A putative phosphoglycerate mutase from M. tuberculosis (Rv3214) has been crystallized. Diffraction data have been collected to 2.15 Å resolution from its selenomethionine-substituted form.
Keywords: Rv3214, phosphoglycerate mutase, Mycobacterium tuberculosis
Abstract
The Mycobacterium tuberculosis open reading frame Rv3214, annotated as a cofactor-dependent phosphoglycerate mutase, has been cloned and expressed as an N-terminally His-tagged protein. Tagged, untagged and selenomethionine-labelled forms of Rv3214 (EntD) have been purified using nickel-affinity chromatography and gel filtration. The selenomethionine-labelled crystals diffracted to 2.15 Å resolution and belong to space group P21, with unit-cell parameters a = 44.36, b = 79.03, c = 52.85 Å, β = 109.11°. There are two molecules of molecular weight 21 948 Da per asymmetric unit.
1. Introduction
Mycobacterium tuberculosis is the causative agent of the disease tuberculosis, which kills 2–3 million people worldwide every year. One third of the world’s population has a latent infection, with 10% of these going on to develop the active form of the disease. The recent re-emergence of the disease can be attributed to the increase in immunocompromization owing to HIV infection and to the evolution of multidrug-resistant strains of the bacterium (World Health Organization, 1998 ▶). The completion of the sequencing and annotation of the M. tuberculosis genome (Cole et al., 1998 ▶) has enabled the targeting of molecules with possible value in novel drug therapies.
Phosphoglycerate mutase (EC 5.4.2.1) is an essential component of the glycolysis and gluconeogenesis pathway (Jedrzejas, 2000 ▶; Fothergill-Gilmore & Watson, 1989 ▶) and exists in cofactor-dependent (dPGM) and cofactor-independent (iPGM) forms (Jedrzejas, 2000 ▶). Both types of enzyme catalyse the reversible conversion of 2-phosphoglycerate to 3-phosphoglycerate (Jedrzejas, 2000 ▶; Fothergill-Gilmore & Watson, 1989 ▶). The cofactor-dependent form only utilizes 2,3-bisphosphoglycerate as cofactor, whereas the cofactor-independent form is dependent only on Mn2+ (Rigden et al., 2003 ▶). The dPGM enzymes are found in vertebrates, budding yeast and eubacterial species, with iPGM being the only phosphoglycerate mutase present in higher plants; some eubacteria possess both forms of the enzyme (Fraser et al., 1999 ▶). Analysis of the M. tuberculosis genome has identified several open reading frames that encode putative phosphoglycerate mutases. These include Rv0489, Rv2419c, Rv3837c and Rv3214. The latter is of particular interest because although it is identified as a potential cofactor-dependent phosphoglycerate mutase, it is also annotated as EntD because of its homology (14% sequence identity) with Escherichia coli EntD. E. coli EntD, for which no three-dimensional structure is yet available, has phosphopantetheinyl transferase activity (Lambalot et al., 1996 ▶) and is involved in the biosynthesis of the iron-acquiring siderophore enterobactin (Gehring et al., 1998 ▶).
Structural analysis has proved useful in helping to define the functions of other phosphoglycerate mutase homologs (Rigden et al., 2001 ▶). To help understand its function and because its possible role in siderophore biosynthesis in M. tuberculosis would make it a potential drug target, we have undertaken determination of the crystal structure of Rv3214 (EntD). Here, we report the cloning, expression and purification of this 22.0 kDa protein (203 residues), leading to its crystallization and preliminary crystallographic analysis.
2. Methods
2.1. Cloning and expression
The open reading frame corresponding to Rv3214 was amplified from genomic DNA from the H37Rv strain of M. tuberculosis using the polymerase chain reaction. It was cloned into the pPROEX HTa N-terminally histidine-tagged expression vector, which incorporates a tobacco etch virus (TEV) protease cleavage site, using the restriction enzymes BamHI and HindIII. DNA sequencing confirmed the integrity of the cloned DNA. Expression was undertaken in the E. coli BL21 (DE3) expression line. A fresh transformation of the EntD construct was used to inoculate an overnight starter culture, which was subsequently used to inoculate 4 l Luria–Bertiani broth. This was incubated at 310 K until the cell density at OD600nm was approximately 0.6. Expression was then induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM and the culture was allowed to incubate at 310 K overnight with shaking at 180 rev min−1 (Sanyo orbital incubator). The selenomethionine (SeMet) labelled protein was expressed using the autoinduction method in the methionine prototroph BL21 (DE3) (Studier, 2005 ▶).
