A trimodular complex comprising the type II cohesin–dockerin interaction from the cellulosome of C. thermocellum has been purified and crystallized by the hanging-drop vapour-diffusion method. A native crystal and a selenomethionine derivative have been analyzed using X-ray diffraction.
Keywords: type II cohesin, dockerin, cellulosome
Abstract
The high-affinity calcium-mediated type II cohesin–dockerin interaction is responsible for the attachment of the multi-enzyme cellulose-degrading complex, termed the cellulosome, to the cell surface of the thermophilic anaerobe Clostridium thermocellum. A trimodular 40 kDa complex comprising the SdbA type II cohesin and the the CipA type II dockerin–X module modular pair from the cellulosome of C. thermocellum has been crystallized. The crystals belong to space group P212121, with unit-cell parameters a = 45.21, b = 52.34, c = 154.69 Å. The asymmetric unit contains one molecule of the protein complex and native and selenomethionine-derivative crystals diffracted to 2.1 and 2.0 Å, respectively.
1. Introduction
Cellulosomes are large extracellular multienzyme complexes responsible for the degradation of crystalline cellulose that have been identified in numerous anaerobic bacteria as well as in ruminal bacteria and anaerobic fungi (for reviews, see Beguin & Lemaire, 1996 ▶; Bayer, Chanzy et al., 1998 ▶; Bayer, Shimon et al., 1998 ▶; Schwarz, 2001 ▶; Doi & Kosugi, 2004 ▶). The cellulosome of the thermophilic anaerobe Clostridium thermocellum was the first to be identified (Lamed, Kenig et al., 1983 ▶; Lamed, Setter et al., 1983 ▶) and comprises two primary subunits. The catalytic subunits, which consist of a variety of cellulases and hemicellulases, are organized on a large multi-modular non-catalytic subunit termed CipA scaffoldin (Gerngross et al., 1993 ▶) to form a single macromolecular machine that is able to efficiently and synergistically break down crystalline cellulose. Attachment of the cellulosome to the bacterial cell surface involves a third cell-surface subunit (Lemaire et al., 1995 ▶; Leibovitz & Beguin, 1996 ▶; Leibovitz et al., 1997 ▶).
Two types of high-affinity calcium-dependent protein–protein interactions between cohesin and calcium-binding dockerin modules are responsible for the assembly of the functional cellulosome complex. The type I interaction anchors the catalytic subunits onto the scaffoldin subunit and involves the nine type I cohesin modules of CipA scaffoldin and the type I dockerin modules of the catalytic subunits; the association of the C-terminal dockerin module of CipA scaffoldin with the cohesin module of the cell-surface subunit constitutes the type II interaction. While sequential comparison, mutagenesis and biophysical studies of the type I and type II cohesin and dockerin modules have provided insight into the regions responsible for the type I and type II interactions and into their specificity (Mechaly et al., 2000 ▶, 2001 ▶; Miras et al., 2002 ▶; Schaeffer et al., 2002 ▶; Handelsman et al., 2004 ▶; Jindou, Kajino et al., 2004 ▶; Jindou, Soda et al., 2004 ▶), it is recent detailed structural studies that have dramatically moved this field forward. These include the crystal structures of two C. thermocellum (Shimon et al., 1997 ▶; Tavares et al., 1997 ▶) and one C. cellulotyticum type I cohesin modules (Spinelli et al., 2000 ▶), a C. thermocellum type I dockerin NMR solution structure (Lytle et al., 2001 ▶), the NMR backbone sequential assignment and secondary-structure determination of a C. thermocellum type II cohesin module (Smith et al., 2002 ▶), a crystal structure of a type II cohesin from Acetivibrio cellulolyticus (Noach et al., 2003 ▶) and the recent crystal structure of a C. thermocellum type I cohesin–dockerin complex (Carvalho et al., 2003 ▶). However, only a direct comparison of the type I complex with a type II cohesin–dockerin complex structure will unequivocally define the underlying structural elements responsible for the type I/type II specificity. In the present communication, we describe the purification, crystallization and preliminary X-ray characterization of a 359-residue 39.3 kDa trimodular complex comprising the C. thermocellum type II SdbA cohesin–CipA dockerin interaction.
