Strongly diffracting crystals of a methanol-induced corrinoid protein from M. thermoacetica have been obtained.
Keywords: corrinoid, methyltransferases
Abstract
A corrinoid protein was induced and overexpressed in methanol-grown cells of the thermophilic anaerobic bacterium Moorella thermoacetica. The protein was purified from cytosolic extracts. After screening for crystallization conditions and optimization, crystals were obtained that diffracted strongly on a rotating-anode X-ray source. A diffraction data set was collected and processed including reflections to 1.9 Å resolution. Reflections were indexed in a primitive orthorhombic cell with unit-cell parameters a = 55.69, b = 62.74, c = 34.54 Å. N-terminal amino-acid sequencing indicates that the crystals contain a C-terminal fragment of the protein.
1. Introduction
Moorella thermoacetica, formerly known as Clostridium thermoaceticum, is a thermophilic anaerobic acetogenic bacterium which converts C1 compounds such as CO2, CO, formate and methanol to acetate using the autotrophic acetyl-CoA pathway (Ljungdahl, 1986 ▶; Wood & Ljungdahl, 1991 ▶; Drake & Daniel, 2004 ▶). This involves a total synthesis of one mole of acetate from two moles of CO2. One mole of CO2 is reduced via formate and tetrahydrofolate intermediates to 5-methyltetrahydrofolate, the methyl group of which is transferred onto the Co atom of a corrinoid iron–sulfur protein (CFeSP), forming a Co-methylcorrinoid (Ragsdale et al., 1987 ▶). The final step, the formation of acetyl-CoA from CO2, CoA and the methyl group of the methylcorrinoid, is catalyzed by the bifunctional Ni–Fe–Cu enzyme carbon monoxide dehydrogenase/acetyl-CoA synthase (CODH/ACS; Ragsdale & Kumar, 1996 ▶; Lindahl, 2002 ▶; Doukov et al., 2002 ▶).
Derivatives of vitamin B12, including methylcorrinoids, play important roles in bacterial C1 metabolism (Ludwig & Matthews, 1997 ▶; Ragsdale, 1999 ▶; Banerjee & Ragsdale, 2003 ▶). The cobalamin-dependent methionine synthase is the most studied methyltransferase (Goulding et al., 1997 ▶; Matthews, 2001 ▶). Other corrinoid-dependent transferases have been isolated from acetogens (Lu et al., 1993 ▶), several methanogens (van der Meijden et al., 1984 ▶; Kremer et al., 1993 ▶; Sauer & Thauer, 1998 ▶; Ding et al., 2002 ▶) and methylotrophs (Coulter et al., 1999 ▶; Studer et al., 1999 ▶). Methyl donors include methyltetrahydrofolate, methyltetrahydromethanopterin, methanol, acetate, methylamines, methyl ethers and halomethanes.
The best characterized corrinoid protein from M. thermoacetica is the CFeSP and, as discussed above, it transfers the methyl group of methyltetrahydrofolate to the CODH/ACS. It is an αβ dimer having two subunits of 33 and 55 kDa. The smaller subunit carries the corrinoid 5-methoxybenzimidazolylcobamide, whereas the larger subunit contains the [4Fe–4S] cluster (Ragsdale et al., 1987 ▶; Lu et al., 1993 ▶). A second corrinoid protein designated MtvC (Naidu & Ragsdale, 2001 ▶) is part of a three-component vanillate O-demethylase system. This enzyme system may have a broad specificity and be involved in the transfer of methyl groups from a number of methoxylated aromatic compounds functioning as methyl donors (Daniel et al., 1991 ▶). A similar system has been described for the acetogen Acetobacterium dehalogenans (Kaufmann et al., 1998 ▶; Engelmann et al., 2001 ▶). A third corrinoid protein of 27 kDa has also been purified from M. thermoacetica, but its function has not been ascertained (Ljungdahl et al., 1973 ▶; Ragsdale et al., 1987 ▶).
