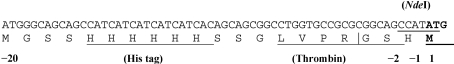Adenosine kinase from M. tuberculosis has been overexpressed, purified and crystallized in the presence of adenosine. Structure determination using molecular replacement with diffraction data collected at 2.2 Å reveals a dimeric structure.
Keywords: adenosine kinase, Mycobacterium tuberculosis
Abstract
Adenosine kinase from Mycobacterium tuberculosis is the only prokaryotic adenosine kinase that has been isolated and characterized. The enzyme catalyzes the phosphorylation of adenosine to adenosine monophosphate and is involved in the activation of 2-methyladenosine, a compound that has demonstrated selective activity against M. tuberculosis. The mechanism of action of 2-methyladenosine is likely to be different from those of current tuberculosis treatments and this compound (or other adenosine analogs) may prove to be a novel therapeutic intervention for this disease. The M. tuberculosis adenosine kinase was overexpressed in Escherichia coli and the enzyme was purified with activity comparable to that reported previously. The protein was crystallized in the presence of adenosine using the vapour-diffusion method. The crystals diffracted X-rays to high resolution and a complete data set was collected to 2.2 Å using synchrotron radiation. The crystal belonged to space group P3121, with unit-cell parameters a = 70.2, c = 111.6 Å, and contained a single protein molecule in the asymmetric unit. An initial structural model of the protein was obtained by the molecular-replacement method, which revealed a dimeric structure. The monomers of the dimer were related by twofold crystallographic symmetry. An understanding of how the M. tuberculosis adenosine kinase differs from the human homolog should aid in the design of more potent and selective antimycobacterial agents that are selectively activated by this enzyme.
1. Introduction
Nucleoside analogs are an important class of drugs that are used in the treatment of viral infections and cancer. Recently, we have identified several nucleoside analogs that are selectively active against Mycobacterium tuberculosis, a human pathogen causing tuberculosis (TB; Chen et al., 2002 ▶; Barrow et al., 2003 ▶). One of these agents, 2-methyladenosine, proved to be activated by adenosine kinase (AK, ATP:adenosine 5′-phosphotransferase; EC 2.7.1.20) that was endogenous to the mycobacterial cells (Barrow et al., 2003 ▶; Chen et al., 2002 ▶; Parker et al., 2004 ▶). AK catalyzes the phosphorylation of adenosine and is an important enzyme in the regulation of cellular levels of adenosine and its nucleotides. AK from a variety of sources exhibits a relatively broad substrate specificity, tolerating modifications in both the sugar and base moieties, and converts many analogs to cytotoxic compounds (Barrow et al., 2003 ▶; Miller et al., 1979 ▶; Parker et al., 2004 ▶; Willis et al., 1978 ▶; Yamada et al., 1981 ▶). The M. tuberculosis AK (Mtb-AK) gene has recently been identified and cloned, and its gene product has been characterized biochemically (Long et al., 2003 ▶). Mtb-AK activity was required for the phosphorylation of 2-methyladenosine and for its antitubercular activity (Barrow et al., 2003 ▶; Parker et al., 2004 ▶). Therefore, Mtb-AK plays an important role in the bioactivation of this agent and offers a promising target for therapeutic development against TB.
The current treatment of active TB is a four-drug regimen comprising isoniazid, rifampin, pyrazinamide and ethambutol for a period of at least six months (Cohn et al., 1990 ▶; East African–British Medical Research Councils, 1974a ▶,b ▶). These drugs inhibit cell-wall formation and RNA synthesis of the mycobacterium (Chopra & Brennan, 1997 ▶). The failure of patients to complete six months of this therapy has led to the emergence of multi-drug-resistant (MDR) strains of M. tuberculosis (Bloom & Murray, 1992 ▶; Cohn et al., 1997 ▶; Ramaswamy & Musser, 1998 ▶). TB is a major opportunistic infection associated with HIV/AIDS; many latent TB infections were reactivated when patients were infected with HIV. The synergy of TB with HIV infection claims millions of lives annually. Thus, efforts to discover new drugs to combat TB are warranted. Nucleoside analogs may prove to be particularly useful in treating MDR TB, since their mechanism of action is likely to be different from those of existing therapies. To understand the substrate specificity of Mtb-AK and the structural basis for the selective activation of nucleoside analogs by the enzyme, efforts have been made to overexpress, purify and crystallize the enzyme for three-dimensional structure determination by X-ray crystallography.