2.2. Purification
The cells from the expression culture were harvested by centrifugation at 4600g for 10 min and resuspended in buffer A (50 mM Tris–HCl pH 7.5, 50 mM NaCl). The resuspended cells were lysed using a cell disruptor (Constant Cell Disruption Systems) at a pressure of 124 MPa. The cell debris was removed by centrifugation at 30 000g for 30 min. The N-terminal His tag was utilized as a purification aid in nickel-affinity chromatography; Rv3214 was eluted using a 0–1.0 M imidazole gradient. The fractions containing Rv3214 were digested overnight with TEV protease to remove the N-terminal His tag; for crystallization trials with the His-tagged protein, this step was omitted. The untagged Rv3214 was then separated from the His-tagged TEV protease and possible undigested tagged protein by carrying out nickel-affinity purification as before. The untagged Rv3214 was then concentrated and applied onto a Superdex 200 10/30 column (Amersham Pharmacia) equilibrated with buffer A. The peak corresponding to Rv3214 was collected and run once more over the gel-filtration column. Dynamic light scattering (DynaPro, Protein Solutions) was carried out in order to assess the quality of the purified protein for crystallization. SeMet-labelled protein was also purified in this manner, with the addition of 2 mM β-mercaptoethanol to all buffers. The protein was concentrated to 11 mg ml−1 in a solution comprising 50 mM Tris–HCl, 50 mM NaCl pH 7.5 for the native and the same solution supplemented with 2 mM β-mercaptoethanol for the SeMet protein.
2.3. Crystallization
Crystal screening was carried out using the Multiprobe II HT EX (Perkin Elmer Life Sciences) and Honeybee (Cartesian Dispensing Systems) robot technology based on a sitting-drop format with 100 nl protein and 100 nl reservoir solution using Intelliplates (Art Robbins Instruments) and a 75 µl volume of well solution and incubated at 291 K. Initial screens included Hampton Crystal Screens 1 and 2 (Hampton Research), a systematic PEG–pH screen (Kingston et al., 1994 ▶), the PEG/Ion screen (Hampton Research) and the Footprint Screen (Stura et al., 1992 ▶). Initial crystallization conditions were optimized using a fine screen around the original conditions in which the pH was varied between 6.0 and 7.0, the lithium sulfate concentration between 0.2 and 2.4 M and the MPD concentration between 4 and 12%(v/v). This optimization was carried out in 96-well sitting-drop crystallization plates comprised of CrystalClear strips (Hampton Research).
3. Results and discussion
Rv3214 in all three forms was purified to near-complete purity as determined by SDS–PAGE analysis (results not shown). Dynamic light scattering showed that in each case the protein existed as a dimer in solution and was of a quality suitable for crystallization (see Table 1 ▶).
Table 1. Dynamic light-scattering results relative to predicted molecular weight in post-gel-filtration buffer.
The polydispersity index Cp/Rh measures the dispersity of the peak relative to the hydrodynamic radius Rh.
| Sample | Molecular weight of monomer (kDa) | Experimental molecular weight (kDa) | Cp/Rh (%) |
|---|---|---|---|
| His-tagged Rv3214 | 25.29 | 66.4 | 24.6 |
| Rv3214, tag removed | 21.95 | 46.7 | 9.7 |
| Rv3214, SeMet-labelled, tag removed | 21.95 | 38.1 | 26.1 |
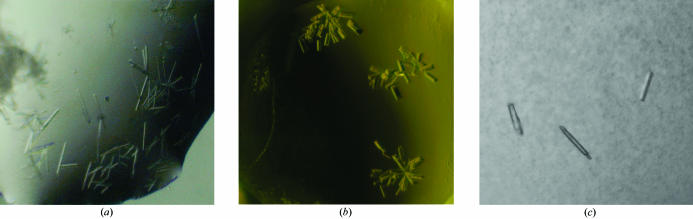
The His-tagged Rv3214 crystallized in 68% 2,4-dimethylpentanediol (MPD), 0.1 M HEPES pH 7.0 (condition 1), the untagged protein gave crystals in 74% MPD, 0.2 M MgCl2, 0.1 M HEPES pH 7.5 (condition 2) and the SeMet-labelled protein gave crystals in 2.0 M Li2SO4, 6% MPD and 0.1 M imidazole–HCl pH 7.0 (condition 3) (Fig. 1 ▶). The SeMet-Rv3214 crystals took approximately 21 d to form and achieved dimensions of 0.01 × 0.01 × 0.05 mm.
Figure 1.
(a) Native crystals formed in condition 1 from His-tagged EntD. (b) Native crystals formed in condition 2 from EntD with the His tag cleaved. (c) Crystals of SeMet-labelled EntD with the His tag removed formed in condition 3 at pH 7.0.
