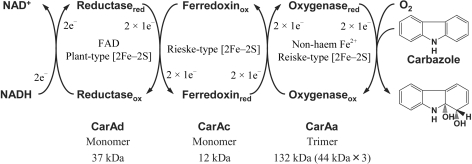
The electron-transfer complex between the terminal oxygenase and ferredoxin of carbazole 1,9a-dioxygenase was crystallized and diffraction data were collected to 1.90 Å resolution.
Keywords: angular dioxygenases, carbazole, electron-transfer complexes, Rieske non-haem iron oxygenase systems, Rieske-type ferredoxins, Rieske-type proteins
Abstract
Carbazole 1,9a-dioxygenase, which consists of an oxygenase component (CARDO-O) and the electron-transport components ferredoxin (CARDO-F) and ferredoxin reductase (CARDO-R), catalyzes dihydroxylation at the C1 and C9a positions of carbazole. The electron-transport complex between CARDO-O and CARDO-F crystallizes at 293 K using hanging-drop vapour diffusion with the precipitant PEG MME 2000 (type I crystals) or PEG 3350 (type II). Blossom-shaped crystals form from a pile of triangular plate-shaped crystals. The type I crystal diffracts to a maximum resolution of 1.90 Å and belongs to space group P21, with unit-cell parameters a = 97.1, b = 89.8, c = 104.9 Å, α = γ = 90, β = 103.8°. Diffraction data for the type I crystal gave an overall R merge of 8.0% and a completeness of 100%. Its V M value is 2.63 Å3 Da−1, indicating a solvent content of 53.2%.
1. Introduction
Rieske non-haem iron oxygenase systems (ROSs) are the initial catalysts in the degradation pathways of various environmentally significant aromatic compounds, including dioxins, polychlorinated biphenyls and crude-oil components such as polycyclic aromatic hydrocarbons and carbazole (Wittich, 1998 ▶; Bressler & Fedorak, 2000 ▶; Nojiri & Omori, 2002 ▶; Habe & Omori, 2003 ▶; Furukawa et al., 2004 ▶). In the initial dioxygenation of carbazole and dioxins, one carbon that is bonded to the heteroatom and its adjacent carbon in the aromatic ring are both hydroxylated. This reaction, called angular dioxygenation, is catalyzed by a limited number of ROSs, which are called angular dioxygenases (Nojiri & Omori, 2002 ▶).
Since the car operon containing the genes encoding the angular dioxygenase system carbazole 1,9a-dioxygenase (CARDO; Fig. 1 ▶) was first isolated from a carbazole-degrader, Pseudomonas resinovorans strain CA10 (Sato et al., 1997 ▶), we have studied the enzymatic function of this novel enzyme system extensively (Nojiri & Omori, 2002 ▶; Inoue et al., 2004 ▶). We have purified and characterized each component of CARDO of strain CA10 (CARDOCA10): the terminal oxygenase component (CarAaCA10 trimer; CARDO-OCA10) and the electron-transport components ferredoxin (CarAcCA10 monomer; CARDO-FCA10) and ferredoxin reductase (CarAdCA10 monomer; CARDO-RCA10) (Nam et al., 2002 ▶). A single subunit of CARDO-OCA10 contains a Rieske [2Fe–2S] cluster and a non-haem iron. CARDO-FCA10 and CARDO-RCA10 each contain a Rieske [2Fe–2S] cluster and a plant-type [2Fe–2S] cluster and an FAD, respectively. Although these characteristics of the electron-transport components suggest that the CARDOCA10 system can be classified as a class III ROS according to Batie’s classification (Batie et al., 1991 ▶), CARDO is a novel ROS owing to several important distinguishing characteristics. For example, the amino-acid sequence of CarAaCA10 shares rather low homology (<19% identity) with the corresponding proteins of other ROSs and shows greatest homology (35% identity) with the monooxygenase component (OxoO) from Pseudomonas putida strain 86 (Rosche et al., 1995 ▶), except for other CarAa proteins from different bacterial sources. In addition, CARDO-OCA10 consists of only one catalytic subunit with an α3 configuration, which is a typical feature of class IA ROSs in Batie’s system. Therefore, the electron-transport mechanism in CARDOCA10 has several unusual features. In addition, studies of its substrate specificity have shown that CARDOCA10 has a broad substrate range and that it catalyzes not only angular dioxygenation but also other oxygenation reactions, such as lateral dioxygenation (e.g. 1,2-diydroxylation of naphthalene) and monooxygenation (9-hydroxylation of fluorene and sulfoxidation of dibenzothiophene) (Nojiri et al., 1999 ▶; Takagi et al., 2002 ▶). CARDOCA10 also attacks some chlorinated dioxin congeners (Habe et al., 2001 ▶). Considering these unique properties of the CARDO system, structure-based elucidation of CARDO function is of great interest.