Mtb-AK contains 324 amino acids with a molecular weight of 35.5 kDa and forms a dimer in solution (Long et al., 2003 ▶). The enzyme belongs to the phosphofructokinase B (PfkB) family of carbohydrate and nucleoside kinases, a family of structurally related enzymes that includes ribokinase, fructokinase and hexokinase (Spychala et al., 1996 ▶). AKs from various mammalian and parasitic sources have been characterized, and the crystal structures of AKs from human (Mathews et al., 1998 ▶) and Toxoplasma gondii (Schumacher et al., 2000 ▶) have been reported. However, the enzyme is not commonly found in bacteria. From the results of sequence and phylogenic analyses, Mtb-AK showed very low sequence homology (less than 20% sequence identity) with AKs from eukaryotic cells and was more closely related to ribokinases from other sources (Long et al., 2003 ▶). Sequence-similarity searches over DNA sequence databases have shown that Mtb-AK is highly homologous to some genes, putatively identified as carbohydrate kinases or sugar kinases, from several other Gram-positive and Gram-negative bacteria, suggesting that AK may exist in other bacteria. Structural comparison of this unique enzyme with the human AK could shed light on the molecular basis for the selective antimycobacterial activity of 2-methyladenosine and could aid in the design of more potent and selective antimycobacterial adenosine analogs. Structural insights will likely provide an understanding of the enzyme’s function and its general reaction mechanism in prokaryotic cells.
2. Experimental
2.1. Cloning and protein expression of Mtb-AK
The gene encoding AK was amplified by PCR from the genomic DNA of the M. tuberculosis strain H37Rv using two primers for upstream and downstream sequences: 5′-GGACGGAGATCATATGACGATCGCGGTAACC-3′ (upstream) and 5′-CCACACGGTGGAATTCCGCGTCTGCTCGGC-3′ (downstream). The DNA was subcloned using NdeI and EcoRI restriction sites into the pET28a vector (Novagen) to generate a recombinant vector containing a 5′ sequence encoding a 20-amino-acid N-terminal His tag and a thrombin-cleavage site (Fig. 1 ▶)
Figure 1.
The His tag sequence in the recombinant vector.
The entire coding sequence was verified by DNA sequencing. The resulting plasmid was transformed into competent cells of Escherichia coli strain BL21*(DE3) and the transformed cells were selected on LB agar plates containing 50 µg ml−1 kanamycin. For protein expression, the E. coli cells containing the recombinant vector were inoculated and grown at 310 K for about 6 h in 50 ml LB medium containing kanamycin. Isopropyl β-d-thiogalactopyranoside (IPTG), the expression inducer, was added to a concentration between 0.05 and 1 mM when the optical absorbance of the culture reached 0.7–0.8 at 600 nm and the cultures were grown at different temperatures from 290 to 310 K for an additional 6–40 h. Cells were harvested by centrifugation, resuspended and lysed using the BugBuster Protein Extraction Reagent (Novagen) containing benzonase. The cell lysate was separated by centrifugation into soluble and insoluble fractions. These samples were analyzed for protein expression by protein gel electrophoresis. It was found that the bacterial cell expressed mostly soluble AK protein when the expression temperature was controlled at about 295 K with 0.2 mM IPTG and an expression time of about 20 h. Large-scale cell culture was performed under the optimized conditions and harvested cell pellets were stored at 193 K.
2.2. Protein purification
Cells were resuspended in the lysis buffer (50 mM Tris–HCl pH 8.0, 50 mM NaCl, 1 mM TCEP) supplemented with a protease-inhibitor cocktail and 1 µg ml−1 benzonase and were lysed by two passages through a French press. Cell debris was removed by centrifugation and the clear supernatant was passed through a nickel-chelating Sepharose column (Amersham Biosciences) equilibrated with the lysis buffer. The column was washed extensively with lysis buffer containing 50 mM imidazole for the removal of non-specific binding proteins. The bound protein was eluted using a 300 ml linear gradient of 50–500 mM imidazole. Elution fractions were analyzed by SDS–PAGE and the fractions containing Mtb-AK were pooled and buffer-exchanged to reduce the concentration of imidazole. Further purification was performed with a strong anion-exchange column (Mono-Q, Amersham Biosciences) and a gel-filtration column (Superdex 75, Amersham Biosciences). Eluted fractions from each column were analyzed for kinase activity and protein purity by gel electrophoresis using both native and denatured conditions. Active enzyme fractions were pooled and concentrated. The protein concentration was determined as described by Lowry et al. (1951 ▶), with BSA as the standard. The protein buffer was exchanged gradually during concentration into 20 mM Tris–HCl pH 8.0, 20 mM NaCl, 10 mM KCl, 5 mM MgCl2, 1 mM DTT and 5% glycerol (referred to as protein buffer). The additives MgCl2, DTT and glycerol were added to stabilize the protein in solution.