We report here the purification of a corrinoid-containing polypeptide containing one mole of corrinoid from M. thermoacetica grown on methanol. Based on its N-terminal amino-acid sequence, the newly isolated protein was found to be encoded by orf1948 of contig 303 of the M. thermoacetica genome, which encodes a protein of 210 amino-acid residues with a weight of 22 329 Da. [A draft of annotated nucleotide sequences of the M. thermoacetica (ATCC39073) genome completed by The Joint Genome Institute, Department of Energy is available at http://genome.jgi-psf.org/draft_microbes/mooth/mooth.draft.html.] Based on its sequence, orf1948 appears to encode a homolog of MtaC, which is a corrinoid protein and a component of the methanol:CoM methyltransferase system of methane-producing archaea (van der Meijden et al., 1983 ▶; Ding et al., 2002 ▶). This system consists of three components: MtaA, MtaB and MtaC. MtaB catalyzes the transfer of the methyl group from methanol to the corrinoid cofactor of MtaC, while MtaA catalyzes the transfer of the methyl group from the corrinoid of MtaC to CoM. Sequence homologs of mtaA and mtaB are also present in the M. thermoacetica genome. Based on these similarities and the induction and overexpression in the presence of methanol, we propose a role of the orf1948 gene product in the synthesis of acetyl-CoA from methanol in M. thermoacetica. Strongly diffracting crystals of the protein have been obtained.
2. Materials and methods
2.1. Protein isolation
The corrinoid protein was purified from cytosolic extracts from cells grown on methanol with CO2 in the gas phase (Das et al., 2005 ▶). Briefly, frozen cell paste (15 g wet weight) of M. thermoacetica was suspended in 30 ml 50 mM Tris pH 8.0 containing 1 mM phenylmethylsulfonyl fluoride (PMSF). The cells were broken by passing the cell suspension through a French pressure cell. Cell extracts were centrifuged at 100 000g for 1 h and the pellet was discarded. Ammonium sulfate was added to the supernatant to a final saturation of 40%. The mixture was stirred at 277 K for 1 h and then centrifuged at 20 000g. The pellet was discarded and the dark red supernatant (approximately 30 ml) was dialyzed against 2 l 20 mM sodium phosphate pH 7.2 with three changes at 6 h intervals. The dialyzed sample was then loaded onto a HiPrep column (16/10 QXL, Amersham Biosciences, Piscataway, NJ, USA) and washed with six bed volumes of 50 mM sodium phosphate pH 6.8. Proteins bound to the column were eluted with a 0–500 mM NaCl gradient. Fractions with a maximum absorption at 357 nm were collected, pooled and dialyzed against 20 mM HEPES pH 7.6 containing 100 mM NaCl overnight. The sample was concentrated to a volume of approximately 4 ml (7 mg ml−1 protein) and then loaded onto a HiLoad gel-filtration column (16/60 Superdex 75 prep-grade from Amersham Biosciences) equilibrated with the same buffer. The protein was eluted with the same HEPES buffer. Fractions with an absorption maximum at 357 nm were collected and combined. Metal-content analysis was performed by inductively coupled plasma emission.
2.2. Crystallization
Initial screening was carried out using 384 conditions selected from seven commercial sparse-matrix screens [Crystal Screen, Crystal Screen 2, MembFac, Crystal Screen Cryo and PEG/Ion (Hampton Research, Aliso Viejo, CA, USA), Wizard I and II (Emerald Biostructures, Bainbridge Island, WA, USA)] and MP1, a custom-designed sparse-matrix screen, with a protein sample concentrated to 7 mg ml−1 in 20 mM HEPES pH 7.6 and 100 mM NaCl. Screening was performed using a Cartesian robot (Genomic Solutions, Ann Arbor, MI, USA) with the sitting-drop method, a reservoir of 0.1 ml crystallization reagent and a mixture of 0.2 µl each of protein solution and crystallization reagent in the crystallization drop. The optimization grid-screening trays were set up using the modified microbatch method (Chayen et al., 1990 ▶; D’Arcy et al., 1996 ▶), mixing 0.5 µl each of protein solution and crystallization reagent using an ORYX 1-6 protein crystallization robot (Douglas Instruments Ltd, East Garston, UK), optimizing an original condition composed of 0.1 M NaCl, 0.1 M HEPES pH 7.5 and 1.6 M (NH4)2SO4. A crystallization condition containing 0.1 M HEPES pH 6.7 and 1.6 M (NH4)2SO4 was further optimized with additive screening by addition of 0.2 µl reagent to the microbatch crystallization drop. All crystallization trials were carried out at 291 K. Diffraction-quality crystals were obtained by modified microbatch crystallization from a drop that was made up from the following components: (i) 0.5 µl of a solution containing 1.6 M (NH4)2SO4 and 0.1 M HEPES pH 6.7, (ii) 0.5 µl protein solution and (iii) 0.2 µl 30%(v/v) 1,6-hexanediol in water.
