Crystals of the cytoplasmic domain of FlhA, a membrane-protein component of the bacterial flagellar type III protein-export apparatus from Salmonella, have been obtained and characterized by X-ray diffraction.
Keywords: flagellar type III export apparatus, FlhA
Abstract
The axial components of the bacterial flagellum and the scaffolding proteins for its assembly are exported through the flagellar-specific type III protein-export apparatus, which is believed to be located on the cytoplasmic surface of the basal body. FlhA is an essential component of the type III export apparatus of Salmonella and consists of two major portions: an N-terminal transmembrane domain and a C-terminal cytoplasmic domain (FlhAC). FlhAC and a 38 kDa fragment of FlhAC (FlhAC38K) were purified and crystallized. The crystals were obtained by the sitting-drop vapour-diffusion technique with PEG 8000 as a precipitant. FlhAC crystals grew in the tetragonal space group I41/I43, with unit-cell parameters a = b = 216.6, c = 65.0 Å. FlhAC38K was crystallized in an orthorhombic form, with unit-cell parameters a = 53.0, b = 93.1, c = 186.5 Å. X-ray diffraction data from crystals of FlhAC and the SeMet derivative of FlhAC were collected to 2.9 and 3.2 Å, respectively.
1. Introduction
The flagellum is a long filamentous organelle used for bacterial motility. It consists of a basal body, a short curved segment called the hook and a long helical filament. The basal body is a reversible rotary motor and generates torque. The hook is a universal joint that transmits the motor torque to the long helical propeller in its different orientations. The filament is a thin helical structure that typically grows to around 15 µm in length and rapidly rotates as a propeller for cell locomotion in viscous environments.
The basal body spans the inner and outer cell membranes, while the hook and the filament are external to the cell. Therefore, most of the flagellar component proteins have to be exported from the cytoplasm for flagellar assembly. These proteins are exported across the cell membranes by the flagellar protein-export apparatus, which is a member of the type III export system. The export apparatus is believed to be located within the C ring, a large cytoplasmic structure of the flagellar basal body. Previous genetic and biochemical studies have demonstrated that the export apparatus consists of six integral membrane components (FlhA, FlhB, FliO, FliP, FliQ and FliR) and three cytoplasmic components (FliH, FliI and FliJ; Minamino & Macnab, 1999 ▶, 2000a ▶). FliI is an ATPase and presumably pushes the export-substrate proteins out through the central pore of the basal body by utilizing the energy of ATP hydrolysis (Dreyfus et al., 1993 ▶; Fan & Macnab, 1996 ▶). FliH regulates the ATPase activity of FliI and contributes to the interaction between FliI and FlhA or FliI and FlhB (Auvray et al., 2002 ▶; González-Pedrajo et al., 2002 ▶; Minamino & Macnab, 2000b ▶; Minamino et al., 2003 ▶). FliJ acts as a general chaperone for the export-substrate proteins (Minamino et al., 2000 ▶). FlhB functions as the substrate-specificity switch for the export apparatus during the flagellar assembly (Minamino & Macnab, 2000a ▶; Zhu et al., 2002 ▶; Fraser et al., 2003 ▶).
FlhA consists of 692 amino-acid residues with a molecular weight of 75 kDa. flhA-deletion mutants cannot produce the flagellar axial structure including the rod, the hook and the filament (Kubori et al., 1992 ▶). There are also mutations in flhA that allow FliI to bind to the export apparatus in the absence of FliH (Minamino et al., 2003 ▶). These reports indicate that FlhA is an essential component of the export mechanism. FlhA can be divided into two major domains: a hydrophobic N-terminal transmembrane domain (FlhATM, 34.5 kDa) and a hydrophilic C-terminal cytoplasmic domain (FlhAC, 40.5 kDa). FlhAC consists of two distinct portions: a compact 38 kDa fragment, FlhAC38K, and a linker region connecting FlhATM and FlhAC38K. FlhAC38K, which contains amino-acid residues 352–629 of FlhA, is directly involved in the export of flagellar proteins, because temperature-sensitive flhA mutations and the bypass mutation described above lie within FlhAC. The linker region is thought to be important for stabilizing the interactions between FlhAC38K and the cytoplasmic components of the export apparatus, such as FliI, FliH, FliJ and the cytoplasmic domain of FlhB (Saijo-Hamano et al., 2004 ▶).
Genetic and biochemical studies have revealed the properties of these proteins (Minamino & Namba, 2004 ▶). However, the export mechanism is not clear except that energy released by ATP hydrolysis by FliI is used for the export process. To elucidate the protein-export mechanism, the atomic structures of the component proteins are essential. Here, we report an X-ray crystallographic study of FlhAC.