An attempt was made to remove the His tag by treating the sample with biotinylated thrombin for about 16 h. The His tag, thrombin and uncut Mtb-AK were removed using a streptavidin agarose column and an Ni-chelating Sepharose column. However, the cleavage of the His tag was partial. Most of the protein recovered after gel filtration retained the His tag.
2.3. Adenosine kinase assay
The enzyme activity of Mtb-AK was assayed by monitoring the phosphorylation of [3H]-adenosine with a DEAE cellulose filter disk as described previously (Long et al., 2003 ▶). Briefly, phosphorylation of [3H]-adenosine was quantified by the amount of [3H]-AMP bound to a DE-81 cellulose disk following the reaction. The reaction was started by the addition of enzyme to the reaction solution, which contained 50 mM Tris–HCl pH 8.0, 10 mM KCl, 10 mM MgCl2, 5 mM ATP and 20 µM [3H]-adenosine (10 µCi ml−1). The reaction mixture was incubated for the desired time at 310 K and the reaction was stopped by the addition of 10 µl 0.1 M EDTA. The reaction mixture was applied to DEAE cellulose disks in 50 µl aliquots and the disks were washed with 1 mM ammonium acetate pH 5.0, rinsed with 95% ethanol and then dried. The filter disks were transferred to scintillation vials with 10 ml of Complete Counting Cocktail (Research Products International) and radioactivity was detected with a Packard Tri-Carb model 1900 TR liquid-scintillation analyzer. The control reaction contained the same amount of all components without the enzyme.
2.4. Protein crystallization
Crystallization of Mtb-AK was carried out by the sitting-drop vapor-diffusion method. The purified protein sample was used with adenosine at a final concentration of 1 mM. After incubation on ice for about 30 min, the protein solution was mixed with an equal volume of the crystallization solution (∼2 µl) and the drop of mixture was equilibrated with 200 µl crystallization solution in a sealed chamber. The initial crystallization condition screening was performed at both room temperature and 277 K with the sparse-matrix Crystallization Screen Kits from Hampton Research. Crystals were observed in two preliminary conditions: (i) 30%(w/v) PEG MME 5K as precipitant and 100 mM MES pH 6.5 buffer and (ii) 20% PEG 4K and 100 mM sodium citrate pH 5.6. The protein concentration was varied from 5 to 30 mg ml−1. These conditions were fine-tuned by optimizing the pH value of the buffer and the concentration of the precipitant and additives. Large crystals were produced from the final crystallization solutions containing 28–30% PEG MME 5K, 200 mM ammonium sulfate and 100 mM MES pH 6.3–6.5 and are referred to as form 1 crystals. Many small thin-plate crystals (form 2) were observed in solutions at lower pH values (5.7–6.1). The protein concentration used in the final conditions was 10 mg ml−1 in protein buffer (see §2.2) with the presence of 1 mM adenosine.
2.5. Data collection, processing and analysis
Diffraction data were collected at the Northeastern Collective Access Team (NE-CAT) beamline 8-BM at the Advanced Photon Source, Argonne National Laboratory. A crystal soaked in a solution containing 20% glycerol as the cryoprotectant and 35% PEG MME 5K was mounted and rapidly frozen in a nitrogen-gas stream at a temperature of 100 K using a cryocooling device from Oxford Cryosystems. Intensity data were recorded using an ADSC Quantum 315 CCD area detector with an oscillation angle of 0.75–1.0° per frame and an exposure time of 10 s. The crystal-to-detector distance was set at 350 mm. The diffraction data were indexed, integrated and scaled using HKL2000 (Otwinowski & Minor, 1997 ▶). The resulting reflection data were then placed on an absolute scale with TRUNCATE (French & Wilson, 1978 ▶). Further data analysis was performed using programs from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). These data analyses allowed the determination of the unit-cell parameters and showed the space group to be either P3121 or P3221. The ambiguity in the correct choice of enantiomorph was eventually resolved by the molecular-replacement method.