2.3. Data collection
A crystal measuring 0.07 × 0.08 × 0.2 mm was separated from a cluster of crystals, recovered from the crystallization drop using a fiber loop (Teng, 1990 ▶) and immersed for 3 s into a 1 µl drop of a 4:1 mixture of 1.6 M (NH4)2SO4/0.1 M HEPES pH 6.7 and 2 M Li2SO4. Still in the loop, it was dragged along the surface of a microscopic cover slide to remove excess liquid and flash-frozen (Hope, 1988 ▶) in liquid nitrogen before the data-collection process.
Diffraction data were collected with a Smart 6000 CCD detector (Bruker AXS) on an FRD rotating copper-anode source equipped with HiRes2 optics (Rigaku/MSC). Two passes of 600 oscillation images (Δω = −0.3°) were recorded at ϕ = 115 and 15°. The crystal-to-detector distance was 80 mm; κ = 54.79° and 2θ = 12°. Data processing was carried out using the PROTEUM suite.
2.4. N-terminal peptide sequencing
Several protein crystals were dissolved in 40 µl 0.1 M HEPES pH 6.7 and submitted to the Microchemical and Proteomics Facility at Emory University (Atlanta, GA, USA) for sequencing.
3. Results and discussion
3.1. Protein purification
The corrinoid protein was purified from M. thermoacetica cytosolic extracts in three steps: ammonium sulfate precipitation, ion-exchange chromatography and gel filtration. The protein eluted from the gel-filtration column was found to be homogeneous on SDS–PAGE (not shown). Using size-exclusion chromatography on a Superose 12 column, the purified protein eluted as a single peak with an apparent approximate molecular weight of 25 kDa (not shown), suggesting that the purified protein is a monomer. The purified protein had a metal content of 0.9 mol cobalt per mole of polypeptide, with significantly lower amounts of other metals (not shown), including iron (less than 0.1 mol per mole of polypeptide). UV–visible spectra of the purified protein exhibited absorption at 357 and 542 nm in the oxidized form which shifted to 363 and 551 nm following treatment with cyanide and to 369 and 580 nm after boiling with cyanide (not shown), which released the corrinoid from the protein. These features are similar to those reported for the 27 kDa protein (Ljungdahl et al., 1973 ▶).
3.2. Crystallization
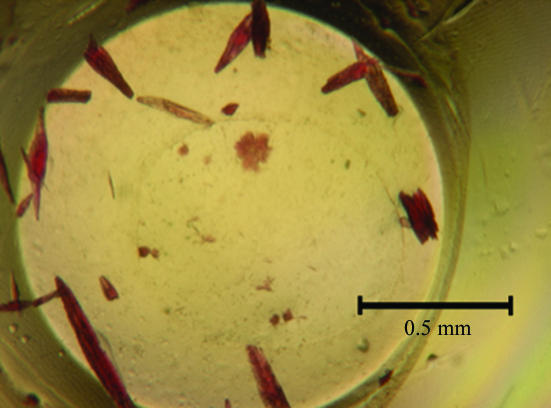
This study confirmed earlier observations in our laboratory that conditions identified during high-throughput screening with vapor-diffusion methods translate well into optimization trials using the modified microbatch method. Initial screening and grid optimization yielded crystals that were too small to be mounted. Addition of 1,6-hexanediol produced crystals of satisfactory size after nearly one month of incubation. As no separate single crystals could be located in the crystallization drop (Fig. 1 ▶), a rod-shaped piece was carefully cleaved from a cluster of crystals.
