Abstract
We have synthesized DNA segments with different handedness, twisting and radii of curvature, and have analyzed the effect of untwisting on them. The results indicate that the dynamic behavior of curved DNA upon untwisting is strongly determined by the initial sequence-dependent DNA trajectory. In particular, DNA with the same radii but with opposite handedness of superhelix twisting can show very different conformational responses to ethidium bromide untwisting. Upon treatment with ethidium bromide, right-handed superhelixes decrease their twist and increase the planarity of the superhelix, while left-handed superhelixes increase twisting and decrease their degree of planarity.
Sequence-dependent curvature of DNA (or bent DNA) has been associated with important biological processes (1–8). Our understanding of the role of curved DNA in these processes could be expanded by considering the sequence-dependent geometrical features of DNA and structural origin of DNA bending (7–12). Curved DNA does not correspond to a single shape but rather to a continuum of different three-dimensional (3D) shapes, characterized by different radii, handedness, and twisting of curved segments (11–13).
In the present work, we have analyzed some aspects of the relation between sequence-dependent DNA shape and ethidium bromide (EB)-induced untwisting of DNA. We show that inherent sequence-dependent structural features of DNA such as handedness and twisting of curved segments can be dramatically changed by an in vivo-relevant degree of DNA untwisting.
MATERIALS AND METHODS
Design and Synthesis of Well Defined Shapes of DNA.
DNA molecules containing bent sequence elements were designed using data from the literature (10, 11, 14–19). Ligation of monomer units was done using the following sequence design: (i) planar curved DNA [21-mer (AAAAAGGCCCAAAAAGGGCCC)]; (ii) superhelix DNAs with similar radii of curvature, but with opposite handedness [20-mer (AAAAAGGGCCAAAAAGGGCC) versus 22-mer (AAAAAGGGCCCAAAAAAGGCCC), and 19-mer (AAAAAGGCCAAAAAGGCCC) versus 23-mer (AAAAAGGGCCCAAAAAAGGGCCC)]; (iii) superhelix DNAs with the same handedness, but with different radii and macro-twisting of the superhelix (19-mer versus 20-mer and 22-mer versus 23-mer, as above). As a straight DNA control, we used (CA)n sequence repeat lengths (19- to 23-mers). All oligonucleotides were synthesized using an Applied Biosystems model 381 A Oligonucleotide Synthesizer and were purified by 8% polyacrilamide gel electrophoresis. The purified oligonucleotides were 5′ end-labeled as described (19), complementary strands were mixed, heated to 90°C, and slowly cooled to form hybrids. The ligations were done as described (20, 21).
Gel Mobility Assay.
Electrophoresis was done using 8% polyacrylamide gels (20). The applied voltage was 5 V/cm. In the “untwisting assay” the electrophoresis was done in the presence of 0.005 mg/ml of EB in the gel and the gel-running buffer. The buffer in the electrode compartments was recirculated, as described by Diekmann (20).
Cyclization Experiments.
The experiments were done as described (21, 22). Circles were detected by exonuclease III digestion (23). “Untwisting assay” was done using different EB concentrations in 10× increments from 0.01 μg/ml to 1 mg/ml.
RESULTS
Our experimental design was based on differential phasing of GGCC and AAAAA sequence elements. These sequence motifs have a high net roll-angle difference (9–11, 18, 19, 24), and one can produce a variety of different 3D trajectories using the same sequence elements (12–14, 17, 25, 26). Curved monomer units that are smaller than the helical pitch (10.5 bp on average) will form inherently left-handed superhelixes after concatemerization, and the handedness of the superhelix will be the opposite if the curved monomer units are longer than the helical pitch. Each type of designed monomer unit was ligated to give multimers of various lengths. To monitor the untwisting-induced change in the 3D shape of curved DNA, we used gel mobility and cyclization assays. Both assays were done under stress-free (no EB) and untwisting (with EB) conditions (see Materials and Methods).