2.6. Structural analysis
Molecular replacement was used to determine the initial phases. Self-rotation function maps were calculated in spherical polar angles using the CCP4 program POLARRFN to determine possible molecular symmetry. Reflection data within the range 15.0–2.6 Å and an integration radius of 25 Å were used in the self-rotation function calculation. Cross-rotation function and translation-function searches were carried out using the program AMoRe (Navaza, 2001 ▶). Mtb-AK has only about 18% or less sequence identity to other known adenosine kinases or sugar kinases. Available structures of related proteins, such as adenosine kinases from human (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1bx4; Mathews et al., 1998 ▶) and T. gondii (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1dmg; Schumacher et al., 2000 ▶), 2-keto-3-deoxygluconate kinase from Thermus thermophilus (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1v1s; Ohshima et al., 2004 ▶) and ribokinases from E. coli (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1rkd; Sigrell et al., 1998 ▶) and Thermotoga maritima (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1vm7; Joint Center For Structural Genomics, unpublished work), were used as search probes in the initial molecular-replacement experiments. The search probe used to obtain the molecular-replacement solution was designed based on comparative structural analysis of known structures of adenosine kinase and other sugar kinases. In this model, only the conserved core structure of 2-keto-3-deoxygluconate kinase was used in the search probe, which contained residues 3–10, 34–47, 50–73, 127–132, 139–164, 184–193, 214–228 and 269–284. Both P3121 and P3221 space groups were tried in the translation-function search. Use of P3121 in the search yielded the solution, indicating this enantiomorph to be the correct space group of the form 1 Mtb-AK crystal. CNS (Brünger et al., 1998 ▶) and the molecular-graphics program O (Jones et al., 1991 ▶) were used for crystal packing and geometric analysis. CNS was also used for model refinement.
3. Results and discussion
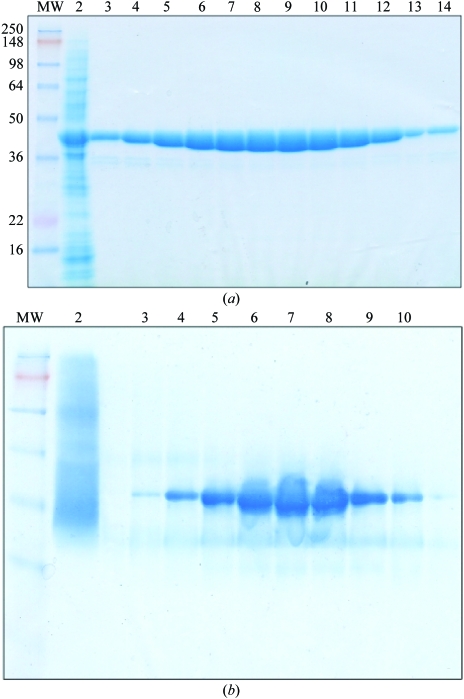
Mtb-AK was expressed in E. coli as a soluble protein when the expression temperature was reduced to 291–295 K. The expression yield was about 20 mg per litre of cell culture. The protein was purified by Ni2+-affinity chromatography as shown by SDS–PAGE (Fig. 2 ▶ a). The protein samples of the eluted fractions migrated as a single major band corresponding to a molecular weight of between 36 and 50 kDa, in agreement with the calculated molecular weight of 38.0 kDa (including the 20-amino-acid affinity tag). The protein gel under native conditions showed a clear major band (Fig. 2 ▶ b), indicating that the protein was well folded with a uniform conformation in solution. Attempts were made to cleave the His tag with thrombin. However, only a small portion of the protein had the His tag removed. The protein sample was further purified by ion-exchange and gel-filtration chromatography to remove minor contaminant proteins. A kinase activity assay of the purified His-tagged protein showed that the recombinant Mtb-AK expressed from E. coli was active, with a specific activity of about 1200 µmol mg−1 min−1. This activity was similar to the activity of the enzyme purified from mycobacterial cells (Long et al., 2003 ▶), which indicated that the His tag did not affect the enzyme activity.