2. Expression, purification and complex formation
Expression and purification of the CipA type II dockerin–X module modular pair (DocX) involved the growth of transformed cells in 1 l LB medium supplemented with 100 µg l−1 ampicillin in a 2.8 l Fernbach flask at 310 K with shaking to an OD600 of 0.6. IPTG was added to a final concentration of 1 mM and growth was continued for an additional 4 h. The cells were harvested by centrifugation, resuspended in a volume of buffer A (25 mM Tris–HCl pH 7.4, 250 mM NaCl, 8 M urea) equal to ten times their packed weight and disrupted by sonication (3 × 30 s) at room temperature. Cell debris was removed by centrifugation for 1 h at 120 000g in a Beckman Ti70 ultracentrifuge rotor. The supernatant solution was collected and applied onto a column packed with 2 ml Ni2+-chelating Sepharose beads (Amersham Pharmacia Biotech) equilibrated with buffer A. The column was washed with 20 ml buffer A containing 20 mM imidazole and bound protein was eluted with buffer A containing 300 mM imidazole in 1 ml fractions. Fractions containing protein were determined by SDS–PAGE and subsequently pooled and dialyzed in buffer B (25 mM Tris–HCl pH 7.4, 50 mM NaCl, 10 mM EDTA). The refolded protein sample was applied onto a Hi-Load 16/60 Superdex 75 size-exclusion column (Amersham Pharmacia Biosciences) equilibrated with buffer B and was eluted in 2 ml fractions using the same buffer. Fractions containing protein were pooled and concentrated to a final volume of 5 ml using a Millipore Amicon 10 kDa centrifugal device and were stored at 253 K. Recombinant C. thermocellum SdbA type II cohesin was expressed and purified as described previously (Smith et al., 2002 ▶). Expression and purification of seleno-l-methionine (SeMet) labeled SdbA type II cohesin and DocX was performed in a manner similar to that previously described for the uniform 15N-labeling of SdbA type II cohesin (Smith et al., 2002 ▶), with the exception that in the current study the expression plasmids were transformed into the auxotrophic Escherichia coli strain DL41 and the medium was supplemented with 50 mg SeMet rather than 1 g 15N-NH4Cl (Cambridge Isotope Laboratories). Typical yields were 40 and 20 mg per litre for the native and SeMet-labeled SdbA type II cohesin modules and 30 and 15 mg per litre for the native and SeMet-labeled DocX, respectively.
Formation of the SdbA type II cohesin–DocX complex involved the combination of purified SdbA type II cohesin and DocX in a 1.3:1 molar ratio in the presence of calcium in 25 mM Tris–HCl pH 7.5, 50 mM NaCl, 10 mM CaCl2, 5 mM DTT (buffer C). The 1:1 SdbA type II cohesin–DocX complex was separated from excess type II cohesin by application onto a Hi-Load 16/60 Superdex 75 size-exclusion column (Amersham Pharmacia Biosciences) equilibrated with buffer C and elution in 2 ml fractions using the same buffer. Fractions containing the complex were analyzed by SDS–PAGE, pooled, concentrated to a final concentration of 50 mg l−1 using a Millipore Amicon 10 kDa centrifugal device and stored at 277 K. The current protocol for complex formation is in contrast to that used for the type I cohesin–dockerin complex, in which the two modules were coexpressed from a single plasmid and the complex was purified by nickel-affinity and anion-exchange chromatography (Carvalho et al., 2003 ▶).
3. Crystallization
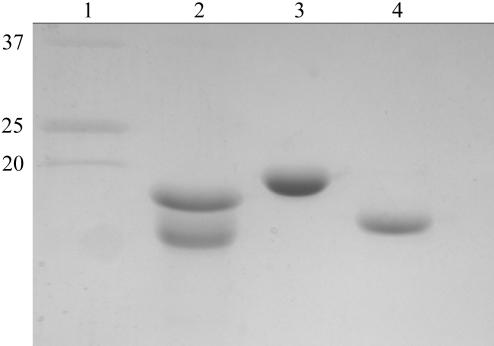
The native SdbA type II cohesin–DocX complex crystals were formed by the hanging-drop technique, in which a 4 µl 1:1(v:v) drop of 25 mg ml−1 type II complex and 0.1 M sodium acetate pH 5.0, 1.4 M ammonium sulfate, 6% dioxane was equilibrated with 0.1 M sodium acetate pH 5.0, 1.4 M ammonium sulfate, 6% dioxane well solution. The native crystals formed in approximately 30 d after plating at room temperature (Fig. 1 ▶). Sample crystals were washed three times with 20 µl well solution, dissolved in 5 µl 25 mM Tris pH 7.5, 50 mM NaCl, 5 mM EDTA, 1 mM DTT and analyzed by SDS–PAGE to ensure that the crystal lattice comprised the SdbA type II cohesin–DocX complex (Fig. 2 ▶). In contrast, the SeMet-labeled complex crystals were formed from a 4 µl 1:1(v:v) drop of 25 mg ml−1 type II complex and 0.1 M sodium acetate pH 4.25, 1.3 M ammonium sulfate, 7% dioxane equilibrated with a well solution consisting of 0.1 M sodium acetate pH 4.25, 1.3 M ammonium sulfate, 7% dioxane. These crystals grew at a significantly faster rate, with growth completion in 7 d at room temperature. The native and SeMet-derivative crystals are isomorphous, as shown in Table 1 ▶.