Gel Mobility Assay.
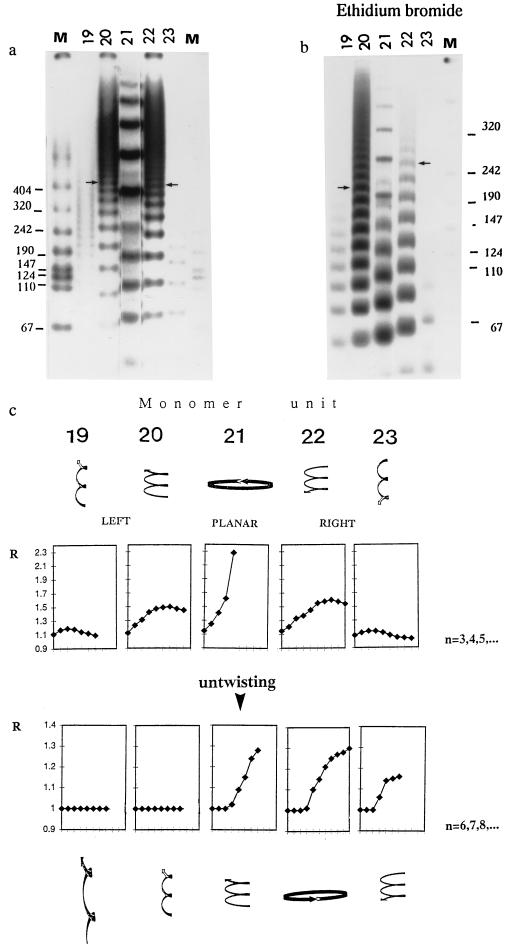
The differences in 3D trajectory of curved DNA resulted in differences in electrophoretic mobility as illustrated in Fig. 1. Under stress-free conditions, the 3D trajectory formed by multimers with the same radii of curvature but with opposite handedness of twisting, showed a similar mobility anomaly, expressed as the coefficient of retardation (R). (Compare 20-mers to 22-mers and 19-mers to 23-mers, Fig. 1 a and c Upper.) The “plateau region” of the R versus n plots (13), where “n” represents the multimer of ligated monomer units, presented in Fig. 1c, indicates the onset of one full turn of superhelix. The “plateau region” starts near 100 bp for ligated 19- and 23-mers and around 180 bp for 20- and 22-mers, before untwisting (Fig. 1c Upper). Thus the 19- and 23-mers show smaller “plateau values” of R versus n, than 20- and 22-mers. This indicates that 19-mers and 23-mers have smaller radii but a higher twisting of superhelixes. To simulate untwisting conditions, EB was introduced in the gel and gel-running buffer. After adding EB, the mobility anomaly between superhelixes, which are designed to have initially the same radii of curvature but the opposite chirality of twisting, was significantly different (compare the position of the arrows in Fig. 1b). Only planar (21-mers) and right-handed curved DNA fragments (22- and 23-mers) were showing a mobility anomaly after untwisting (bottom of Fig. 1c). The left-handed curved 19- and 20-mers did not reveal any retardation for all multimer lengths (R = 1). In the presence of EB the start of the “plateau region” was shifted toward longer DNA lengths compared with the stress-free conditions. Note that the scale of the n values is higher for the bottom (n = 6, 7, 8, … ) compared with the Fig. 1c Upper (n = 3, 4, 5, … ).
Figure 1.