Figure 1.
Crystals of the methanol-induced corrinoid protein from M. thermoacetica.
3.3. X-ray diffraction analysis
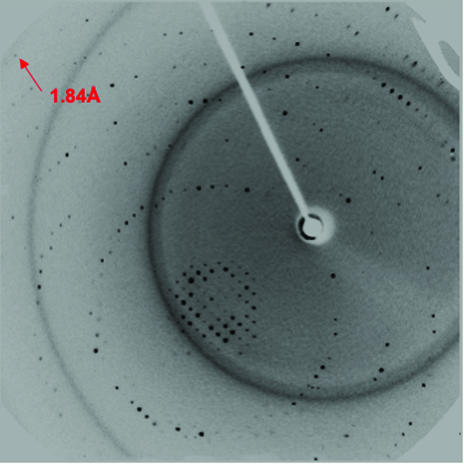
Sufficient cryoprotection was achieved with lithium sulfate based on published recommendations (Rubinson et al., 2000 ▶). The data-processing statistics are given in Table 1 ▶ and show reasonable diffraction to 1.9 Å resolution even on a rotating-anode source (Fig. 2 ▶). Systematic absences suggested that the crystal belongs to space group P21212 (Table 1 ▶). Assuming one molecule per asymmetric unit, the Matthews coefficient (Matthews, 1968 ▶) was calculated to be 1.4 Å3 Da−1, indicating there to be only one molecule per asymmetric unit, corresponding to a solvent content of only 8%. This unusually low value prompted an analysis of the crystals’ peptide composition. In order to confirm the identity of the crystallized peptide, crystals were dissolved and the N-terminal amino-acid sequence was determined. It was found that the crystal contained a fragment with Asp84 at the N-terminus. Assuming that the fragment continues to the C-terminus of the orf1948 gene product, a Matthews coefficient of 2.3 Å3 Da−1 and a solvent content of 45% were calculated. Structure determination is currently in progress.
Table 1. Data-processing statistics.
Values in parentheses refer to the high-resolution shell.
| Resolution range (Å) | 62.5–1.90 (2.04–1.90) |
| Wavelength (Å) | 1.5418 |
| Space group | P21212 |
| Unit-cell parameters (Å) | a = 55.52, b = 62.65, c = 34.45 |
| Total observations | 58770 |
| Unique reflections | 9197 (1153) |
| Completeness (%) | 92.1 (60.3) |
| I/σ(I) | 12.3 (4.9) |
| Rmerge (%) | 4.9 (7.6) |
| Redundancy | 6.4 |
Figure 2.
Oscillation image obtained using an in-house rotating-anode source.
Acknowledgments
We thank the National Institutes of Health, the University of Georgia Research Foundation and Georgia Research Alliance (B-CW) and the US Department of Energy (LGL) for support.
References
- Banerjee, R. & Ragsdale, S. W. (2003). Annu. Rev. Biochem.72, 209–247. [DOI] [PubMed] [Google Scholar]
- Chayen, N. E., Shaw Stewart, P. D., Maeder, D. L. & Blow, D. M. (1990). J. Appl. Cryst.23, 297–302. [Google Scholar]
- Coulter, C., Hamilton, J. T. G., McRoberts, W. C., Kulakov, L., Larkin, M. J. & Harper, D. B. (1999). Appl. Environ. Microbiol.65, 4301–4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, S. L., Keith, E. S., Yang, H., Lin, Y. S. & Drake, H. L. (1991). Biochem. Biophys. Res. Commun.180, 416–422. [DOI] [PubMed] [Google Scholar]
- D’Arcy, A., Elmore, C., Stihle, M. & Johnston, J. E. (1996). J. Cryst. Growth, 168, 175–180.
