The first crystallographic study of a sucrose phosphate synthase from H. orenii, an organism that is both thermophilic and halophilic, is reported. The protein crystal diffracts X-rays to 3.01 Å.
Keywords: sucrose phosphate synthase, Halothermothrix orenii, thermophiles, halophiles, sucrose metabolism
Abstract
This is the first report of the crystallization of a sucrose phosphate synthase (SPS; EC 2.4.1.14). It also constitutes the first study of a sucrose phosphate synthase from a non-photosynthetic thermohalophilic anaerobic bacterium, Halothermothrix orenii. The purified recombinant spsA protein has been crystallized in the monoclinic space group C2, with unit-cell parameters a = 154.2, b = 47.9, c = 72.3 Å, β = 103.16°, using the hanging-drop vapour-diffusion method. The crystal diffracts X-rays to a resolution limit of 3.01 Å. Heavy-metal and halide-soaking trials are currently in progress to solve the structure.
1. Introduction
Halothermothrix orenii is a unique anaerobic bacterium that grows optimally at 333 K in the presence of 10% NaCl. This combination of high temperature and high ionic strength represents one of the most extreme conditions recorded for the growth of any organism thus far (Cayol et al., 1994 ▶). These extreme growth characteristics not only make H. orenii a potentially important source for commercially useful enzymes, but also for evolutionary studies and the study of fundamental aspects of protein structure and function. We have previously reported the characteristics and crystallization of two α-amylases, AmyA and AmyB (Li et al., 2002 ▶; Tan et al., 2003 ▶). Our previous studies of a random genomic DNA-sequence analysis of H. orenii identified an ORF, designated spsA, with significant homology to cyanobacterial sucrose phosphate synthase (SPS; EC 2.4.1.14; Mijts & Patel, 2001 ▶), an enzyme that is only ubiquitous in photosynthetic organisms, including plants and cyanobacteria. SPS, together with sucrose synthase (SS) and sucrose phosphate phosphorylase (SPP), is involved in sucrose synthesis and belongs to a group of enzymes that are collectively known as sucrose-biosynthesis proteins (SBPs). It is thought that plant SPS plays a role in osmotic regulation of intracellular turgor pressure in eukaryotes (Hite et al., 1993 ▶). To the best of our knowledge, there have been no other reports of crystal structures of SBPs, with the exception of an unreleased X-ray structure of an SPP from Synechocystis sp. PCC6803 (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1s2o). As part of our ongoing studies on H. orenii, we have overexpressed and purified the spsA gene product of H. orenii. The biochemical characterization of this purified recombinant spsA confirmed its enzymatic activity, but showed significant differences from the previously reported cyanobacterial and plant SPSs (Huynh, 2004 ▶). Recombinant spsA is a 61.1 kDa protein that has an optimal temperature for activity at 323–328 K and exhibits cross-reactivity with polyclonal antibodies raised against plant SPSs (AgriSera, Sweden). Like its cyanobacterial prokaryotic counterparts, the recombinant enzyme can also accept ADP-glucose and GDP-glucose as the glycosyl donor in place of UDP-glucose. By crystallizing and solving its structure, we hope to gain an insight into the structural and functional characteristics of SPS.
2. Materials and methods
2.1. Expression and purification of recombinant SPS
The spsA gene was amplified by PCR using primers incorporating BamHI and KpnI restriction-enzyme sites at the 5′ and 3′ ends, respectively. The resulting PCR fragment was digested using these restriction enzymes, ligated into the pTrcHisA expression vector (Invitrogen) encoding an N-terminal hexahistidine fusion peptide and transformed into TransforMax Escherichia coli cells (Epicentre). Bacterial clones were cultivated in 1 l LB medium supplemented with 50 µg ml−1 ampicillin at 310 K to an OD600 of 0.6. Recombinant protein expression was induced for 4 h with IPTG to a final concentration of 1 mM. The cells were harvested by centrifugation at 5000g and resuspended in 20 ml 20 mM Tris–HCl pH 8.0, 500 mM NaCl, 1 mg ml−1 lysozyme, 10 U ml−1 DNase I, 0.1 mg ml−1 RNase A, 1 mM PMSF and one Complete EDTA-free protease-inhibitor cocktail tablet (Roche). The mixture was incubated at 310 K for 20 min and sonicated on ice. Final cell lysis was achieved by three rapid freeze–thaw cycles in liquid nitrogen. Cell lysates were cleared by centrifugation at 4000 rev min−1 (Sigma 4K-15) for 30 min and the soluble fraction was collected.
HisLink Protein Purification Resin (Promega) was added to the cleared cell lysate and incubated for 1 h at 277 K with gentle agitation. The mixture was transferred to a chromatography column and washed with at least 50 column volumes of 20 mM Tris–HCl pH 7.5, 500 mM NaCl, 10 mM imidazole. Recombinant SPS was eluted with 20 mM Tris–HCl pH 7.5, 100 mM NaCl, 500 mM imidazole. Simultaneous removal of imidazole and protein concentration was achieved by several rounds of diafiltration using an Amicon Ultra-15 Centrifugal Filter device (Millipore) with 20 mM Tris–HCl pH 7.5, 100 mM NaCl to a final concentration of 10 mg ml−1 (Bradford method).