Figure 2.
Purification of Mtb-AK protein expressed in E. coli and protein electrophoresis under denatured (a) and native conditions (b) in 8–16% polyacrylamide gradient gels. (a) SDS–PAGE. Lane 1, molecular-weight (MW) markers; lane 2, crude cell extract; lanes 3–14, elution fractions from Ni2+-affinity column. (b) Native PAGE. Lane 1, MW markers; lane 2, crude cell extract; lanes 3–10, elution fractions from the same column.
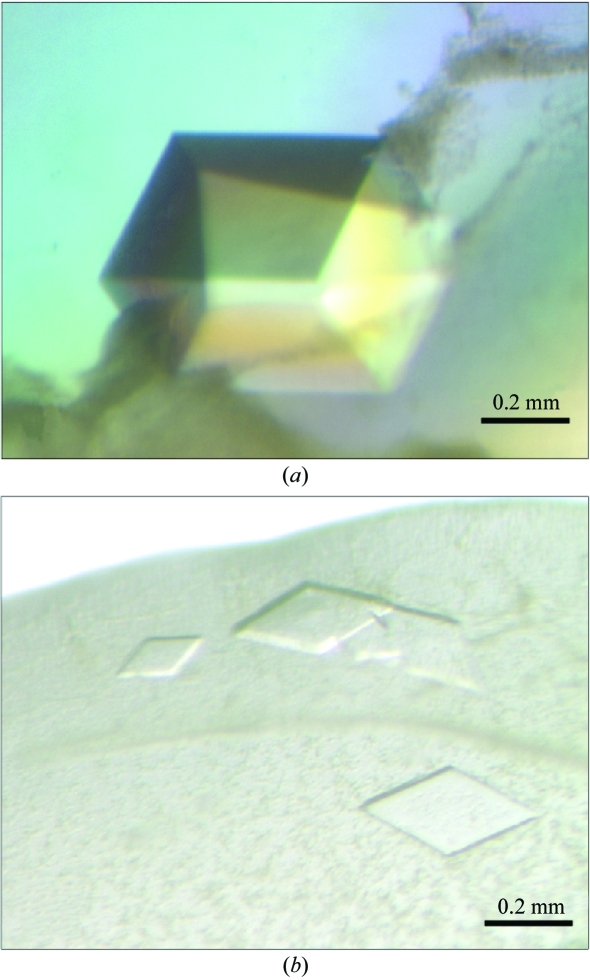
Two crystallization conditions for Mtb-AK were found after initial screening with factorial solution sets, but only one of these conditions yielded large crystals after optimization (Fig. 3 ▶ a). The best crystals with typical dimensions of 0.3 × 0.3 × 0.4 mm (form 1) were found in sitting drops formed at 277 K by mixing 1 µl crystallization solution with the protein sample at 10 mg ml−1 in the protein buffer with the addition of 1 mM adenosine. The crystallization solution contained 28–30%(w/v) PEG MME 5K, 200 mM ammonium sulfate and 100 mM MES pH 6.3–6.5. Many small crystals in different shapes (thin plates, form 2) were found when the pH value was lower than 6.1 (Fig. 3 ▶ b). No crystals were formed when adenosine was not added to the protein solution.
Figure 3.
Mtb-AK crystals. The protein crystallized in different crystal forms when the pH value of the buffer was varied slightly. (a) pH ≃ 6.3–6.5 (form 1), (b) pH < 6.1 (form 2).
Mtb-AK crystals of form 1 diffracted X-rays to 1.9 Å using synchrotron radiation. A complete data set was collected to 2.2 Å and was processed with the HKL2000 program. The highest symmetry was found to be in the primitive trigonal lattice. Data scaling was performed using SCALEPACK (Otwinowski & Minor, 1997 ▶), which confirmed that the crystal belonged to either the space group P3121 or P3221, with unit-cell parameters a = 70.2, c = 111.6 Å. The space group was later determined to be P3121 based on the structure solution solved by the molecular-replacement method. The calculated packing parameter, based on a molecular weight of 38 kDa, indicated the presence of one monomer in the asymmetric unit. The self-rotation function calculated by POLARRFN showed no significant peaks other than the origin peaks, which confirmed that there was one molecule in the asymmetric unit. This corresponds to a Matthews coefficient (V M) of 2.33 Å3 Da−1 and 45.0% solvent content, which are within the expected range (Matthews, 1968 ▶). The statistics of the data collection and processing are shown in Table 1 ▶. Crystals of form 2 formed at low pH diffracted X-rays to 3.0 Å at best and had a unit cell that was different from that of form 1 only in the c axis. The c dimension of the form 2 crystal was 218.0 Å, almost double that of the form 1 crystal.