Figure 1.
Typical crystal formed by native trimodular type II SdbA cohesin–DocX complex. Approximate crystal dimensions are 0.2 × 0.2 × 0.03 mm.
Figure 2.
SDS–PAGE analysis of the native trimodular type II SdbA cohesin–DocX complex crystal composition. Lane 1, molecular-weight markers (kDa); lane 2, native complex crystal; lane 3, purified recombinant SdbA type II cohesin module; lane 4, purified recombinant DocX. The small differences between the molecular weights of the complex components and the individual proteins can be attributed to the different buffer conditions of the samples.
Table 1. Statistical analysis of X-ray diffraction data for native and SeMet complex crystals.
Values in parentheses are for the highest resolution shell.
| Native type II complex | SeMet type II complex | |
|---|---|---|
| X-ray source | CHESS (F2 station) | NSLS (X8C beamline) |
| Wavelength (Å) | 0.9790 | 0.9788 |
| Detector | CCD | CCD |
| Crystal parameters | ||
| Space group | P212121 | P212121 |
| Unit-cell parameters | ||
| a (Å) | 45.57 | 45.21 |
| b (Å) | 52.22 | 52.34 |
| c (Å) | 156.26 | 154.69 |
| Resolution (Å) | 2.1 (2.18–2.10) | 2.0 (2.07–2.00) |
| Mosaicity (°) | 0.53 | 0.65 |
| Solvent content (%) | 48 | 48 |
| Molecules per AU (heterodimers) | 1 | 1 |
| Data processing | ||
| No. observed reflections | 88650 | 300975 |
| No. unique reflections | 22173 | 25648 |
| Completeness (%) | 99.0 (95.8) | 99.6 (96.3) |
| Rsym (%) | 5.9 (18.1) | 10.3 (35.4) |
| Average I/σ(I) | 15.8 (4.8) | 25.81 (3.0) |
4. Data collection and processing
Data sets were collected from both crystal forms using a home source (Rigaku RU-200 generator operated at 50 kV and 100 mA equipped with a MAR imaging plate). A native data set was collected using synchrotron-source X-rays at the F1 station at the Cornell High Energy Synchrotron Source (CHESS) using a Q-210 CCD detector. The SeMet-labeled complex crystal was used to collect a selenium-edge MAD data set at the X8C beamline of Brookhaven National Laboratory (BNL) fitted with a Quantum IV CCD detector. Both native and SeMet-labeled crystals were soaked in their respective well solutions containing an additional 20%(v/v) glycerol as a cryoprotectant, flash-cooled in a cryostream of liquid N2 gas and stored in liquid N2 until data were collected at the synchrotron sources. All data sets were indexed, processed and scaled using DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶).
The native crystal lattice was identified to have a primitive orthorhombic space group, with unit-cell parameters a = 45.57, b = 52.22, c = 156.26 Å, and to contain one molecule of the heterodimeric complex in the asymmetric unit. Scaled data sets collected at CHESS gave 2.1 Å resolution with an R sym of 0.059 (Table 1 ▶). Similarly, the SeMet crystal has unit-cell parameters a = 45.21, b = 52.34, c = 154.69 Å. The scaled data sets were resolved to 2.0 Å resolution with an R sym of 0.103 (Table 1 ▶). Unambiguous identification of the screw-axes symmetry of P212121 was elucidated by systematic absences.
Attempts to solve the structure of the trimodular complex comprising the type II cohesin–dockerin interaction by molecular replacement using the type I cohesin–dockerin complex (Carvalho et al., 2003 ▶) have proven to be unsuccessful, which is not surprising given the low sequence identities between the type I and II dockerin (30%) and cohesin (20%) modules. In addition, the X-module is not present in the type I model. Utilizing the MAD data, we have determined the positions of all Se atoms and structure determination is ongoing.
Acknowledgments
We would like to acknowledge BNL and CHESS for synchrotron data collection. We also wish to thank Melanie Adams, Andrew Wong and Michael Suits for useful discussions and the referees for constructive comments. JA is the recipient of a McLaughlin Graduate Scholarship and ZJ is a Canada Research Chair in Structural Biology and an NSERC Stacie fellow. This research was supported by Canadian Institutes of Health Research operating grants to SPS and ZJ. Infrastructure used in this study was funded through the Canadian Foundation for Innovation and the Ontario Innovation Trust.
