The first crystallographic study of a member of the YaeQ family of proteins, which are conserved in a small group of Gram-negative bacteria, most of which are animal or plant pathogens, is reported. Diffraction data were collected to 1.9 Å resolution and an interpretable electron-density map was obtained.
Keywords: YaeQ, XAC2396
Abstract
Xanthomonas axonopodis pv. citri YaeQ (XAC2396) is a member of a family of bacterial proteins conserved in several Gram-negative pathogens. Here, the cloning, expression, purification and crystallization of the 182-residue (20.6 kDa) YaeQ protein are described. Recombinant YaeQ containing selenomethionine was crystallized in space group P21 and crystals diffracted to 1.9 Å resolution at a synchrotron source. The unit-cell parameters are a = 39.75, b = 91.88, c = 48.03 Å, β = 108.37°. The calculated Matthews coefficient suggests the presence of two YaeQ molecules in the asymmetric unit. Initial experimental phases were calculated by the multiple-wavelength anomalous dispersion technique and an interpretable electron-density map was obtained.
1. Introduction
Xanthomonas is a genus of phytopathogenic bacteria that attack a variety of hosts with economic importance, including citrus fruits, rice, beans, grapes and cotton. Citrus canker is caused by the phytopathogen Xanthomonas axonopodis pv. citri (Xac), a rod-like Gram-negative motile bacterium. The Xac genome (Da Silva et al., 2002 ▶) has a 5 175 554 bp chromosome and two megaplasmids: pXAC33 (33 699 bp) and pXAC64 (64 920 bp). Genome annotation identified over 4400 open reading frames, 2770 of which were assigned putative functions based on similarities with sequences in the public databases (Da Silva et al., 2002 ▶).
Xac YaeQ (XAC2396) is a member of a family of bacterial proteins that are conserved in several Gram-negative pathogens, including Salmonella ssp., Escherichia coli, Pseudomonas ssp., Shigella ssp., Yersinia ssp., Ralstonia ssp., Erwinia ssp., Bordetella ssp., Burkholderia fungorum, Vibrio parahaemolyticus and Chromobacterium violaceum, as well as Azotobacter vinelandii, Dechloromonas aromatica, Shewanella oneidensis, Geobacter metallireducens and G. sulfurreducens. This 182-amino-acid residue protein was selected for structural analysis owing to its well folded and non-aggregated state in solution, both in bacterial lysates and in the purified form (Galvão-Botton et al., 2003 ▶).
It has been suggested that YaeQ is involved in the regulation of transcription of genes encoding virulence factors such as haemolysin and enzymes involved in the production of lipopolysaccharides. This hypothesis was presented as a YaeQ-encoding fragment from Salmonella typhyimurium was shown to complement a defect in the RfaH-dependent expression of the hlyCABD operon (Wong et al., 1998 ▶). However, Vicari & Artsimovitch (2004 ▶) have recently presented evidence that E. coli YaeQ does not complement the absence of RfaH in hlyCABD expression assays in vitro or in vivo. Thus, the precise function of YaeQ and its orthologues remains unknown. The YaeQ family does not show any sequence similarity with proteins of known structure or function. Determination of the three-dimensional structure of YaeQ may provide insights regarding its function. Several recent examples have demonstrated the feasibility of obtaining functional information from structure (Zarembinski et al., 1998 ▶; Bhattacharyya et al., 2002 ▶; Christendat et al., 2002 ▶; Jackson & Russell, 2001 ▶). In this report, we describe the cloning and expression of recombinant Xac YaeQ containing selenomethionine and its crystallization and also the determination of an initial electron-density map of the Xac YaeQ crystal structure by multiwavelength anomalous dispersion (MAD).