Influence of untwisting on the gel mobility anomaly in curved DNA molecules (19- to 23-mers) with well-defined 3D shape: (a) Gel mobility under stress-free conditions (no EB). Monomer units used in ligations (19- to 23-mers) are designated at the top of each gel lane. Lane M is the molecular weight marker VII (Boehringer Mannheim). Arrows denote the similar mobilities of 220-bp left-handed and right-handed curved DNA fragments. (b) Gel mobility under untwisting conditions. Electrophoresis was done in the presence of 0.005 mg/ml of EB in the gel and gel-running buffer. Arrows denote the now quite different mobilities of 220-bp fragments of left-handed and right-handed curved DNA segments. (c) Gel mobility anomaly R versus n for the different monomer units (19- to 23-mers). R is the coefficient of retardation (13). The experimental error in determination of R is estimated as ±0.03. The number of ligated monomer units is denoted by “n.” Note that n starts at 3 (Upper) without untwisting and at 6 (Lower) with untwisting. Note also that the vertical scales of R are different for stress-free versus untwisting conditions. Highly schematized 3D shapes of curved DNA before and after untwisting are depicted at the top and bottom, respectively. Upon untwisting, the right-handed superhelixes (Right) increase their radii but decrease their overall twisting and become more planar. Initially planar curved DNA molecules (Center) are converted to left-handed superhelixes, while left-handed superhelixes (Left) become more twisted, with smaller radii.
Cyclization Assay.
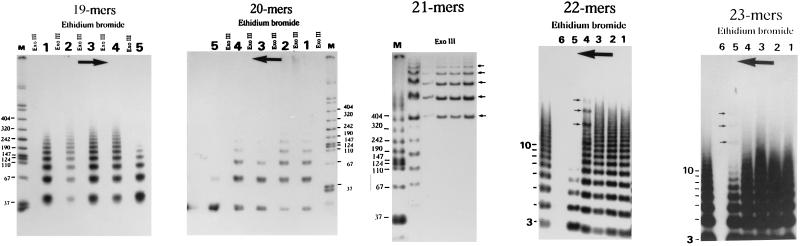
We also compared the pattern of cyclization under stress-free versus untwisting conditions for well defined 3D-curved DNA multimers (Fig. 2). Under stress-free conditions, only curved 21-mers gave small circles, starting at 126 bp (Fig. 2 Middle). There was no detectable cyclization of 3D-curved 19-, 20-, 22-, or 23-mers. Under untwisting conditions, initially curved right-handed superhelixes (22- and 23-mers) did form small circles, starting at 154 bp (see arrows in 22- and 23-mer panels), but not curved left-handed 19- and 20-mers. The cyclization of 23-mers (lane 5 of the 23-mer panel) was possible only under 10 times higher concentration of EB than for 22-mers (lane 4 of the 22-mer panel). Under untwisting conditions, the planar curved 21-mers gave the same pattern of cyclization, but with lower efficiency. The presence of circles was confirmed in all cases by treatment with exonuclease III. Contrary to what was seen with curved DNA, straight (CA)n DNA did not show a differential pattern of cyclization among all ligated sequence repeats (19- to 23-mers), either under stress-free or untwisting conditions (data not shown).
Figure 2.
Chirality-dependent cyclization of right- and left-handed 3D-curved DNA in the presence or absence of EB. Lane M is molecular weight marker VII (Boehringer Mannheim), while the scale 3 to 10 at right denotes the number of ligated monomer units. Lanes 1 to 6 show the effect of increasing concentration of EB on cyclization, in the direction indicated by large arrows. EB concentration increases by a factor of 10 from one lane to the next, from 0.01 μg/ml (lane 1) to 1 mg/ml (lane 6). The presence or absence of circles was additionally confirmed by exonuclease III digestion as shown for 19-, 20-, and 21-mers. The presence of circles is indicated by small arrows next to gel peaks. Note that the 23-mer needs a 10 times higher concentration of EB (lane 5) than does the 22-mer (lane 4) to form circles, and that the 19- and 20-mers were not able to make circular products under the same range of EB concentrations.