2. Materials and methods
2.1. Protein purification
To prepare purified FlhAC and FlhAC38K for crystallization, we slightly modified our previous purification procedures (Saijo-Hamano et al., 2004 ▶). FlhAC from Salmonella with an extra 24 residues including a His tag was overexpressed in Escherichia coli cells and purified as previously described (Saijo-Hamano et al., 2004 ▶). To completely trap the contaminated proteases, FlhAC fractions eluted from an anion-exchange column were incubated with 10 units of a2-macroglobulin (Roche Diagnostics) at 277 K for 1 h. After incubation, a2-macroglobulin was removed from the FlhAC fraction by gel-filtration (HiLoad 26/60 Superdex 75 prep-grade column; Amersham Biosciences) and anion-exchange chromatography (HiTrap Q 5 ml column; Amersham Biosciences).
FlhAC38K was prepared from purified FlhAC by limited proteolysis. The FlhAC solution was mixed with endoproteinase Glu-C (V8 protease; Roche Diagnostics) at a protein:protease ratio of 1000:1 or 2000:1 (w:w) and incubated for 16 h at room temperature. The proteolytic product was then purified by affinity chromatography and anion-exchange chromatography and treated with 30–50 units of a2-macroglobulin. Finally, a2-macroglobulin was removed from the solution by gel-filtration and anion-exchange chromatography. The purity of the sample solution was examined by SDS–PAGE and MALDI–TOF mass spectrometry (Voyager DE/PRO). Purified proteins were dialyzed extensively against 25 mM Tris–HCl buffer pH 8.0 with 2 mM EDTA and concentrated to 6–12 mg ml−1 using Vivaspin 20 (30000 MWCO, Vivascience).
FlhAC with the SeMet label was expressed in B834 (DE3) pLysS carrying pMM104 (Saijo-Hamano et al., 2004 ▶) using M9 medium containing 1 mM MgSO4, 4 g glucose, 40 mg of all l-amino acids except Met, 40 mg seleno-l-Met, 25 mg FeSO4·7H2O, 1.25 mg riboflavin, 1.25 mg niacinamide, 1.25 mg pyridoxine hydrochloride, 1.25 mg thiamine hydrochloride and 100 mg ampicillin per litre. SeMet-labelled FlhAC was purified using the same protocol as for FlhAC.
2.2. Crystallization
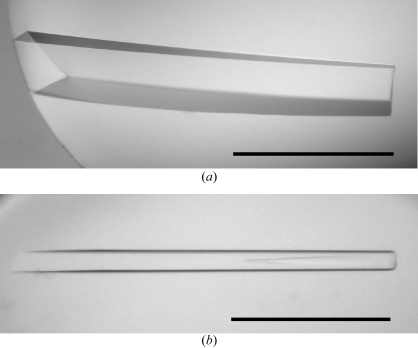
Initial crystallization screening was performed at 293 K by the sitting-drop or hanging-drop vapour-diffusion techniques using the following screening kits: Wizard I and II, Cryo I and II (Emerald Biostructures) and Crystal Screens I and II (Hampton Research). Drops were prepared by mixing 2 µl protein solution with 2 µl reservoir solution. Clusters of small needle-shaped crystals of FlhAC and FlhAC38K were only grown from the sitting-drop plates under various conditions containing PEG 8000 as a precipitant in the pH range 6–9. We optimized the conditions by varying the additives, the concentration of precipitant and the pH using the sitting-drop method. In the initial stage of the screening, both FlhAC and FlhAC38K crystallized with similar morphology under more or less similar conditions, but under optimum conditions the shape of the best crystals of each were completely different. FlhAC grew into large rod-shaped crystals (Fig. 1 ▶ a) with typical dimensions of approximately 0.15 × 0.15 × 1 mm from a solution containing 0.1 M Tris–HCl buffer pH 8.5, 0.5 mM EDTA, 10–15%(v/v) PEG 300, 4–6%(v/v) PEG 8000 and 10–15%(w/v) glycerol at 293 K. The best crystals of FlhAC38K (Fig. 1 ▶ b) were rather small, with typical dimensions of approximately 0.06 × 0.06 × 1 mm. The crystals were obtained at 293 K with a reservoir solution containing 50 mM imidazole–HCl buffer pH 8.0, 5–10%(v/v) PEG 8000, 80–180 mM calcium acetate and 10–15%(w/v) glycerol. Both crystals appeared within two weeks. Crystals of SeMet-derivative protein were obtained using the same conditions as used for the native crystals.
Figure 1.
Crystals of (a) FlhAC and (b) FlhAC38K from S. typhimurium. Scale bar, 0.5 mm.