References
- Bayer, E. A., Chanzy, H., Lamed, R. & Shoham, Y. (1998). Curr. Opin. Struct. Biol.8, 548–557. [DOI] [PubMed] [Google Scholar]
- Bayer, E. A., Shimon, L. J., Shoham, Y. & Lamed, R. (1998). J. Struct. Biol.124, 221–234. [DOI] [PubMed] [Google Scholar]
- Beguin, P. & Lemaire, M. (1996). Crit. Rev. Biochem. Mol. Biol.31, 201–236. [DOI] [PubMed] [Google Scholar]
- Carvalho, A. L., Dias, F. M., Prates, J. A., Nagy, T., Gilbert, H. J., Davies, G. J., Ferreira, L. M., Romao, M. J. & Fontes, C. M. (2003). Proc. Natl Acad. Sci. USA, 100, 13809–13814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi, R. H. & Kosugi, A. (2004). Nature Rev. Microbiol.2, 541–551. [DOI] [PubMed]
- Gerngross, U. T., Romaniec, M. P., Kobayashi, T., Huskisson, N. S. & Demain, A. L. (1993). Mol. Microbiol.8, 325–334. [DOI] [PubMed] [Google Scholar]
- Handelsman, T., Barak, Y., Nakar, D., Mechaly, A., Lamed, R., Shoham, Y. & Bayer, E. A. (2004). FEBS Lett.572, 195–200. [DOI] [PubMed] [Google Scholar]
- Jindou, S., Kajino, T., Inagaki, M., Karita, S., Beguin, P., Kimura, T., Sakka, K. & Ohmiya, K. (2004). Biosci. Biotechnol. Biochem.68, 924–926. [DOI] [PubMed] [Google Scholar]
- Jindou, S., Soda, A., Karita, S., Kajino, T., Beguin, P., Wu, J. H., Inagaki, M., Kimura, T., Sakka, K. & Ohmiya, K. (2004). J. Biol. Chem.279, 9867–9874. [DOI] [PubMed] [Google Scholar]
- Lamed, R., Kenig, R., Setter, E. & Bayer, E. A. (1983). Biotechnol. Bioeng. Symp.13, 163–181.
- Lamed, R., Setter, E. & Bayer, E. A. (1983). J. Bacteriol.156, 828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovitz, E. & Beguin, P. (1996). J. Bacteriol.178, 3077–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovitz, E., Ohayon, H., Gounon, P. & Beguin, P. (1997). J. Bacteriol.179, 2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire, M., Ohayon, H., Gounon, P., Fujino, T. & Beguin, P. (1995). J. Bacteriol.177, 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle, B. L., Volkman, B. F., Westler, W. M., Heckman, M. P. & Wu, J. H. (2001). J. Mol. Biol.307, 745–753. [DOI] [PubMed] [Google Scholar]
- Mechaly, A., Fierobe, H. P., Belaich, A., Belaich, J. P., Lamed, R., Shoham, Y. & Bayer, E. A. (2001). J. Biol. Chem.276, 9883–9888. [DOI] [PubMed] [Google Scholar]
- Mechaly, A., Yaron, S., Lamed, R., Fierobe, H. P., Belaich, A., Belaich, J. P., Shoham, Y. & Bayer, E. A. (2000). Proteins, 39, 170–177. [DOI] [PubMed] [Google Scholar]
- Miras, I., Schaeffer, F., Beguin, P. & Alzari, P. M. (2002). Biochemistry, 41, 2115–2119. [DOI] [PubMed] [Google Scholar]
- Noach, I., Lamed, R., Xu, Q., Rosenheck, S., Shimon, L. J., Bayer, E. A. & Frolow, F. (2003). Acta Cryst. D59, 1670–1673. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Schaeffer, F., Matuschek, M., Guglielmi, G., Miras, I., Alzari, P. M. & Beguin, P. (2002). Biochemistry, 41, 2106–2114. [DOI] [PubMed] [Google Scholar]
- Schwarz, W. H. (2001). Appl. Microbiol. Biotechnol.56, 634–649. [DOI] [PubMed] [Google Scholar]
- Shimon, L. J., Bayer, E. A., Morag, E., Lamed, R., Yaron, S., Shoham, Y. & Frolow, F. (1997). Structure, 5, 381–390. [DOI] [PubMed] [Google Scholar]
- Smith, S. P., Beguin, P., Alzari, P. M. & Gehring, K. (2002). J. Biomol. NMR, 23, 73–74. [DOI] [PubMed] [Google Scholar]
- Spinelli, S., Fierobe, H. P., Belaich, A., Belaich, J. P., Henrissat, B. & Cambillau, C. (2000). J. Mol. Biol.304, 189–200. [DOI] [PubMed] [Google Scholar]
- Tavares, G. A., Beguin, P. & Alzari, P. M. (1997). J. Mol. Biol.273, 701–713. [DOI] [PubMed] [Google Scholar]