DISCUSSION
When DNA curves in three dimensions it departs from the local plane of curvature. In such cases, the plane curvature twists by some angle as the DNA advances through space, so the DNA coils into three dimensions forming a superhelix or supercoil (12). The global 3D trajectory of curved DNA can be monitored by gel mobility (4, 13, 14, 19, 25, 27–31). The relation between the gel mobility anomaly and 3D twisting and/or curving of repetitive sequence DNA trajectory has been established (13). Our gel mobility experiments were designed to show the changes of the well defined shape of 3D-curved DNA upon untwisting. The results obtained are compatible with the conceptual framework proposed by Calladine et al. (13). The R versus n values and the plateau region (Fig. 1c) reflect the radius and the twisting of the 3D-curved DNA trajectory before and after untwisting. Retardation coefficient values (R) for untwisting conditions are smaller than in stress-free conditions, and the plateau region is shifted toward longer DNA lengths. This indicates that EB not only untwists DNA, but also destabilizes the fine stacking interactions that are important for DNA bending (31). EB acts locally between base steps, but affects the global shape of the superhelixes. The general effect of untwisting on 3D-curved DNA is that only 21-, 22-, and 23-mers show gel mobility anomalies, but not 19- and 20-mers.
A cyclization assay was also used to show changes of the well defined 3D shape of curved DNA upon untwisting. However, this assay is less straightforward than the gel mobility assay, since cyclization is affected not only by the sequence-dependent 3D shape but also by sequence-dependent dynamic features of DNA (17, 21, 22, 27, 32–34). The fact that the sequence-dependent flexibility of DNA is anisotropic (32, 33) should not be underestimated. The term “anisotropic flexibility” of DNA (32, 33) means that DNA bends more easily toward the major than toward the minor groove—i.e., that roll angle fluctuations are predominantly positive in sign (33). The size of DNA circles is determined by the value of the net roll angle difference between phased sequence motifs at the moment of cyclization (27). In general, circles are smaller if the net roll angle difference between phased sequence motifs is bigger.
In two recent papers by Dlakic et al. (35, 36), the authors monitored the cyclization of curved DNA with and without A-tracts, under external conditions that increase the flexibility of DNA [the presence of 2-methyl-2,4-pentanediol, MPD (48)]. They observed a differential effect between the two types of curved DNA and concluded that their results demonstrated a sequence-specific straightening of A-tracts by MPD that is not observed with general sequences. This was used to cast doubt on the validity of crystal structures of A-tracts, which have uniformly shown them to be straight and unbent, and to support the “bent-A-tract” model for curvature in phased alternating A-tract/non-A sequences, rather than the “bent general sequence” model. However, the effect of increased DNA flexibility caused by MPD was not considered properly in their model. In the presence of MPD, A-tracts will mostly fluctuate on the positive side of the rolling scale (i.e., toward the major groove), which means that the net roll angle difference between A-tracts and other sequence motifs will be smaller. Therefore, if flexibility of DNA is increased via MPD, this will result in bigger circles only for A-tract-containing curved DNA, exactly what Dlakic et al. observe. Non-A-tract-containing curved DNA will produce the opposite trend—i.e., the circles will be smaller. In that case, the net roll angle difference for non-AA/TT-containing curved sequence motifs can be even higher, because they could either increase or decrease the roll angle values. Again, the results of Dlakic et al. (35, 36) confirm this. In summary, their results must be reinterpreted in the light of MPD-induced changes in differential DNA flexibility, and not simply static bending as they have done.
In our case, since we were comparing curved DNA with the same sequence, we should have the same flexibility between the multimers. Under stress-free conditions only curved 21-mers produced small circles. Under untwisting conditions only curved right-handed superhelixes (22- and 23-mers) and curved planar 21-mers were able to produce small circles (Fig. 2). The curved left-handed 19- and 20-mers did not give circular products in the range that was monitored. The straight (CA)n DNA did not show any differential behavior upon untwisting for all sequence repeat lengths (19- to 23-mers). Taken together, these results indicate that, in our assay, the end-to-end rotation distance between ends of DNA did not affect cyclization as much as the global 3D trajectory of DNA. They confirm the results of the gel mobility assay that indicate a very different configuration of 19- to 20-mers versus 22- to 23-mers upon untwisting.
