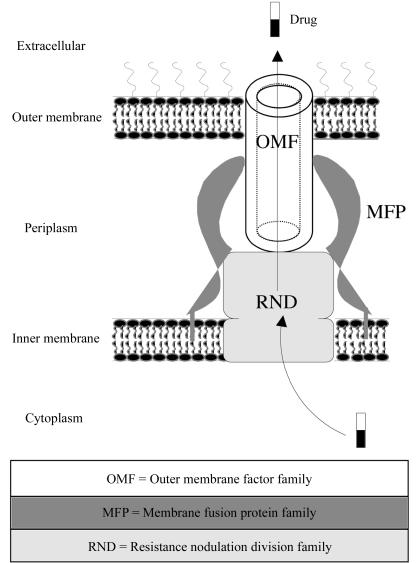
One of the current priorities of hospitals and research institutions is countering the emergence of multidrug-resistant bacteria. Pseudomonas aeruginosa is an opportunistic pathogen and a leading cause of hospital-acquired infections associated with high morbidity and mortality. This organism uses several processes to resist antibiotics, one of which is active efflux at the cellular membrane level (McDermott et al., 2003 ▶). P. aeruginosa is a Gram-negative organism that is naturally not very sensitive to antibiotics. Its complete genome has been sequenced (Stover et al., 2000 ▶), showing 12 distinct genetic efflux-pump systems organized into operons (Schweizer, 2003 ▶), seven of which have been confirmed genetically (Poole et al., 1993 ▶, 1996 ▶; Li et al., 1995 ▶, 2003 ▶; Köhler et al., 1997 ▶; Mine et al., 1999 ▶; Westbrock-Wadman et al., 1999 ▶; Aendekerk et al., 2002 ▶; Chuanchuen et al., 2002 ▶). Each pump is composed of three proteins (Fig. 1 ▶): an inner membrane protein belonging to the RND (resistance nodulation and cell division) family which acts as a proton-driving force pump, an outer membrane protein which belongs to the OMF family (outer membrane factor) and a third protein which connects the other two. The latter is located in the periplasmic compartment and belongs to the MFP (membrane fusion protein) family. These seven different pumps lead to different phenotypes and confer resistance to most β-lactams, quinolones, aminoglycosides, macrolides, fourth-class cephalosporines, trimethoprime sulfamides, tetracyclines, chloramphenicols and erythromycins (Schweizer, 2003 ▶). Our work focuses on the study of two particular pumps: OprM-MexAB and OprN-MexEF. The OprM-MexAB pump is responsible for the efflux of many antibiotics as well as ethidium bromide, homoserine lactone and SDS. The OprN-MexEF pump works on a narrower spectrum of antibiotics. In order to understand the link between the pump architecture and its specificity, determination of the three-dimensional structure of the various components of these pumps has been undertaken.
Figure 1.
Schematic representation of an efflux pump constituted of three different proteins: one outer membrane protein (OMF family), one inner membrane protein acting as a pump (RND family) and one periplasmic protein anchored in the inner membrane (OMF family) connecting the first two.
In this preliminary crystallographic study, results from the outer membrane proteins OprM and OprN are presented. The mature proteins are composed of 468 and 453 amino acids, respectively, with 29% identity. Their folding is thought to be homologous to the TolC structure from Escherichia coli (Koronakis et al., 2000 ▶), although the identity between OprM and TolC is only 18%.
2. Protein cloning, expression and purification
A previously developed protocol for OprM cloning and purification (Charbonnier et al., 2001 ▶) did not yield protein that was amenable to crystallization. Consequently, new constructs were designed. The oprM and oprN genes of P. aeruginosa (PAO1 strain) were generated separately as NdeI-XmaI fragments with a polyhistidine tag at their C-termini by PCR using plasmids pOM1 (mexAB-oprM) and pKMJ002 (mexEF-oprN), respectively, as template DNA (Köhler et al., 1997 ▶). After restriction-enzyme digestion, each gene was cloned under the control of an arabinose-inducible promoter in the expression vector pBAD33-GFP (Guzman et al., 1995 ▶; Benabdelhak et al., unpublished data). PCR primers were as follows. oprM: forward, GGAATTCCATATGAAACGGTCCTTCCTTTCC; reverse, TCCCCCCGGGTCAGTGATGGTGATGGTGATGAGCCTGGGGATCTTCCTTCTTCGCGGTCTG. oprN: forward, GGAATTCCATATGATTCACGCGCAGTCGATCCGGAGCGGG, reverse, TCCCCCCGGGTCAGTGATGGTGATGGTGATGGGCGCTGGGTTGCCAGCCACCCCGAG.