- Das, A., Silaghi-Dumitrescu, R., Ljungdahl, L. G. & Kurtz, D. M. Jr (2005). J. Bacteriol.187, 2020–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y. H., Zhang, S. P., Tomb, J. F. & Ferry, J. G. (2002). FEMS Microbiol. Lett.215, 127–132. [DOI] [PubMed] [Google Scholar]
- Doukov, T. I., Iverson, T. M., Seravalli, J., Ragsdale, S. W. & Drennan, C. L. (2002). Science, 298, 567–572. [DOI] [PubMed] [Google Scholar]
- Drake, H. L. & Daniel, S. L. (2004). Res. Microbiol.155, 869–883. [DOI] [PubMed] [Google Scholar]
- Engelmann, T., Kaufmann, F. & Diekert, G. (2001). Arch. Microbiol.175, 376–383. [DOI] [PubMed] [Google Scholar]
- Goulding, C. W., Postigo, D. & Matthews, R. G. (1997). Biochemistry, 36, 8082–8091. [DOI] [PubMed] [Google Scholar]
- Hope, H. (1988). Acta Cryst. B44, 22–26. [DOI] [PubMed] [Google Scholar]
- Kaufmann, F., Wohlfarth, G. & Diekert, G. (1998). Eur. J. Biochem.253, 706–711. [DOI] [PubMed] [Google Scholar]
- Kremer, J. D., Cao, X. J. & Krzycki, J. (1993). J. Bacteriol.175, 4824–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, P. A. (2002). Biochemistry, 41, 2097–2105. [DOI] [PubMed] [Google Scholar]
- Ljungdahl, L. G. (1986). Annu. Rev. Microbiol.40, 415–450. [DOI] [PubMed] [Google Scholar]
- Ljungdahl, L. G., LeGall, J. & Lee, J. P. (1973). Biochemistry, 12, 1802–1808. [DOI] [PubMed] [Google Scholar]
- Lu, W. P., Schiau, I., Cunningham, J. R. & Ragsdale, S. W. (1993). J. Biol. Chem.268, 5605–5614. [PubMed] [Google Scholar]
- Ludwig, M. L. & Matthews, R. G. (1997). Annu. Rev. Biochem.66, 269–313. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Matthews, R. G. (2001). Acc. Chem. Res.34, 681–689. [DOI] [PubMed] [Google Scholar]
- Meijden, P. van der, Heythuysen, H. J., Pouwels, A., Houwen, F., van der Drift, C. & Vogels, G. D. (1983). Arch. Microbiol.134, 238–242. [DOI] [PubMed] [Google Scholar]
- Meijden, P. van der, te Brömmelstroet, B. W., Poirot, C. M., van der Drift, C. & Vogels, G. D. (1984). J. Bacteriol.160, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu, D. & Ragsdale, S. W. (2001). J. Bacteriol.183, 3276–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale, S. W. (1999). Chemistry and Biochemistry of B12, Vol. 1, edited by R. Banerjee, pp. 633–695. New York: John Wiley & Sons.
- Ragsdale, S. W. & Kumar, M. (1996). Chem. Rev.96, 2515–2539. [DOI] [PubMed] [Google Scholar]
- Ragsdale, S. W., Lindahl, P. A. & Münck, E. (1987). J. Biol. Chem.262, 14289–14297. [PubMed] [Google Scholar]
- Rubinson, K. A., Ladner, J. E., Tordova, M. & Gilliland, G. L. (2000). Acta Cryst. D56, 996–1001. [DOI] [PubMed] [Google Scholar]
- Sauer, K. & Thauer, R. K. (1998). Eur. J. Biochem.253, 698–705. [DOI] [PubMed] [Google Scholar]
- Studer, A., Vuilleumier, S. & Leisinger, T. (1999). Eur. J. Biochem.264, 242–249. [DOI] [PubMed] [Google Scholar]
- Teng, T.-Y. (1990). J. Appl. Cryst.23, 387–391. [Google Scholar]
- Wood, H. G. & Ljungdahl, L. G. (1991). Variations in Autotrophic Life, edited by J. M. Shively & L. L. Barton, pp. 201–250. San Diego: Academic Press.