Bednar et al. (37) recently showed by electron microscopy that intrinsically straight DNA has a persistence length of about 250 bp, versus only 150 bp for DNA of natural origin. It seems that the previous value of 150 bp now includes a contribution from slight random curvature as well as thermal motion, and hence that the DNA is more thermally rigid than was previously thought. Therefore, it seems unlikely that the superhelixes that we observed could result from a high DNA flexibility attributable to thermal motion.
Twisting of superhelixes is determined by the local values of twisting between base pairs. Untwisting modulates the phasing of bent motifs and induces changes in the global 3D shape of DNA. Upon untwisting, the right-handed curved superhelixes will decrease twisting and increase the planarity of 3D curvature, while the left-handed curved superhelixes will increase twisting and decrease planarity of 3D curvature (as illustrated in Fig. 1c). This change in shape upon untwisting results in a stronger gel mobility anomaly and a higher efficiency of cyclization of right-handed superhelixes. The opposite trend is seen for left-handed superhelixes. A similar mechanistic explanation was used to rationalize the temperature-dependent transitions of DNA global shape as measured by gel mobility anomaly and the same reasoning was used in the design of helical phasing experiments (19, 25, 27–29, 30, 31).
The average linking deficit present in vivo is about 1.7 degrees per base step (12). In the case of EB-induced cyclization of curved 22-mers, the value of twist decrease is even less than the linking deficit which is present in vivo. Therefore, our data would suggest that right-handed superhelixes can switch easily to more planar curves, or even to left-handed superhelixes under untwisting conditions in cells. In the recent paper by Hirota and Ohyama (38) it was demonstrated that right-handed superhelixes upstream of a bacterial promoter enhance transcription more than do planar circles or left-handed superhelixes. If this DNA bending pattern is a general feature of DNA upstream of a bacterial promoter, one could envisage that DNA shape transitions could be used as recognition signals for protein binding and as a driving force for transcription and/or replication of DNA (38–43). Right-handed superhelixes could be unwound to left-handed superhelixes by RNA polymerase to store torsional stress, that later could help to unwind the −10 region TATAAAT.
The inherent sequence-dependent deformability/flexibility of DNA (7, 8, 12, 44) and deformations induced by proteins (45–47) both determine the final shape of regulatory DNA segments. The experimental results presented here show that the same degree of untwisting can induce differential changes in the global shape of left-handed and right-handed 3D-curved DNA. These changes will be predetermined by the starting, sequence-dependent, macro-shape of curved DNA. In other words, (i) the distribution of bent sequence motifs that is coded by the sequence and (ii) the degree of untwisting/overtwisting that is imposed by proteins, are both relevant in influencing the overall trajectory of a given segment of DNA in vivo.
Acknowledgments
We thank Dr. Horace Drew (Commonwealth Scientific and Industrial Research Organization, Sydney) for his continual interest during this work. In addition, we thank Ana Savic (Institut for Molecular Genetics and Genetic Engineering and University of Belgrade), Dietrich Suck (European Molecular Biology Laboratory, Heidelberg), J. Clendenning and J. M. Schurr (University of Washington, Seattle), Sandor Pongor (International Center for Genetic Engineering and Biotechnology, Trieste, Italy), Benoit Houle, Mustafa El Hassan, and Graham Dellaire for valuable suggestions during the writing of the manuscript. This work was supported by a grant from the Cancer Research Society to P.C. I.B. is a recipient of Medical Research Council of Canada postdoctoral fellowship and A.B. is a scholar from Medical Research Council–Cancer Research Society.
Footnotes
Abbreviations: 3D, three dimensional; EB, ethidium bromide.
