Abstract
The traditional classification of nucleic acid polymerases as either DNA or RNA polymerases is based, in large part, on their fundamental preference for the incorporation of either deoxyribonucleotides or ribonucleotides during chain elongation. The refined structure determination of Moloney murine leukemia virus reverse transcriptase, a strict DNA polymerase, recently allowed the prediction that a single amino acid residue at the active site might be responsible for the discrimination against the 2′OH group of an incoming ribonucleotide. Mutation of this residue resulted in a variant enzyme now capable of acting as an RNA polymerase. In marked contrast to the wild-type enzyme, the Km of the mutant enzyme for ribonucleotides was comparable to that for deoxyribonucleotides. The results are consistent with proposals of a common evolutionary origin for both classes of enzymes and support models of a common mechanism of nucleic acid synthesis underlying catalysis by all such polymerases.
Keywords: deoxyribonucleotides, DNA synthesis
A key characteristic of nucleic acid polymerases is their traditional classification as either DNA or RNA polymerases, which is determined by a given enzyme’s ability to selectively use either deoxyribonucleotides (dNTPs) or ribonucleotides (rNTPs) as substrates for incorporation into a growing chain (1, 2). This classification, however, may not be as fundamental as originally thought (3–5). Crystallographic studies have demonstrated that DNA and RNA polymerases have remarkable structural similarities (refs. 6–15; reviewed in ref. 16), even though they lack extensive primary sequence homology. Both have a characteristic protein fold forming a nucleic acid binding cleft and a trio of carboxylic acid residues thought to participate directly in catalysis through two bound divalent metal ions. Steady-state analyses further support the notion of a common stepwise polymerization mechanism (17, 18). These observations suggest that it might be possible to convert a DNA polymerase into an RNA polymerase by relatively minor alterations in its structure.
Reverse transcriptases (RTs), encoded by all retroviruses, play a defining role in the retroviral life cycle (refs. 19 and 20; for reviews see ref. 21). The enzyme is responsible for the synthesis of a double-stranded linear DNA copy of the RNA genome, which is subsequently inserted into the host genome to form the integrated proviral DNA.The reverse transcription reaction is complex, requiring RNA-dependent DNA polymerase activity, DNA-dependent DNA polymerase activity, and an associated RNase activity specific for RNA in RNA:DNA hybrid form (22). Although the enzyme can copy either RNA or DNA templates, RT, like all DNA polymerases, can only use deoxyribonucleotides, and not ribonucleotides, as substrates. Studies of the HIV-1 RT have permitted modeling of the position of the incoming nucleotide at the active site (23, 24), with α-helices C and E and β-sheet strands 6 and 9–11, setting the major topology of the dNTP binding site. A recently determined crystal structure of a catalytic fragment of Moloney murine leukemia virus (MMLV) RT at 1.8-Å resolution has made it possible to visualize how such selectivity for deoxyribonucleotides might be achieved: the enzyme is proposed to discriminate against ribonucleotides through an unfavorable interaction between the aromatic ring of Phe-155 and the 2′OH of the incoming rNTP (ref. 14; Fig. 1). Here we report that substitution of this residue by valine, as predicted, does indeed render the enzyme capable of incorporating ribonucleotide substrates into products.
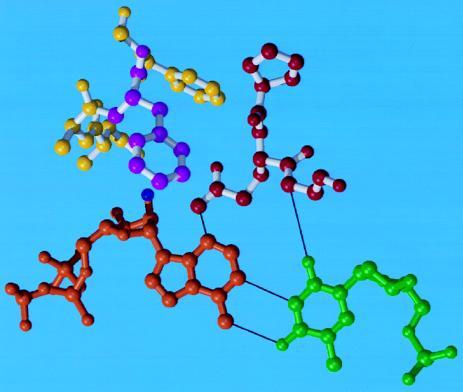
Figure 1.
Modeling of interactions between the MMLV RT, DNA, and rATP at the polymerase active site. A ball-and-stick representation of the minor groove hydrogen-bonding interactions is shown for the modeled ternary complex. Residues 189–191 are shown in red, residues 153, 154, and 156 in yellow, F155 in magenta, rATP in orange, and dT in green. F155 is shown directly below the 2′OH of rATP, serving to discriminate between ribose- and deoxyribose-containing nucleotides.
MATERIALS AND METHODS
Construction of RT Mutants.