Figure 1.
Components and functions of the CARDO system, showing the electron-transfer reactions and conversion of carbazole to an unstable product, 1,9a-dihydroxy-1-hydrocarbazole, which has not been detected directly.
From a survey of the distribution and diversity of this novel enzyme, we have obtained several bacterial strains that have a car operon similar to that of strain CA10. One of these strains, Janthinobacterium sp. strain J3, has a carAa gene that has 47 nucleotide mismatches with the gene of strain CA10, which results in three amino-acid substitutions in its 384-amino-acid length. No functional differences were observed between CARDO-O of strain J3 (CARDO-OJ3) and CARDO-OCA10 in the analysis of substrate specificity using resting cells of CARDO-expressing Escherichia coli (data not shown). Therefore, we tried to determine the crystal structure of the CARDO components using proteins of strains CA10 and J3. Recently, we succeeded in determining the crystal structures of CARDO-FCA10 (Nam et al., 2005 ▶) and CARDO-OJ3 (Nojiri et al., 2005 ▶). In this study, as a first attempt to reveal the electron-transfer mechanism of CARDO, we report the crystallization and preliminary X-ray diffraction studies of the complex of CARDO-OJ3 with CARDO-FCA10.
2. Materials and methods
2.1. Protein expression, purification and characterization
In this study, we used CARDO-OJ3 and CARDO-FCA10 as CARDO components because expression and purification procedures for their crystallization have been established and because the crystal structures of both components have been determined (Nam et al., 2005 ▶; Nojiri et al., 2005 ▶). Both proteins were expressed in E. coli strains as six-histidine-tagged proteins at their C-terminal regions. A thorough description of the expression and purification of CARDO-OJ3 will be provided elsewhere (Nojiri et al., 2005 ▶). CARDO-FCA10 was expressed and purified as described previously (Nam et al., 2002 ▶), except that 20 mM Tris–HCl buffer pH 7.5 containing 0.5 M NaCl was used to resuspend the E. coli cells.
The two proteins were purified separately using metal-chelation chromatography and were then mixed and subjected to gel-filtration chromatography using Superdex-200 (26 × 600 mm; Amersham Bioscience, NJ, USA). As the two proteins were eluted separately with gel-filtration chromatography, the respective fractions were collected. Each protein was concentrated separately and buffer-exchanged with 50 mM Tris–HCl pH 7.5 using Vivaspin 20 membranes (10 000 MWCO; Sartorius KK, Gottingen, Germany) or Amicon Centriprep YM-10 membranes (Millipore, MA, USA). Protein concentrations were estimated with a protein assay kit (Bio-Rad, CA, USA; Bradford, 1976 ▶) using BSA as a standard.
The electron transferability between the purified CARDO-OJ3 and CARDO-FCA10 was confirmed by detecting oxygenase activity for carbazole of reconstituted CARDO (Nam et al., 2002 ▶).
Western blot analysis was performed using the ECL Plus Western Blotting Detection System (Amersham Bioscience) according to the manufacturer’s instructions. Denatured samples of dissolved crystals were separated using 15% SDS–PAGE and electrophoretically transferred onto Trans-Blot Transfer Medium Nitrocellulose membranes (Bio-Rad). The CarAcCA10 protein was detected immunologically using a mouse polyclonal antibody as the primary antibody and a horseradish-peroxidase-labelled antibody as the secondary antibody. The CarAcCA10 protein signals were visualized using the Luminescent Image Analyzer LAS-1000 Plus (Fuji Photo Film, Tokyo, Japan).