Table 1. Summary of data-collection and processing statistics of the Mtb-AK crystal.
Values in parentheses are for the last resolution shell (2.3–2.2 Å).
| Space group | P3121 |
| Unit-cell parameters | |
| a (Å) | 72.2 |
| c (Å) | 111.6 |
| Resolution (Å) | 50.0–2.2 |
| Wavelength (Å) | 1.00928 |
| Observations | 235757 |
| Unique reflections | 18930 |
| Completeness (%) | 98.0 (72.0) |
| Redundancy | 7.5 (4.5) |
| Rmerge (%) | 3.2 (19.6) |
| I/σ(I) | 23 (3.5) |
| Solvent content (%) | 45.0 |
| VM (Å3 Da−1) | 2.33 |
| Monomers per AU | 1 |
Molecular replacement was applied to obtain initial protein phases for the diffraction data using the available structures of related proteins as a search probe, including adenosine kinases, 2-keto-3-deoxygluconate kinase and ribokinases. The overall sequence identity of Mtb-AK to these proteins is less than 20%; however, the sequence identity of the catalytic core domain is about 30% among these proteins. Although these carbohydrate kinases share low sequence homology, they have a similar fold, especially in the catalytic domain. After we failed to find a solution with these protein structures, a model was designed by removing the non-conserved segments based on the structural alignment. This model contains only the main-chain atoms of the core domain and the Cβ atom if the amino acid is larger than glycine. With this designed model, a rotation search using AMoRe in the resolution shell 15–5.5 Å provided a peak above the background. A translation search with this orientation revealed the solution when space group P3121 was used, which indicated the true space group of the form 1 AK crystal. This unique solution had a correlation coefficient of 0.455 and an R factor of 48%.
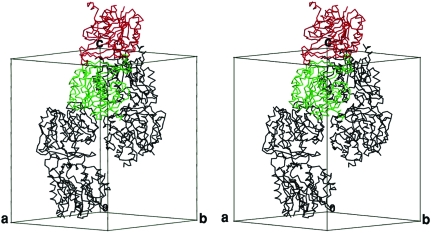
By applying the transformation matrix of the top solution to either the designed search model or the original structure of 2-keto-3-deoxygluconate kinase, an initial model was generated and there were no violations of crystal packing observed when the crystallographic symmetry operations were applied. Preliminary refinement of the initial model using the protocols of rigid-body and positional refinements reduced both the conventional R and free R values significantly. These results indicated that the model obtained by molecular replacement was the correct solution. Two molecules related by twofold crystallographic symmetry interact through the smaller lid domain, mainly in the β-sheet structure (Fig. 4 ▶). This dimer interaction is similar to that observed in ribokinase and 2-keto-3-deoxygluconate kinase. This structural evidence for Mtb-AK as a dimer is consistent with the published results (Long et al., 2003 ▶), although the detailed interactions are yet to be revealed. Interestingly, other AK enzymes with known structures have an α/β-structure in the small lid domain and function as monomers. It is possible that the Mtb-AK is structurally more similar to ribokinases than to AKs. Full crystal structure determination of Mtb-AK is in progress, which will reveal the nature of the dimer interaction, the conformation of the active site and the structural basis for substrate specificity.
Figure 4.
Stereoview of the molecular packing in the unit cell of the form 1 Mtb-AK crystal. Three dimers are shown in Cα backbone structure. The monomers of each molecular dimer are related by twofold crystallographic symmetry (as highlighted in red and green colors). This figure was generated by MOLSCRIPT (Kraulis, 1991 ▶).
In summary, we successfully expressed and purified recombinant Mtb-AK and obtained crystals of the protein in the presence of adenosine that diffracted X-rays to high resolution. An initial structural model was obtained using the molecular-replacement method and revealed a dimeric form of the protein. The high-resolution structure of Mtb-AK will reveal the protein–ligand interactions and substrate specificity that can be exploited for the design of adenosine analogs as potential TB therapeutic agents.