References
- 1.Lewin B. Cell. 1994;79:397–406. doi: 10.1016/0092-8674(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 2.Travers A A. Nature (London) 1989;341:184–185. doi: 10.1038/341184a0. [DOI] [PubMed] [Google Scholar]
- 3.Felsenfeld G. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 4.Hagerman P J. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 5.Luetke K H, Sadowski P D. J Mol Biol. 1995;251:493–506. doi: 10.1006/jmbi.1995.0451. [DOI] [PubMed] [Google Scholar]
- 6.Milot E, Belmaaza A, Wallenburg C, Gusew N, Bradley W E C, Chartrand P. EMBO J. 1992;11:5063–5070. doi: 10.1002/j.1460-2075.1992.tb05613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Travers A A, Klug A. In: DNA Topology and Its Biological Effects. Cozzarelli N R, Wang J C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 57–106. [Google Scholar]
- 8.Dickerson R E. Methods Enzymol. 1992;211:67–111. doi: 10.1016/0076-6879(92)11007-6. [DOI] [PubMed] [Google Scholar]
- 9.Grzeskowiak K, Goodsell D S, Kaczor-Grzeskowiak M, Cascio D, Dickerson R E. Biochemistry. 1993;32:8923–8931. doi: 10.1021/bi00085a025. [DOI] [PubMed] [Google Scholar]
- 10.Goodsell D S, Kopka M L, Cascio D, Dickerson R E. Proc Natl Acad Sci USA. 1993;90:2930–2934. doi: 10.1073/pnas.90.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodsell D S, Dickerson R E. Nucleic Acids Res. 1994;22:5497–5503. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calladine C R, Drew H R. Understanding DNA: The Molecule and How it Works. London: Academic; 1992. [Google Scholar]
- 13.Calladine C R, Drew H R, McCall M J. J Mol Biol. 1988;201:127–137. doi: 10.1016/0022-2836(88)90444-5. [DOI] [PubMed] [Google Scholar]
- 14.Goodsell D S, Kaczor-Grzeskowiak M, Dickerson R E. J Mol Biol. 1994;239:79–96. doi: 10.1006/jmbi.1994.1352. [DOI] [PubMed] [Google Scholar]
- 15.Mahtab R, Rogers J P, Singleton C P, Murphy C J. J Am Chem Soc. 1996;118:7028–7032. [Google Scholar]
- 16.Mahtab R, Rogers J P, Murphy C. J Am Chem Soc. 1995;117:9099–9100. [Google Scholar]
- 17.Ulanovsky L, Bodner M, Trifonov E N, Choder M. Proc Natl Acad Sci USA. 1986;83:862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson H C M, Finch J T, Luisi B F, Klug A. Nature (London) 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 19.Brukner I, Sanchez R, Suck D, Pongor S. EMBO J. 1995;14:1812–1818. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diekmann S. Methods Enzymol. 1992;212:30–46. doi: 10.1016/0076-6879(92)12004-a. [DOI] [PubMed] [Google Scholar]
- 21.Hagerman P J, Ramadevi V A. J Mol Biol. 1990;212:351–362. doi: 10.1016/0022-2836(90)90130-E. [DOI] [PubMed] [Google Scholar]
- 22.Crothers D M, Drak J, Kahn J D, Levene S D. Methods Enzymol. 1992;212:3–29. doi: 10.1016/0076-6879(92)12003-9. [DOI] [PubMed] [Google Scholar]