2.2. Crystallization
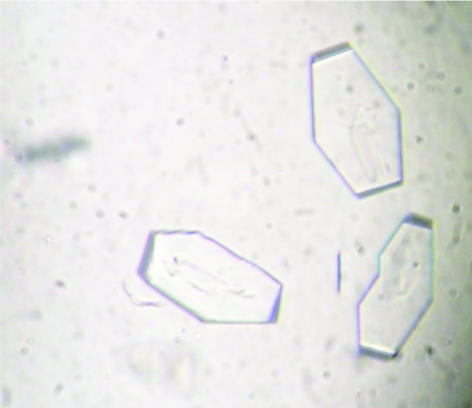
Initial crystallization conditions were screened by the hanging-drop vapour-diffusion technique using the JBScreen Mixed crystallography reagent kit (Jena Bioscience). 2 µl SPS solution was mixed with 2 µl reservoir solution and equilibrated against 0.75 ml reservoir solution at 293 K. Single crystals appeared after 2 d in a solution containing 16% PEG 4000, 0.1 M Tris–HCl pH 8.5, 0.2 M magnesium chloride. The condition was further optimized and crystals were obtained reproducibly with 15% PEG 3350, 0.1 M Tris–HCl pH 8.0, 0.2 M magnesium chloride with maximum dimensions of 0.5 × 0.35 mm after 1 d (Fig. 1 ▶).
Figure 1.
Plate-like crystals of H. orenii SPS. The typical dimensions of diffraction-quality crystals are approximately 0.5 × 0.35 mm.
2.3. Data collection and analysis
Crystals were picked up from the crystallization drop using nylon loops and flash-cooled in liquid nitrogen. X-ray diffraction data were collected using an in-house Rigaku RU-H3R generator and Rigaku R-AXIS IV++ detector. All data were indexed, integrated and scaled using the programs DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
The spsA gene from H. orenii was successfully cloned and overexpressed and the recombinant SPS protein was purified. The enzyme was confirmed to be active at elevated temperatures and exists as a monomer. The SPS crystals diffract X-rays to 3.01 Å and belong to the monoclinic space group C2, with unit-cell parameters a = 154.2, b = 47.9, c = 72.3 Å, β = 103.16°. Crystal parameters and crystallographic data statistics are summarized in Table 1 ▶. The Matthews coefficient (V M; Matthews, 1968 ▶) of 2.13 Å3 Da−1 for this crystal corresponds to an estimated solvent content of 42.2% and a monomer in the asymmetric unit.
Table 1. Diffraction data statistics.
Values in parentheses refer to the highest resolution shell (3.12–3.01 Å).
| Radiation source | Rigaku RU-H3R generator and R-AXIS IV++ detector |
| Wavelength (Å) | 1.5418 |
| No. imaging frames | 360 |
| Crystal oscillation range (°) | 0–360 |
| Unit-cell parameters (Å, °) | a = 154.2, b = 47.9, c = 72.3, β = 103.16 |
| Space group | C2 |
| Mosaicity of crystal (°) | 0.86 |
| Resolution range (Å) | 50–3.01 |
| Total No. reflections | 49,015 |
| No. unique reflections | 17882 |
| Redundancy | 2.7 (2.2) |
| Completeness (%) | 89.7 (80.6) |
| Rsym† (%) | 0.054 (0.130) |
R
sym = 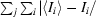
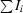 .
.
No other protein structures with sequence identity greater than 30% to SPS have been reported to date. As a consequence, molecular-replacement modelling is not an option that we can use to solve the structure and therefore attempts at heavy-atom screening for MAD analysis are under way. In addition, we hope to further optimize the cryoprotectant to reduce the relatively poor crystal mosaicity. The structure, once solved, will establish the structural fold for this family of enzymes and provide further insight into the molecular mechanism of thermostability in sucrose phosphate synthase.
References
- Cayol, J. L., Ollivier, B., Patel, B. K., Prensier, G., Guezennec, J. & Garcia, J. L. (1994). Int. J. Syst. Bacteriol.44, 534–540. [DOI] [PubMed] [Google Scholar]
- Hite, D., Outlaw, W. H. Jr & Tarczynski, M. C. (1993). Plant Physiol.101, 1217–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, F. (2004). BSc (Hons) thesis. Griffith University, Brisbane, Australia.
- Li, N., Patel, B. K., Mijts, B. N. & Swaminathan, K. (2002). Acta Cryst. D58, 2125–2126. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Mijts, B. N. & Patel, B. K. (2001). Extremophiles, 5, 61–69. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Tan, T. C., Yien, Y. Y., Patel, B. K., Mijts, B. N. & Swaminathan, K. (2003). Acta Cryst. D59, 2257–2258. [DOI] [PubMed] [Google Scholar]