2.2. Crystallization
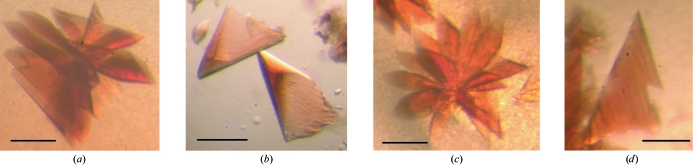
For crystallization experiments, purified CARDO-OJ3 and CARDO-FCA10 proteins were mixed in an approximate molar ratio of 1:6 or 1:3. The concentration of the protein mixture in 50 mM Tris–HCl pH 7.5 was adjusted to 15–30 mg ml−1. The proteins were crystallized using the hanging-drop vapour-diffusion method at 293 K. Drops containing 3 µl protein solution and 3 µl mother liquor were equilibrated against 700 µl reservoir solution. The initial crystallization was screened using Crystal Screen I/II, Grid Screens (PEG 6000 and MPD), Index Screens (Hampton Research, CA, USA), Wizard I/II (Jena Bioscience, Jena, Germany) and JBScreen (Emerald Biostructures, WA, USA). The initial crystallization trials showed that protein crystals appeared under a variety of conditions. After optimizing the crystallization parameters, protein concentration and molar ratio in the protein mixture, blossom-shaped crystals built up from piles of triangular plates were obtained with two reservoir solutions using VDX Plates (Hampton Research) and 20 mg ml−1 protein solution (CARDO-OJ3:CARDO-FCA10 = 1:6) at 293 K (drops contained 3 µl protein solution and 3 µl precipitating solution). Type I crystals (Figs. 2 ▶ a and 2 ▶ b) were formed with 1–2 mM sodium dithionite and 14%(v/v) PEG MME 2000 in 0.05 M Bis-Tris pH 6.5 and type II crystals (Figs. 2 ▶ c and 2 ▶ d) were formed with 0.1 M ammonium acetate and 12.5%(v/v) PEG 3350 in 0.05 M Bis-Tris pH 5.5. Growth of type I and type II crystals was observed after two weeks and 2–3 d, respectively. The triangle plate-shaped crystal pieces grew to maximum dimensions of 0.3 × 0.2 × 0.05 mm after four weeks and one week for types I and II, respectively.
Figure 2.
Photographs of the CARDO-OJ3 and CARDO-FCA10 complex crystals. The lines indicate 0.2 mm. (a) Type I crystals grown in 1–2 mM sodium dithionite, 14%(v/v) PEG MME 2000 and 0.05 M Bis-Tris pH 6.5. (b) Triangular plate-shaped crystals; pieces of the blossom-like crystals shown in (a). (c) Type I crystals grown in 0.1 M ammonium acetate, 12.5%(v/v) PEG 3350 and 0.05 M Bis-Tris pH 5.5. (d) Triangular plate-shaped crystals; pieces of the blossom-like crystals shown in (c).
2.3. Data collection
Owing to the sensitivity of the crystals to X-ray radiation, cryocooling was necessary during data collection to avoid crystal decay at room temperature. Both crystals were directly transferred into cryoprotectant solution (reservoir solution containing 20% glycerol), mounted in a nylon loop and flash-frozen in a nitrogen stream at 100 K. Diffraction experiments were conducted at beamlines AR-NW12 and BL-5A, Photon Factory, Tsukuba, Japan. Diffraction data were gathered with a wavelength of 1.0 Å using a Quantum 210 CCD X-ray detector (ADSC, CA, USA) at AR-NW12 and a Quantum 315 CCD X-ray detector (ADSC, CA, USA) at BL-5A. For the type I crystal, two data sets were collected initially using a single crystal in 0.5° oscillation steps over a range of 180° with 15 s exposure per frame and in 0.5° oscillation steps over a range of 270° with 2 s exposure per frame to cover the low-resolution diffraction; the two data sets were scaled and merged into a single data set using the HKL2000 program suite (Otwinowski & Minor, 1997 ▶). The type II crystal data set was collected in 0.5° oscillation steps over a range of 180° with 30 s exposure per frame. All diffraction images were indexed, integrated and scaled using the HKL2000 program suite.