Plasmids were expressed in E. coli strain C43 (DE3) (Miroux & Walker, 1996 ▶). Cultures were grown at 303 K in LB medium containing 2 µg ml−1 chloramphenicol until A 600 = 2. Cells were induced by the addition of 2 mM arabinose and harvested 2 h post-induction. The cell pellet was resuspended in 20 mM Tris–HCl pH 8, 5 mM MgCl2 and 50 units of benzonase (Promega). Cells were lysed with a French press at 69 MPa and then centrifuged twice for 30 min at 8500g to remove inclusion bodies and unbroken cells. The cytoplasmic fraction was applied to a step gradient of sucrose (0.5 and 1.5 M) and then centrifuged for 3 h at 200 000g and 277 K. The pellet corresponding to the outer membrane fraction was resuspended in 20 mM Tris–HCl pH 8, 10%(v/v) glycerol, 2%(w/v) octyl-β-d-glucoside (βOG; Anatrace, USA) and then stirred overnight at 296 K. The solubilized membrane proteins were recovered by centrifugation for 30 min at 50 000g before loading onto an Ni–NTA resin column pre-equilibrated in 20 mM Tris pH 8, 10%(v/v) glycerol, 0.9%(w/v) βOG (buffer A). The column was washed with buffer A plus 10 mM imidazole before eluting with a linear gradient of imidazole (10–400 mM) at a flow rate of 5 ml min−1. The fractions containing the OprM or OprN protein, eluted between 100 and 250 mM imidazole, were pooled and exchanged for a suitable buffer. Finally, each protein was concentrated to 6 mg ml−1 on an Amicon Ultra centrifugal filter (molecular-weight cutoff 30 kDa; Millipore) in the presence of 10% glycerol, 20 mM Tris–HCl pH 8.0, 0.9% βOG.
All purification steps were analyzed on SDS–polyacrylamide gels (Laemmli, 1970 ▶). In the final step, the protein samples were loaded with and without heating in order to reveal different oligomeric forms of the proteins, which both appeared as trimers. The quality of the proteins was also analyzed by Western blotting using an anti-histidine antibody.
3. Crystallization
Crystallization trials were performed using the hanging-drop vapour-diffusion method, equilibrating drops of various volumes against 500 µl reservoir solution. Initial crystallization attempts were performed at 291 K using Memfac and Crystal Screens I and II (Hampton Research), which led to microcrystalline material under several conditions.
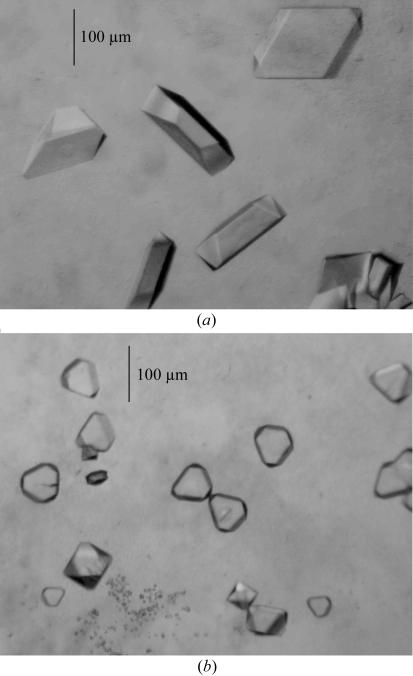
For OprM, optimization of the crystallization conditions was performed varying the pH, ionic strength, precipitant concentration, temperature, cationic compounds, additives and protein:reservoir volume ratio. Because OprM crystals break when transferred into cryoprotectant solutions, various cryoprotectants at different concentrations were tested as additives in the reservoir solution. The optimized crystallization conditions for OprM were 100 mM sodium acetate pH 4.5, 6%(w/v) PEG 20 000, 300 mM ammonium citrate, 25–30%(v/v) glycerol and 0.9%(w/v) β-OG at 298 K. In each hanging drop, the protein and reservoir were mixed in the ratio 3:1.5 µl. Both PEG 20 000 and ammonium citrate were required for crystallization. Variation of the concentration of PEG or citrate affects two perpendicular directions of the growing crystal. Crystals of 80 µm appear in 4 days but then require two months to reach 250 µm in the longest dimension (Fig. 2 ▶ a).
Figure 2.