2.3. Data collection and processing
All diffraction experiments were performed at 100 K. Because the crystallization solutions for both FlhAC and FlhAC38K contained at least 10%(w/v) glycerol, crystals were directly transferred into liquid nitrogen for freezing and mounted in a nitrogen-gas flow at 100 K. X-ray diffraction data were collected at SPring-8 beamlines BL41XU and BL38B1 using MAR CCD (MAR Research) and ADSC Quantum 4R CCD detectors (Area Detector Systems Corporation), respectively. MAD experiments for SeMet-labelled FlhAC crystals were also carried out at the SPring-8 beamline BL41XU. Wavelengths for the MAD measurements were selected based on XAFS measurements. Diffraction data were indexed, integrated and scaled using the programs MOSFLM (Leslie, 1992 ▶) and SCALA from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
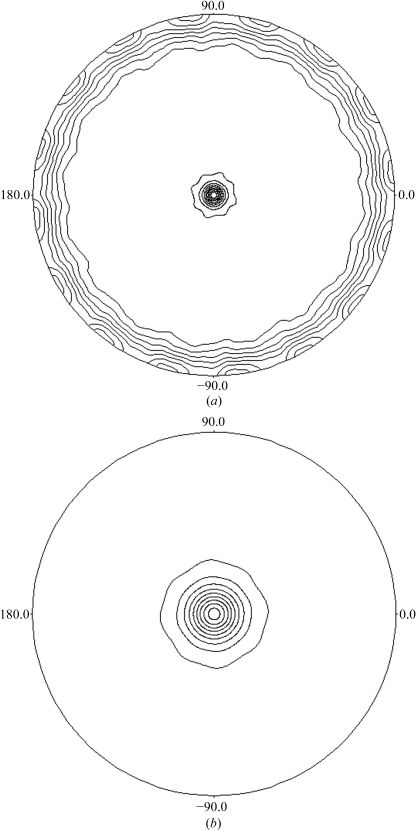
FlhAC crystals diffracted to 2.9 Å resolution. The crystals belong to the tetragonal space group I41/I43, with unit-cell parameters a = b = 216.6, c = 65.0 Å. From calculation of the Matthews coefficient (V M; Matthews, 1968 ▶), the crystals are expected to contain two to four molecules in an asymmetric unit, corresponding to a solvent content of 73–46%. The self-rotation map calculated using the program POLARRFN (Collaborative Computational Project, Number 4, 1994 ▶) revealed strong peaks in the 45 and 180° rotation sections, indicating the presence of two independent non-crystallographic twofold symmetry axes in the crystal ab plane and a 45° non-crystallographic rotation axis parallel to the crystallographic c axis (Fig. 2 ▶). These results imply that two FlhAC dimers related by 45° non-crystallographic rotational axis parallel to the c axis are packed in the asymmetric unit. Diffraction data statistics are summarized in Table 1 ▶.
Figure 2.
Stereographic projection maps of the self-rotation function calculated using data from the FlhAC native crystal. (a) κ = 180° and (b) κ = 45° section.
Table 1. Summary of the data statistics for FlhAC crystals.
Values in parentheses indicate statistics for the highest resolution shell.
| SeMet derivative | ||||
|---|---|---|---|---|
| Native | Peak | Edge | Remote | |
| Space group | I41/I43 | |||
| Unit-cell parameters (Å) | a = b = 216.6, c = 65.0 | |||
| Wavelength (Å) | 0.9243 | 0.9794 | 0.9796 | 0.9843 |
| Resolution (Å) | 2.9 (2.95–2.9) | 3.2 (3.35–3.2) | 3.2 (3.35–3.2) | 3.2 (3.35–3.2) |
| Observations | 271138 | 236744 | 286291 | 286824 |
| Unique reflections | 32969 | 25279 | 25298 | 25281 |
| Completeness (%) | 97.4 (97.4) | 100 (100) | 100 (100) | 100 (100) |
| Redundancy | 8.2 (7.9) | 9.4 (9.5) | 11.3 (11.4) | 11.3 (11.4) |
| I/σ(I) | 2.5 (1.7) | 2.6 (1.4) | 2.6 (1.4) | 2.5 (1.3) |
| Rsym (%) | 11.3 (46.1) | 12.8 (47.6) | 11.6 (52.2) | 10.6 (57.2) |
| Rano (%) | 6.2 (13.3) | 4.3 (14.0) | 3.1 (17.5) | |
FlhAC38K crystallized in the orthorhombic form, with unit-cell parameters a = 53.0, b = 93.1, c = 186.5 Å. The Matthews coefficient indicated one to two molecules in the asymmetric unit, corresponding to a solvent content of 77–55%. The crystal diffracted to 3.0 Å resolution. Only about half the reflections were collected. Therefore, we could not determine the space group of this crystal.