The light-scattering results (Table 1 ▶) correlate with the apparent quality of the crystals obtained for different Rv3214 species (Fig. 1 ▶). Untagged Rv3214 visibly gave the best crystals, but these were very fragile and disintegrated during mounting in loops. The SeMet-labelled protein proved to differ in its properties from the native forms to such an extent that crystallization screening was once again carried out ab initio. In contrast to the wild-type Rv3214 crystallization conditions, which were very rich in MPD, crystals of the SeMet-labelled protein formed in a 2.0 M salt solution at a much lower percentage of MPD, which greatly improved their handling. The SeMet-labelled crystals were soaked transiently in reservoir solution supplemented with 20% ethylene glycol as a cryoprotectant, flash-cooled and stored in liquid nitrogen.
Data collection was carried out at 113 K on beamline 8.2 at the Advanced Light Source (Lawrence Berkeley Laboratory, CA, USA). Diffraction data were collected from the SeMet-labelled protein crystal to 2.2 Å, with a crystal-to-detector distance of 249 mm, and were processed using DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶). The crystal was monoclinic, with space group P21 and unit-cell parameters a = 44.36, b = 79.03, c = 52.85 Å, β = 109.11°. Assuming the presence of a dimer (two molecules of 22.0 kDa) in the asymmetric unit, the Matthews coefficient V M is 2.00 Å3 Da−1, consistent with a solvent content of about 37.7%. Data-collection statistics are shown in Table 2 ▶.
Table 2. Data-collection statistics for SeMet-substituted Rv3214 crystal.
Values shown in parentheses are for the outermost shell: 2.25–2.15 Å for the peak wavelength and 2.69–2.6 Å for the remote wavelength.
| Peak wavelength | Remote wavelength | |
|---|---|---|
| Wavelength (Å) | 0.91840 | 0.97972 |
| Space group | P21 | |
| Unit-cell parameters (Å, °) | a = 44.36, b = 79.03, c = 52.85, β = 109.11 | |
| Resolution (Å) | 50–2.15 | 50–2.6 |
| Completeness (%) | 95.1 (70.2) | 96.2 (75.5) |
| Observed reflections | 151474 | 67903 |
| Unique reflections | 22262 | 10306 |
| 〈I/σ(I)〉 | 57.3 (14.9) | 10.1 (1.4) |
| Rmerge† | 0.105 (0.334) | 0.157 (0.588) |
R
merge = 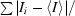
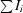 .
.
The SeMet-substituted crystals are of sufficient quality to enable us to undertake the structure solution of Rv3214 by multiwavelength anomalous diffraction (MAD) methods using the three methionine residues in each molecule. The solution of this structure will enable us to confirm the annotation of the open reading frame and then proceed to functional studies to assess the value of this protein as a possible drug target.
References
- Cole, S. T. et al. (1998). Nature (London), 393, 537–544. [Google Scholar]
- Fothergill-Gilmore, L. A. & Watson, H. C. (1989). Adv. Enzymol. Relat. Areas Mol. Biol.62, 227–313. [DOI] [PubMed] [Google Scholar]
- Fraser, H. I., Kvaratskhelia, M. & White, M. F. (1999). FEBS Lett.455, 344–348. [DOI] [PubMed] [Google Scholar]
- Gehring, A. M., Mori, I. & Walsh, C. T. (1998). Biochemistry, 37, 2648–2659. [DOI] [PubMed] [Google Scholar]
- Jedrzejas, M. J. (2000). Prog. Biophys. Mol. Biol.73, 263–287. [DOI] [PubMed] [Google Scholar]
- Kingston, R. L., Baker, H. M. & Baker, E. N. (1994). Acta Cryst. D50, 429–440. [DOI] [PubMed] [Google Scholar]
- Lambalot, R. H., Gehring, A. M., Flugel, R. S., Zuber, P., LaCelle, M., Marahiel, M. A., Reid, R., Khosla, C. & Walsh, C. T. (1996). Chem. Biol.3, 923–936. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Rigden, D. J., Bagyan, I., Lamani, E., Setlow, P. & Jedrzejas, M. J. (2001). Protein Sci.10, 1835–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigden, D. J., Littlejohn, J. E., Henderson, K. & Jedrzejas, M. J. (2003). J. Mol. Biol.328, 909–920. [DOI] [PubMed] [Google Scholar]
- Studier, F. W. (2005). Protein Expr. Purif.41, 207–234. [DOI] [PubMed] [Google Scholar]
- Stura, E. A., Nemerow, G. R. & Wilson, I. A. (1992). J. Cryst. Growth, 122, 273–285.
- World Health Organization (1998). The World Health Report 1998. Geneva, Switzerland: World Health Organization.