(a) Crystals of OprM obtained in 100 mM sodium acetate pH 4.5, 6%(w/v) PEG 20 000, 300 mM ammonium citrate, 25–30% glycerol and 0.9% β-OG at 298 K. The protein and reservoir solutions were mixed in the ratio 3:1.5 µl. (b) Crystals of OprN obtained in 100 mM ADA pH 6.5, 0.8 M ammonium sulfate and 0.9% β-OG at 291 K.
The best crystals of OprN appeared using 100 mM ADA pH 6.5, 0.8 M ammonium sulfate and 0.9%(w/v) β-OG at 291 K, with maximum dimensions of 100 × 100 × 30 µm (Fig. 2 ▶ b). Unlike with OprM, co-crystallization in the presence of various additives or cryoprotectants in the crystallization solution hampered the growth of OprN crystals. Therefore, cryoprotectant was added prior to diffraction.
4. Preliminary X-ray data
Several crystals were tested using the PX beamline at SLS (Switzerland) or the ID14, ID29 and BM30 beamlines at ESRF (Grenoble, France). The best results were obtained using the ID29 beamline equipped with an ADSC detector with an exposure time of 10 s per degree. OprM crystals were flash-cooled directly from the crystallization drop. Prior to flash-cooling the OprN crystals, 25%(v/v) glycerol was directly added to the drop.
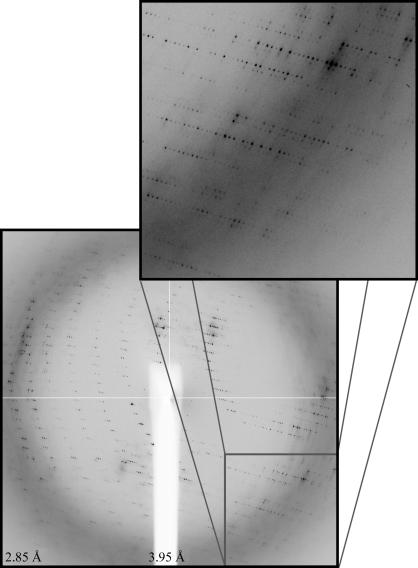
Both OprM and OprN crystals belong to space group C2, with nearly identical unit-cell parameters: a = 152.6, b = 87.9, c = 355.9 Å, β = 98.9° and a = 151.3, b = 87.6, c = 356.5 Å, β = 98.1°, respectively. OprM diffraction (Fig. 3 ▶) extends to 3.4 Å. The OprN diffraction pattern is quite similar, although limited to a maximum of 4 Å resolution. The rapid decay of the flash-cooled crystals of OprN prevented a complete data set from being recorded. This might be because of either their smaller size or inadequate cryoprotection of the crystals.
Figure 3.
OprM diffraction recorded using the ID29 beamline (ESRF, France). The frame was recorded with an oscillation of 1° per 10 s. The resolution limits at the detector edges are indicated. Enlargement of the square zone allows the reflections along the large c unit-cell axis to be distinguished.
A complete data set to 3.8 Å was recorded for OprM using the ID29 beamline (ESRF, France). X-ray data were processed using MOSFLM from the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶) with a mean refined mosaicity of 0.6° over the complete data set. Subsequent scaling and merging of intensities were carried out using SCALA. Data statistics are summarized in Table 1 ▶. Although reflections extend to 3.4 Å, the processing was limited to 3.8 Å owing to the anisotropy of the diffraction pattern. Because of the size of the c unit-cell parameter, the detector was set at a rather large distance (400 mm) from the crystal in order to separate each spot during the integration process. The R merge value for all resolution ranges is mainly a consequence of overlap of some integration boxes in the c* direction.
Table 1. Data-collection statistics for OprM crystals.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.0052 |
| Crystal-to-detector distance (mm) | 400 |
| Exposure time per degree (s) | 10 |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 152.6, b = 87.9, c = 355.9, β = 98.9 |
| Resolution (Å) | 88–3.8 (4.01–3.80) |
| No. of measured observations | 131301 (17427) |
| No. of unique reflections | 40566 (5260) |
| Data completeness (%) | 87.7 (77.9) |
| Data redundancy | 3.2 (3.3) |
| Rmerge† (%) | 12.2 (31.2) |
| I/σ(I) | 8.5 (3.3) |
R
merge = 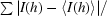
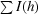 , where I(h) is the observed intensity and 〈I(h)〉 is the mean intensity of reflection h over all measurements of I(h).
, where I(h) is the observed intensity and 〈I(h)〉 is the mean intensity of reflection h over all measurements of I(h).