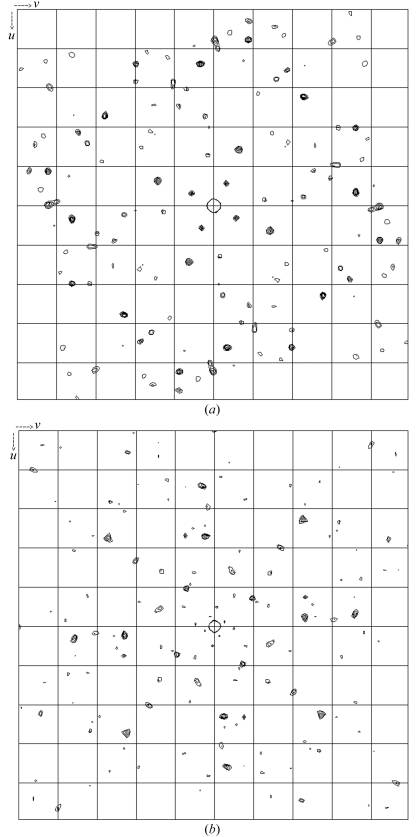
The SeMet FlhAC crystals grew to a similar size to the native FlhAC crystal and diffracted to 3.2 Å resolution. The unit-cell parameters of the crystals were same as the native crystals. Statistics of the diffraction data at three different wavelengths are summarized in Table 1 ▶. Bijvoet and dispersive difference Patterson maps of the SeMet derivative showed clear peaks on the Harker section (Fig. 3 ▶) and so did the isomorphous difference Patterson map, suggesting phasing with these data to be quite promising.
Figure 3.
(a) Bijvoet and (b) dispersive Patterson maps calculated using data from the FlhAC SeMet derivative crystal. The Se peaks are shown in the w = 0.25 Harker section.
Acknowledgments
We thank S. Nagashima, H. Matsunami, R. Iwase, M. Shimada and the staff at SPring-8 beamlines BL41XU and BL38B1 for help in data collection. This research was partly supported by grants to KI from the ‘National Project on Protein Structural and Functional Analyses’, a Grant-In-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, a Grant-In-Aid for Scientific Research on Priority Areas by MEXT to KN and USPHS grant AI12202 to RMM.
References
- Auvray, F., Ozin, A. J., Claret, L. & Hughes, C. (2002). J. Mol. Biol.318, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Dreyfus, G., Williams, A. W., Kawagishi, I. & Macnab, R. M. (1993). J. Bacteriol.175, 3131–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, F. & Macnab, R. M. (1996). J. Biol. Chem.271, 31981–31988. [DOI] [PubMed] [Google Scholar]
- Fraser, G. M., Hirano, T., Ferris, H. U., Devgan, L. L., Kihara, M. & Macnab, R. M. (2003). Mol. Microbiol.48, 1043–1057. [DOI] [PubMed] [Google Scholar]
- González-Pedrajo, B., Fraser, G. M., Minamino, T. & Macnab, R. M. (2002). Mol. Microbiol.45, 967–982. [DOI] [PubMed] [Google Scholar]
- Kubori, T., Shimamoto, N., Yamaguchi, S., Namba, K. & Aizawa, S. (1992). J. Mol. Biol.226, 433–446. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Minamino, T., Chu, R., Yamaguchi, S. & Macnab, R. M. (2000). J. Bacteriol.182, 4207–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino, T., Gonzalez-Pedrajo, B., Kihara, M., Namba, K. & Macnab, R. M. (2003). J. Bacteriol.185, 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino, T. & Macnab, R. M. (1999). J. Bacteriol.181, 1388–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino, T. & Macnab, R. M. (2000a). Mol. Microbiol.35, 1052–1064. [DOI] [PubMed] [Google Scholar]
- Minamino, T. & Macnab, R. M. (2000b). Mol. Microbiol.37, 1494–1503. [DOI] [PubMed] [Google Scholar]
- Minamino, T. & Namba, K. (2004). J. Mol. Microbiol. Biotechnol.7, 5–17. [DOI] [PubMed] [Google Scholar]
- Saijo-Hamano, Y., Minamino, T., Macnab, R. M. & Namba, K. (2004). J. Mol. Biol.343, 457–466. [DOI] [PubMed] [Google Scholar]
- Zhu, K., Gonzalez-Pedrajo, B. & Macnab, R. M. (2002). Biochemistry, 41, 9516–9524. [DOI] [PubMed] [Google Scholar]