3. Results and discussion
Before crystallization, the purified CARDO-OJ3 was confirmed to retain its angular dioxygenation activity for carbazole when the electron-transfer proteins CARDO-FCA10 and CARDO-RCA10 were supplied (data not shown). This result clearly indicates that CARDO-OJ3 can receive electrons from CARDO-FCA10.
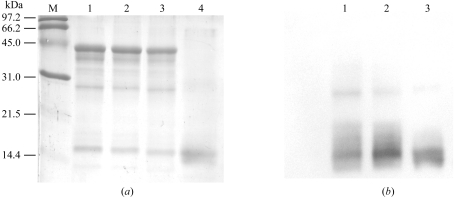
To ensure that the two types of crystal consisted of both CARDO-OJ3 and CARDO-FCA10, SDS–PAGE followed by Western blot analysis was performed using the dissolved crystals. The SDS–PAGE analysis clearly indicated that both types of crystals contained CarAaJ3 (CARDO-OJ3 monomer; Figs. 3 ▶ a and 3 ▶ b, lanes 2 and 3). This result was further confirmed by Western blot analysis, which showed clear detection of CarAaJ3 with ∼44 kDa molecular weight (data not shown). On the other hand, because the degradation peptide that originated from CarAaJ3 had a molecular size similar to that of CarAcCA10 (Fig. 3 ▶ a, lanes 1, 2 and 3), we could not conclude that both types of crystals contained CarAcCA10 (CARDO-FCA10). Western blots using anti-CarAcCA10 antibody indicated that both crystals contained CarAcCA10 protein, although the bands at about 30 kDa reacted slightly with anti-CarAcCA10 antibody (Fig. 3 ▶ b).
Figure 3.
SDS–PAGE and Western blot analysis of the dissolved crystals. (a) SDS–PAGE of the dissolved crystals (5 µg each). Lane M, low-molecular-weight markers; lane 1, dissolved crystals of CARDO-OJ3; lane 2, dissolved type I crystals of the complex; lane 3, dissolved type II crystals of the complex; lane 4, dissolved crystals of CARDO-FCA10. (b) Western blots using anti-CarAcCA10 antibody. Lane numbers are the same as in (a).
The merged data sets for the type I and II crystals were collected to 1.90 and 2.05 Å resolution, respectively. The data-collection and processing statistics are summarized in Table 1 ▶. The space groups of both crystals were determined to be P21, with similar unit-cell parameters. During purification using gel-filtration chromatography, the molecular weights of CARDO-OJ3 and CARDO-FCA10 were calculated to be 132 and 13 kDa (data not shown), suggesting that CARDO-OJ3 and CARDO-FCA10 have α3 homotrimeric and monomeric configurations, respectively. These quaternary structures suggested by biochemical experiments are in accordance with the crystal structures of CARDO-OJ3 and CARDO-FCA10 as single proteins (Nam et al., 2005 ▶; Nojiri et al., 2005 ▶). The trimeric structure of CARDO-OJ3 has three completely separate active sites (catalytic non-haem irons), suggesting that three regions capable of interacting with CARDO-FCA10 exist around three Rieske [2Fe–2S] clusters in CARDO-OJ3. Therefore, assuming one molecule of CARDO-OJ3 and three molecules of CARDO-FCA10 per asymmetric unit, the Matthews coefficient V M (Matthews, 1968 ▶) is 2.63 Å3 Da−1 (corresponding to a solvent content of 53.2%). Consequently, the asymmetric unit in the crystal may contain one complex that is a trimer of a heterodimer consisting of CARDO-OJ3 and CARDO-FCA10 molecules.
Table 1. Crystal parameters and data-collection statistics.