Acknowledgments
X-ray diffraction data were collected at beamline 8-BM in the facilities of the Northeast Collaborative Access Team (NE-CAT) at the Advanced Photon Source, Argonne National Laboratory. We thank Drs Jun Wang and Craig M. Ogata for their assistance and help during the data collection at the synchrotron facilities. We thank Dr Zhican Qu for participation in synchrotron data collection and careful reading of this manuscript. This work was supported in part by two NIH grants: AI55344 to RL and AI43241 to WBP.
References
- Barrow, E. W., Westbrook, L., Bansal, N., Suling, W. J., Maddry, J. A., Parker, W. B. & Barrow, W. W. (2003). J. Antimicrob. Chemother.52, 801–808. [DOI] [PubMed] [Google Scholar]
- Bloom, B. R. & Murray, C. J. (1992). Science, 257, 1055–1064. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Chen, C. K., Barrow, E. W., Allan, P. W., Bansal, N., Maddry, J. A., Suling, W. J., Barrow, W. W. & Parker, W. B. (2002). Microbiology, 148, 289–295. [DOI] [PubMed] [Google Scholar]
- Chopra, I. & Brennan, P. (1997). Tuber. Lung Dis.78, 89–98. [DOI] [PubMed] [Google Scholar]
- Cohn, D. L., Bustreo, F. & Raviglione, M. C. (1997). Clin. Infect. Dis.24, Suppl. 1, S121–S130. [DOI] [PubMed] [Google Scholar]
- Cohn, D. L., Catlin, B. J., Peterson, K. L., Judson, F. N. & Sbarbaro, J. A. (1990). Ann. Intern. Med.112, 407–415. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- East African-British Medical Research Councils (1974a). Lancet, 2, 237–240. [PubMed] [Google Scholar]
- East African-British Medical Research Councils (1974b). Lancet, 2, 1100–1106. [PubMed] [Google Scholar]
- French, S. & Wilson, K. (1978). Acta Cryst. A34, 517–525. [Google Scholar]
- Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kraulis, P. J. (1991). J. Appl. Cryst.24, 946–950. [Google Scholar]
- Long, M. C., Escuyer, V. & Parker, W. B. (2003). J. Bacteriol.185, 6548–6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. (1951). J. Biol. Chem.193, 265–275. [PubMed] [Google Scholar]
- Mathews, I. I., Erion, M. D. & Ealick, S. E. (1998). Biochemistry, 37, 15607–15620. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Miller, R. L., Adamczyk, D. L. & Miller, W. H. (1979). J. Biol. Chem.254, 2339–2345. [PubMed] [Google Scholar]
- Navaza, J. (2001). Acta Cryst. D57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Ohshima, N., Inagaki, E., Yasuike, K., Takio, K. & Tahirov, T. H. (2004). J. Mol. Biol.340, 477–489. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Parker, W. B., Barrow, E. W., Allan, P. W., Shaddix, S. C., Long, M. C., Barrow, W. W., Bansal, N. & Maddry, J. A. (2004). Tuberculosis, 84, 327–336. [DOI] [PubMed] [Google Scholar]
- Ramaswamy, S. & Musser, J. M. (1998). Tuber. Lung Dis.79, 3–29. [DOI] [PubMed] [Google Scholar]
- Schumacher, M. A., Scott, D. M., Mathews, I. I., Ealick, S. E., Roos, D. S., Ullman, B. & Brennan, R. G. (2000). J. Mol. Biol.298, 875–893. [DOI] [PubMed] [Google Scholar]
- Sigrell, J. A., Cameron, A. D., Jones, T. A. & Mowbray, S. L. (1998). Structure, 6, 183–193. [DOI] [PubMed] [Google Scholar]
- Spychala, J., Datta, N. S., Takabayashi, K., Datta, M., Fox, I. H., Gribbin, T. & Mitchell, B. S. (1996). Proc. Natl Acad. Sci. USA, 93, 1232–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, R. C., Carson, D. A. & Seegmiller, J. E. (1978). Proc. Natl Acad. Sci. USA, 75, 3042–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, Y., Goto, H. & Ogasawara, N. (1981). Biochim. Biophys. Acta, 660, 36–43. [DOI] [PubMed] [Google Scholar]