- 23.Drew H R, Travers A A. J Mol Biol. 1986;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- 24.Brukner I, Dlakic M, Savic A, Susic S, Pongor S, Suck D. Nucleic Acids Res. 1993;21:1025–1029. doi: 10.1093/nar/21.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drak J, Crothers D. Proc Natl Acad Sci USA. 1991;88:3074–3078. doi: 10.1073/pnas.88.8.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revet B, Brahms S, Brahms G. Proc Natl Acad Sci USA. 1995;92:7535–7539. doi: 10.1073/pnas.92.16.7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calladine C R, Drew H R. J Mol Biol. 1996;257:479–485. doi: 10.1006/jmbi.1996.0177. [DOI] [PubMed] [Google Scholar]
- 28.Shlyakhtenko L S, Lyubchenko Y L, Chernov B K, Zhurkin V B. Mol Biol (Moscow) 1990;24:79–95. [PubMed] [Google Scholar]
- 29.Hagerman P J. Biochemistry. 1985;24:7033–7036. doi: 10.1021/bi00346a001. [DOI] [PubMed] [Google Scholar]
- 30.Diekmann S. Nucleic Acids Res. 1987;15:247–265. doi: 10.1093/nar/15.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diekmann S, Mazzarelli J M, McLaughlin L W, von Kitzing E, Travers A A. J Mol Biol. 1992;225:729–738. doi: 10.1016/0022-2836(92)90397-3. [DOI] [PubMed] [Google Scholar]
- 32.Hagerman P J. Annu Rev Biochem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 33.Olson W K, Marky N L, Jernigan R L, Zhurkin V B. J Mol Biol. 1993;232:530–554. doi: 10.1006/jmbi.1993.1409. [DOI] [PubMed] [Google Scholar]
- 34.Dlakic M, Harrington R. J Biol Chem. 1995;270:29945–29952. doi: 10.1074/jbc.270.50.29945. [DOI] [PubMed] [Google Scholar]
- 35.Dlakic M, Harrington R. Proc Natl Acad Sci. 1996;93:3847–3852. doi: 10.1073/pnas.93.9.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dlakic M, Park K, Griffith J D, Harvey S C, Harrington R E. J Biol Chem. 1996;271:17911–17919. doi: 10.1074/jbc.271.30.17911. [DOI] [PubMed] [Google Scholar]
- 37.Bednar J, Furrer P, Katritch V, Stasiak A Z, Dubochet J, Stasiak A. J Mol Biol. 1995;254:579–594. doi: 10.1006/jmbi.1995.0640. [DOI] [PubMed] [Google Scholar]
- 38.Hirota Y, Ohyama T. J Mol Biol. 1995;254:566–578. doi: 10.1006/jmbi.1995.0639. [DOI] [PubMed] [Google Scholar]
- 39.Lilley D. Nature (London) 1986;320:487–488. doi: 10.1038/320487a0. [DOI] [PubMed] [Google Scholar]
- 40.Travers A A. Cell. 1990;60:177–180. doi: 10.1016/0092-8674(90)90729-x. [DOI] [PubMed] [Google Scholar]
- 41.Gartenberg M R, Crothers D M. J Mol Biol. 1991;219:217–230. doi: 10.1016/0022-2836(91)90563-l. [DOI] [PubMed] [Google Scholar]
- 42.Levigne M, Herbert M, Kolb A, Buc H. J Mol Biol. 1992;224:293–306. doi: 10.1016/0022-2836(92)90995-v. [DOI] [PubMed] [Google Scholar]
- 43.Ner S S, Travers A A, Churchill M E A. Trends Biochem Sci. 1994;19:185–187. doi: 10.1016/0968-0004(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 44.Lipanov A, Kopka M L, Kaczor-Grzeskowiak M, Quantina J, Dickerson R E. Biochemistry. 1993;32:1373–1389. doi: 10.1021/bi00056a024. [DOI] [PubMed] [Google Scholar]
- 45.Kim Y, Geiger J H, Hahn S, Sigler P B. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 46.Kim J L, Burley S K. Nat Struct Biol. 1994;1:638–653. doi: 10.1038/nsb0994-638. [DOI] [PubMed] [Google Scholar]
- 47.Lutter L C, Halvorson H R, Calladine C R. J Mol Biol. 1996;261:620–633. doi: 10.1006/jmbi.1996.0488. [DOI] [PubMed] [Google Scholar]
- 48.Dickerson R E, Goodsell D, Kopka M L. J Mol Biol. 1996;256:108–125. doi: 10.1006/jmbi.1996.0071. [DOI] [PubMed] [Google Scholar]