The data were collected on AR-NW12 and BL-5A at the Photon Factory, Tsukuba, Japan. Values in parentheses are for the highest resolution shell.
| Crystal type | Type I | Type II |
|---|---|---|
| Beamline | AR-NW12 | BL-5A |
| Wavelength (Å) | 1.0 | 1.0 |
| Space group | P21 | P21 |
| Unit-cell parameters (Å, °) | a = 97.1, b = 89.8, c = 104.9, α = γ = 90.0, β = 103.8 | a = 98.2, b = 90.0, c = 105.1, α = γ = 90.0, β = 104.0 |
| Resolution range (Å) | 50.0–1.90 (1.97–1.90) | 50.0–2.05 (2.12–2.05) |
| Total No. of reflections | 1131785 | 402412 |
| No. of unique reflections | 138930 (13797) | 109845 (10911) |
| Completeness (%) | 100 (100) | 99.6 (99.7) |
| Average I/σ(I) | 47.8 (3.3) | 31.7 (3.7) |
| Rmerge (%) | 8.0 (41.9) | 7.2 (37.5) |
| Multiplicity | 8.1 (3.7) | 3.7 (3.7) |
We are now attempting to determine the structure of this complex by the molecular-replacement method using the determined structures of CARDO-OJ3 and CARDO-FCA10 and the refinement is now in progress. Until now, various types of electron transport between proteins have been reported, but there are only a few reports of their three-dimensional structures. Three structures of complexes of ferredoxins with their reductases have been determined: the complexes of Anabaena and maize ferredoxin with their respective ferredoxin:NADP+ oxidoreductase in oxygenic photosynthesis (Morales et al., 2000 ▶; Kurisu et al., 2001 ▶) and the complex of mitochondria adrenodoxin with adrenodoxin reductase in steroid biosynthesis (Müller et al., 2001 ▶). To the best of our knowledge, the complex structure of the terminal oxygenase with its electron-donor protein has not been determined yet, except for the complex structure of the haem- and FMN-binding domains of the bacterial cytochrome P450BM-3, a prototype for the complex between eukaryotic microsomal P450 monooxygenase and P450 reductase (Sevrioukova et al., 1999 ▶). Therefore, the complex structure of CARDO-OJ3 with CARDO-FCA10 will represent the first structure of a biologically relevant complex between an oxygenase and its electron donor, ferredoxin.
Several reports have indicated that while the reductase components of ROSs can be replaced by unrelated reductases, the ferredoxin components cannot be replaced (Subramanian et al., 1981 ▶, 1985 ▶; Haiger & Gibson, 1990 ▶; Fukuda et al., 1994 ▶). In the CARDOCA10 system, CARDO-RCA10 can be replaced by an unrelated reductase, such as the ferredoxin reductase from spinach, but CARDO-FCA10 is indispensable for electron transfer to CARDO-OCA10, suggesting that there is a specific interaction between CARDO-OCA10 and CARDO-FCA10 (Nam et al., 2002 ▶). As no functional difference has been observed between CARDO-OJ3 and CARDO-OCA10 in several experiments, the forthcoming structure of the CARDO-OJ3 and CARDO-FCA10 complex will provide insight into the structural basis of the specific interaction between the CARDO components. In addition, as the first structural example of an ROS electron-transport complex structure, it will also provide useful information for understanding the general architecture of the component interactions and electron transport in this important class of multicomponent oxygenase systems.
Acknowledgments
Synchrotron radiation was used for this work with the approval of the Photon Factory Advisory Committee and KEK (High Energy Accelerator Research Organization), Tsukuba (proposal No. 2003G124). This work was supported in part by PROBRAIN (Promotion of Basic Research Activities for Innovative Bioscience).
References
- Batie, C. J., Ballou, D. P. & Correll, C. C. (1991). Chemistry and Biochemistry of Flavoenzymes, Vol. 3, edited by F. Muller, pp. 543–556. Boca Raton, FL, USA: CRC Press.