Attempt to solve the OprM structure by molecular replacement using AMoRe (Navaza, 2001 ▶) with the TolC structure (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1ek9) as a search model failed. During the course of our work, the OprM structure was published in space group R32 (Akama et al., 2004 ▶). It is worth noting that the same problem was encountered in the R32 space group, leading the authors to use the SIRAS method to solve the structure. In fact, the identity between OprM and TolC is only 18%.
The newly published OprM coordinates (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1wp1) were used successfully with Phaser (Storoni et al., 2004 ▶). The solution appeared by introducing only one molecule of the trimeric OprM, positioning two trimers in the asymmetric unit. This corresponds to a solvent content of 63.7%.
Refinement of the structure in the C2 space group is under way. In this space group, the three molecules of the trimer are not forced to be equivalent, owing to the absence of crystallographic threefold symmetry, leaving open the possibility of different conformers.
Acknowledgments
This work and HB were supported by the 5th Framework European Program, contract No. QLR-2000-01339. The authors thank the staff at ESRF ID14, ID29, BM30A and the SLS PX beamlines. mexAB-OprM and mexEF-OprN plasmids were a generous gift from Thilo Khöler at the department of Genetics and Microbiology, CMU 1, Geneva, Switzerland.
References
- Aendekerk, S., Ghysels, B., Cornelis, P. & Baysse, C. (2002). Microbiology, 148, 2371–2381. [DOI] [PubMed] [Google Scholar]
- Akama, H., Kanemaki, M., Yoshimura, M., Tsukihara, T., Kashiwagi, T., Yoneyama, H., Narita, S., Nakagawa, A. & Nakae, T. (2004). J. Biol. Chem.279, 52816–52819. [DOI] [PubMed] [Google Scholar]
- Charbonnier, F., Kohler, T., Pechere, J. & Ducruix, A. (2001). Protein Expr. Purif.23, 121–127. [DOI] [PubMed] [Google Scholar]
- Chuanchuen, R., Narasaki, C. T. & Schweizer, H. P. (2002). J. Bacteriol.184, 5036–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Guzman, L.-M., Belin, D., Carson, M. J. & Beckwith, J. (1995). J. Bacteriol.177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler, T., Michea-Hamzehpour, M., Henze, U., Gotoh, N., Curty, L. & Pechere, J. (1997). Mol. Microbiol.23, 345–354. [DOI] [PubMed] [Google Scholar]
- Koronakis, V., Sharff, A., Koronakis, E., Luisi, B. & Hughes, C. (2000). Nature (London), 405, 914–919. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Li, X.-Z., Nikaido, H. & Poole, K. (1995). Antimicrob. Agents Chemother.39, 1948–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Mima, T., Komori, Y., Morita, Y., Kuroda, T., Mizushima, T. & Tsuchiya, T. (2003). J. Antimicrob. Chemother.52, 572–575. [DOI] [PubMed] [Google Scholar]
- McDermott, P. F., Walker, R. D. & White, D. G. (2003). Int. J. Toxicol.22, 135–143. [DOI] [PubMed] [Google Scholar]
- Mine, T., Morita, Y., Kataoka, A., Mizushima, T. & Tsuchiya, T. (1999). Antimicrob. Agents Chemother.43, 415–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux, B. & Walker, J. (1996). J. Mol. Biol.260, 289–298. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (2001). Acta Cryst. D57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Poole, K., Krebes, K., McNally, C. & Neshat, S. (1993). J. Bacteriol.175, 7362–7372. [DOI] [PMC free article] [PubMed]
- Poole, K., Gotoh, N., Tsujimoto, H., Zhao, Q., Wada, A., Yamasaki, T., Neshat, S., Yamagishi, J., Li, X. Z. & Nishino, T. (1996). Mol. Microbiol.21, 713–724. [DOI] [PubMed] [Google Scholar]
- Schweizer, H. P. (2003). Genet. Mol. Res.2, 48–62. [PubMed] [Google Scholar]
- Storoni, L. C., McCoy, A. J & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed] [Google Scholar]
- Stover, C. K. et al. (2000). Nature (London), 406, 959–964. [Google Scholar]
- Westbrock-Wadman, S., Sherman, D. R., Hickey, M. J., Coulter, S. N., Zhu, Y. Q., Warrener, P., Nguyen, L. Y., Shawar, R. M., Folger, K. R. & Stover, C. K. (1999). Antimicrob. Agents Chemother.43, 2975–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]