2. Cloning and expression of YaeQ
The YaeQ gene (XAC2396) was amplified by PCR from Xac genomic DNA using the following primers that were designed based on the published Xac genome sequence (Da Silva et al., 2002 ▶): 5′-CATGCCATGGCTCATATGGCCCTCACCGCCA-3′ and 5′-GGAATTCAAGCTTTCATTCGGCCGGGGCT-3′. The PCR product was digested with HindIII and NdeI and subcloned into the pET-3a vector (Studier et al., 1990 ▶) previously digested with the same endonucleases. Selenomethionine-containing YaeQ was expressed in E. coli strain BL21(DE3)pLysS (Studier et al., 1990 ▶) by growing a 500 ml culture in M9 medium to an optical density (600 nm) of 0.8, at which point 100 mg l−1 lysine, 100 mg l−1 phenylalanine, 100 mg l−1 threonine, 50 mg l−1 isoleucine, 50 mg l−1 valine and 60 mg l−1 selenomethionine were added (adapted from Berne et al., 1999 ▶). After 15 min, heterologous protein expression was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and the cells were grown for 4 h before harvesting and storage at 203 K.
3. Protein purification
Cells from 1 l culture were resuspended in 25 ml 50 mM Tris–HCl pH 8.0, 25% sucrose, 1 mM EDTA, 1 mM PMSF and lysed using a French press. 2 U ml−1 DNAse and 4 mM magnesium chloride were added to the soluble fraction, followed by incubation on ice for 30 min. This mixture was applied onto a Q-Sepharose Fast Flow (FF) Hiload 16/10 column (Amersham Pharmacia) previously equilibrated with 50 mM Tris–HCl pH 7.0, 1 mM EDTA and 14 mM β-mercaptoethanol. Bound proteins were eluted using a 0–300 mM NaCl gradient over 12 column volumes. Fractions containing YaeQ were concentrated using an Amicon system with a 10 kDa membrane and then further purified by gel filtration on a Superdex 75 prep-grade column (Amersham Pharmacia) equilibrated with 5 mM Tris–HCl pH 7.0. The molecular mass of the purified protein, as determined by MALDI–TOF mass spectrometry, was 20 982.749 ± 5 for the selenomethionine protein and 20 668.582 ± 5 for the methionine-containing protein. The difference in mass between the two preparations demonstrated that all seven methionines were exchanged for selenomethionines. The total protein yield was 46.5 mg l−1, with approximately 99% purity.
4. Crystallization
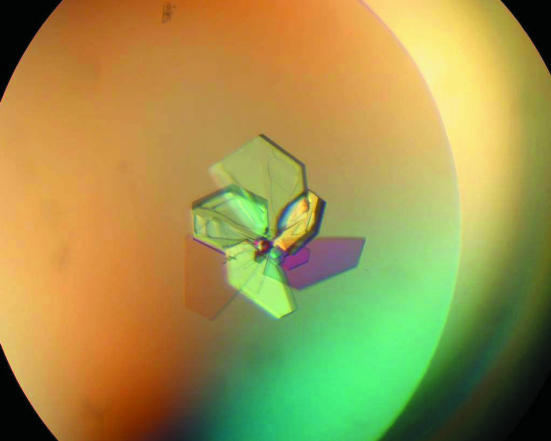
YaeQ crystals (Fig. 1 ▶) were grown using the hanging-drop vapour-diffusion technique at 291 K. Initial crystallization conditions of the methionine-containing protein were screened by the sparse-matrix sampling approach. Crystals were obtained in several conditions using the Crystal Screen (Hampton Research) and Clear Strategy (Molecular Dimensions Limited) screening kits. Optimization was then pursued by varying the precipitant concentration and the buffer pH. Suitable crystals for diffraction experiments were obtained by mixing equal volumes (1 µl) of 10 mg ml−1 protein solution (in 5 mM Tris–HCl pH 7.0) with reservoir solution, which consisted of either (i) 30–32%(w/v) PEG 4000, 0.2 M ammonium acetate and 0.1 M Tris–HCl pH 8.5–9.0 or (ii) 30%(w/v) PEG 8000, 0.1 M sodium cocadylate pH 6.7 and 0.2 M sodium acetate. Hanging drops were then equilibrated against 0.4 ml reservoir solutions. Crystals appeared within a few days and grew to mature size within a few weeks. Both methods gave the same crystal form.
Figure 1.
Typical crystals of YaeQ. The approximate dimensions of individual plates are 0.40 × 0.40 × 0.03 mm.