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed] [Google Scholar]
- Bressler, D. C. & Fedorak, P. M. (2000). Can. J. Microbiol.46, 397–409. [PubMed] [Google Scholar]
- Fukuda, M., Yasukouchi, Y., Kikuchi, Y., Nagata, Y., Kimbara, K., Horiuchi, H., Takagi, M. & Yano, K. (1994). Biochem. Biophys. Res. Commun.202, 850–856. [DOI] [PubMed] [Google Scholar]
- Furukawa, K., Suenaga, H. & Goto, M. (2004). J. Bacteriol.186, 5189–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habe, H., Chung, J.-S., Lee, J.-H., Kasuga, K., Yoshida, T., Nojiri, H. & Omori, T. (2001). Appl. Environ. Microbiol.67, 3610–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habe, H. & Omori, T. (2003). Biosci. Biotechnol. Biochem.67, 225–243. [DOI] [PubMed] [Google Scholar]
- Haiger, B. E. & Gibson, D. T. (1990). J. Bacteriol.172, 465–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., Widada, J., Nakai, S., Endoh, T., Urata, M., Ashikawa, Y., Shintani, M., Saiki, Y., Yoshida, T., Habe, H., Omori, T. & Nojiri, H. (2004). Biosci. Biotechnol. Biochem.68, 1467–1480. [DOI] [PubMed] [Google Scholar]
- Kurisu, G., Kusunoki, M., Katoh, E., Yamazaki, T., Teshima, K., Onda, Y., Kimata-Ariga, Y. & Hase, T. (2001). Nature Struct. Biol.8, 117–121. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Morales, R., Charon, M.-H., Kachalova, G., Serre, L., Medina, M., Gómez-Moreno, C. & Frey, M. (2000). EMBO Rep.1, 271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, J. J., Lapko, A., Bourenkov, G., Ruckpaul, K. & Heinemann, U. (2001). J. Biol. Chem.276, 2786–2789. [DOI] [PubMed] [Google Scholar]
- Nam, J.-W., Noguchi, H., Fujimoto, Z., Mizuno, H., Ashikawa, Y., Abo, M., Fushinobu, S., Kobashi, K., Wakagi, T., Iwata, K., Yoshida, T., Habe, H., Yamane, H., Omori, T. & Nojiri, H. (2005). Proteins, 58, 779–789. [DOI] [PubMed] [Google Scholar]
- Nam, J.-W., Nojiri, H., Noguchi, H., Uchimura, H., Yoshida, T., Habe, H., Yamane, H. & Omori, T. (2002). Appl. Environ. Microbiol.68, 5882–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojiri, H., Ashikawa, Y., Noguchi, H., Nam, J.-W., Urata, M., Fujimoto, Z., Uchimura, H., Terada, T., Nakamura, S., Shimizu, K., Yoshida, T., Habe, H. & Omori, T. (2005). Submitted. [DOI] [PubMed]
- Nojiri, H., Nam, J.-W., Kosaka, M., Morii, K., Takemura, T., Furihata, K., Yamane, H. & Omori, T. (1999). J. Bacteriol.181, 3105–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojiri, H. & Omori, T. (2002). Biosci. Biotechnol. Biochem.66, 2001–2016. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Rosche, B., Tshisuaka, B., Fetzner, S. & Lingens, F. (1995). J. Biol. Chem.270, 17836–17842. [DOI] [PubMed] [Google Scholar]
- Sato, S., Nam, J.-W., Kasuga, K., Nojiri, H., Yamane, H. & Omori, T. (1997). J. Bacteriol.179, 4850–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova, I. F., Li, H., Zhang, H., Petrson, J. A. & Poulos, T. L. (1999). Proc. Natl Acad. Sci. USA, 96, 1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, V., Liu, T.-N., Yeh, W.-K., Narro, M. & Gibson, D. T. (1981). J. Biol. Chem.256, 2723–2730. [PubMed] [Google Scholar]
- Subramanian, V., Liu, T.-N., Yeh, W.-K., Serdar, C. M., Wakett, L. P. & Gibson, D. T. (1985). J. Biol. Chem.260, 2355–2363. [PubMed] [Google Scholar]
- Takagi, T., Nojiri, H., Yoshida, T., Habe, H. & Omori, T. (2002). Biotechnol. Lett.24, 2099–2106.
- Wittich, R.-M. (1998). Appl. Microbiol. Biotechnol.49, 489–499. [DOI] [PubMed] [Google Scholar]