5. Data collection and preliminary structure analysis
Initial X-ray diffraction data from the methionine-containing protein were collected at the D03B beamline of the Laboratório Nacional de Luz Síncrotron, Campinas, Brazil using a MAR CCD detector (data not shown). MAD X-ray diffraction data for selenomethionine-containing crystals were collected at the protein crystallography beamline 8.3.1 of the Advanced Light Source (ALS), Berkeley, California using a ADSC CCD detector. SeMet-YaeQ crystals equilibrated against 32% PEG 4000, 0.1 M Tris–HCl pH 9.0, 0.2 M ammonium acetate were flash-frozen in liquid nitrogen and maintained at 100 K in a nitrogen-gas stream during data acquisition (this crystallization solution was sufficient to cryoprotect the crystals). MAD data sets were collected using a single crystal at three wavelengths, 0.97952, 0.97962 and 1.00000 Å, corresponding to the peak, inflection (approximate) and remote points of the fluorescence spectrum, respectively. The peak data set was collected initially and the inflection and remote data sets were then both collected from another region of the same crystal. The exposure time for each image of the peak data set (5 s) was ten times that for the inflection and remote data sets (0.5 s). The oscillation range for each image was 1°. The diffraction patterns from these data sets extended to approximately 1.9 and 2.4 Å resolution, respectively (Table 1 ▶). The crystal belongs to space group P21, with unit-cell parameters a = 39.75, b = 91.88, c = 48.03 Å, β = 108.37°. The Matthews coefficient (V M = 2.0 Å3 Da−1 and 38.4% solvent content) suggests that there are two protein molecules per asymmetric unit.
Table 1. Recombinant SeMet-labelled Xac YaeQ crystal parameters and data-reduction statistics.
Values in parentheses refer to the highest resolution shell.
| Space group | P21 | ||
| Unit-cell parameters | |||
| a (Å) | 39.75 | ||
| b (Å) | 91.88 | ||
| c (Å) | 48.03 | ||
| β (°) | 108.37 | ||
| Data-set statistics | Peak | Inflection | Remote |
| Resolution range (Å) | 37.80–1.90 (1.97–1.90) | 46.13–2.40 (2.53–2.40) | 45.64–2.40 (2.53–2.40) |
| No. of observed reflections | 99056 (8105) | 49405 (7232) | 51465 (7478) |
| No. of unique reflections | 24507 (2207) | 12499 (1816) | 12469 (1799) |
| 〈I/σ(I)〉† | 25.1 (2.1) | 11.1 (2.0) | 13.0 (2.2) |
| Multiplicity | 4.0 (3.7) | 4.0 (4.0) | 4.1 (4.2) |
| Multiplicity, anomalous† | 2.1 (1.9) | 2.1 (2.0) | 2.1 (2.0) |
| Completeness (%) | 95.0 (86.7) | 97.5 (96.9) | 97.5 (96.7) |
| Completeness, anomalous† (%) | 93.5 (82.8) | 94.0 (94.8) | 94.4 (95.1) |
| R (%) | 3.2 (34.9) | 6.0 (35.2) | 5.1 (31.7) |
| No. of images | 200 | 195 | 199 |
| λ (Å) | 0.97952 | 0.97962 | 1.00000 |
Each member of a Friedel pair is counted as a separate reflection.
Diffraction images data were indexed, integrated, scaled and merged using the HKL2000 package (Otwinowski & Minor, 1997 ▶) for the peak data set and the programs MOSFLM (Leslie, 1992 ▶) and SCALA (Collaborative Computational Project, Number 4, 1994 ▶) for the inflection and remote data sets. The Friedel mates were scaled separately during data processing. Data sets were merged using CAD (Collaborative Computational Project, Number 4, 1994 ▶). Details of data-acquisition and data-processing statistics are shown in Table 1 ▶.
The program SHELXD (Sheldrick, 1998 ▶) was used to locate 14 selenium sites in the asymmetric unit of the crystal (seven selenomethionines per protein molecule are expected) using the anomalous differences from the peak data set. However, only the coordinates of 12 major selenium sites were used to calculate MAD phases using the program SHARP (de La Fortelle & Bricogne, 1997 ▶). Experimental phases obtained by SHARP were improved by density-modification protocols using the programs SOLOMON (Abrahams, 1997 ▶) and DM (Collaborative Computational Project, Number 4, 1994 ▶; Cowtan, 1994 ▶). The resulting electron-density map was used by the program ARP/wARP (Perrakis et al., 1999 ▶) to produce a preliminary polyalanine model that included 285 of the 364 amino-acid residues found within the asymmetric unit of the crystal. On the basis of this preliminary model, the non-crystallographic symmetry relating the two monomers was determined and used for phase improvement and model building. Interpretation of electron-density maps and construction of missing residues is being performed using the program O (Jones et al., 1991 ▶). Structural refinement of the XAC2396 atomic model is currently under way using REFMAC (Murshudov et al., 1997 ▶) and CNS (Brünger et al., 1998 ▶).
Acknowledgments
We would like to thank the ALS 8.3.1 beamline staff and Dr Mark Glover (University of Alberta) and members of his laboratory for the opportunity to collect MAD data at the ALS. We thank the laboratory of Dr Paolo di Mascio (Instituto de Química, Universidade de São Paulo) for MALDI–TOF MS analysis. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) through grant 01/07534-3, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (Brazil) and Associação Brasileira de Tecnologia de Luz Síncrotron (ABTLuS). CRG is a FAPESP fellow and LMPG was supported with a PhD studentship from FCT/MCT (Portugal).
References
- Abrahams, J. P. (1997). Acta Cryst. D53, 371–376. [DOI] [PubMed] [Google Scholar]
- Berne, P. F., Doublié, S. & Carter, C. W. Jr (1999). Crystallization of Nucleic Acids and Proteins: A Practical Approach, 2nd ed., edited by A. Ducruix & R. Giegé, pp. 45–73. Oxford University Press.
- Bhattacharyya, S., Habibi-Nazhad, B., Amegbey, G., Slupsky, C. M., Yee, A., Arrowsmith, C. & Wishart, D. S. (2002). Biochemistry, 41, 4760–4770. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Christendat, D., Saridakis, V., Kim, Y., Kumar, P. A., Xu, X., Semesi, A., Joachimiak, A., Arrowsmith, C. H. & Edwards, A. M. (2002). Protein Sci.11, 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Cowtan (1994). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.31, 34–38
- Da Silva, A. C. R. et al. (2002). Nature (London), 417, 459–463. [Google Scholar]
- Galvão-Botton, L. M. P., Katsuyama, A. M., Guzzo, C. R., Almeida, F. C. L., Farah, C. S. & Valente, A. P. (2003). FEBS Lett.552, 207–213. [DOI] [PubMed] [Google Scholar]
- Jackson, R. M. & Russell, R. B (2001). Comput. Chem.26, 31–39. [DOI] [PubMed] [Google Scholar]
- Jones, T. A., Zhou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- La Fortelle, E. de & Bricogne, G. (1997). Methods Enzymol.244, 472–494. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Perrakis, A., Morris, R. M. & Lamzin, V. S. (1999). Nature Struct. Biol.6, 458–463. [DOI] [PubMed] [Google Scholar]
- Sheldrick, G. M. (1998). Direct Methods for Solving Macromolecular Structures, edited by S. Fortier, pp. 401–411. Dordrecht: Kluwer Academic Publishers.
- Studier, F. W., Rosenberg, A. H., Dunn, J. J. & Dubendorff, J. W. (1990). Methods Enzymol.185, 60–89. [DOI] [PubMed] [Google Scholar]
- Vicari, D. & Artsimovitch, I. (2004). Mol. Genet. Genomics, 272, 489–496. [DOI] [PubMed] [Google Scholar]
- Wong, K. R., Hughes, C. & Koronakis, V. (1998). Mol. Gen. Genet.257, 693–696. [DOI] [PubMed] [Google Scholar]
- Zarembinski, T. I., Hung, L. W., Mueller-Dieckmann, H. J., Kim, K. K., Yokota, H., Kim, R. & Kim, S.-H. (1998). Proc. Natl Acad. Sci. USA, 95, 15189–15193. [DOI] [PMC free article] [PubMed] [Google Scholar]