The RNase H-defective MMLV reverse transcriptase construct (RT-WT-H) has been described previously (25). RT-F155V-H was constructed by replacing a KpnI–SalI fragment of RT-WT-H (nucleotides 261-1108) with KpnI–AflII and AflII–SalI PCR-derived MMLV RT fragments. Primer F155V-sense (5′-ATATAGCTTAAGGATGCCGTTTTCTGCCTGAGACTCCAC-3′), bearing the mutant valine codon (in boldface type; nucleotides 463–465) and silent mutations creating an AflII site (underlined), and a downstream primer were used to generate the 0.2-kb AflII–SalI PCR fragment, while the F155V-antisense primer (5′-ATATAGCTTAAGATCAAGCACAGTGTACCA-3′), bearing silent mutations to create an AflII site (underlined), and an upstream primer were used to generate the 0.6-kb KpnI–AflII PCR fragment. RT-F155Y-H, in which Phe-155 was substituted by tyrosine, was constructed by replacing the 0.2-kb AflII–SalI fragment of RT-F155V-H with a 0.2-kb AflII–SalI PCR fragment containing a TAT tyrosine codon and an AflII site introduced by the sense primer.
Enzyme Purification.
Recombinant RT enzymes were expressed in Escherichia coli DH5α and partially purified with DE52 resin as described (26) for use in homopolymer assays. For all other assays, enzymes were purified to near-homogeneity by chromatography on DE52 cellulose (Whatman), P11 phosphocellulose (Whatman), and MonoS (Pharmacia) fast protein liquid chromatography (FPLC).
Homopolymer Substrate Assays.
Typical assays were performed using ≈40 ng of enzyme (as determined by immunoblot comparison with pure RT standards) in 50 μl of RT reaction buffer (27) containing 60 mM Tris·HCl (pH 8.0), 75 mM NaCl, 0.7 mM MnCl2, 5 mM DTT, 12 μg/ml homopolymer template, 6 μg/ml oligonucleotide primer, 10 μCi/ml (1 Ci = 37 GBq) 32P-labeled nucleotide, and 12 μM unlabeled nucleotide substrate.
Measurement of Enzyme Kinetics.
Purified enzyme was added to substrates in reaction buffer to initiate the reaction. At each time point, 10 μl of reaction solution was removed and stopped by addition of EDTA. Samples were spotted on DE81 paper (Whatman) and washed with 2× standard saline citrate, followed by scintillation counting. Radioactivity retained on the paper, in comparison with total radioactivity in each sample, was used to determine the amount of dTTP incorporated into the product. Parameters were determined by double reciprocal plot.
Single Nucleotide Extension Assay.
Oligonucleotide C14 (5′-GGTTCCTACCGGCC-3′) was end labeled with [γ-32P]ATP using polynucleotide kinase (New England Biolabs) according to the manufacturer’s specifications. The radiolabeled product oligonucleotide (*C14) was purified by G25 spin column (Boehringer Mannheim) and annealed to G17 (3′-CCAAGGATGGCCGGATC-5′) at room temperature for 0.5 hr. Primer extension was initiated by adding 3 μg of purified enzyme to 60 μl of reaction buffer containing 60 mM Tris·HCl (pH 8.0), 75 mM NaCl, 0.7 mM MnCl2, 5 mM DTT, 0.1 μM *C14/G17, and unlabeled nucleotide substrate at the indicated concentration. At each time point, 10 μl of the reaction was taken out and mixed with 10 μl of stop solution (80% formamide, 0.1% xylene cyanole, 0.1% bromophenol blue, and 0.1 M EDTA). The extension products were resolved by electrophoresis on a 23% urea polyacrylamide gel and detected by autoradiography.
Ribonucleotide Incorporation by RT-F155V-H Using Heteropolymeric Templates.
A 0.3-kb PCR fragment, generated by the F155V-antisense primer and the upstream primer, was cloned into pBluescript (Stratagene) by blunt-end ligation, oriented such that the antisense primer sequence was near the T7 promotor in the vector. The plasmid was linearized and transcribed to generate a 0.32-kb RNA fragment by in vitro run-off transcription using T7 RNA polymerase (Boehringer Mannheim) according to the manufacturer’s instructions. A 0.3-kb single strand DNA was generated by asymmetric PCR using an excess of the F155V antisense primer. The upstream primer was annealed to either template, at concentrations of 40 nM primer and 50 nM template, and extended by 3 μg of purified enzyme for 30 min at 37°C in 60 μl of reaction buffer containing 60 mM Tris·HCl (pH 8.0), 75 mM NaCl, 7.5 mM MgCl2, 5 mM DTT, 500 μM dNTPs, 1 unit/ml RNasin, and 50 μCi of[32P]rNTP. The extended products were precipitated, resuspended in 10 μl of stop solution, and resolved by electrophoresis on a 5% urea polyacrylamide gel, followed by autoradiography.
RNA Synthesis by RT-F155V-H.
Primer oligonucleotide P17 (5′-AAGCCCCACATACAGAG-3′) was end labeled and annealed to template oligonucleotide T28 (3′-TTCGGGGTGTATGTCTCTGACAACCTGG-5′) as described above. The primer (0.1 μM) was extended by 3 μg of RT-F155V-H per 60 μl using four dNTPs (500 μM each) or rNTPs (500 μM each) as substrates. Products were processed and analyzed as described above.
RESULTS AND DISCUSSION
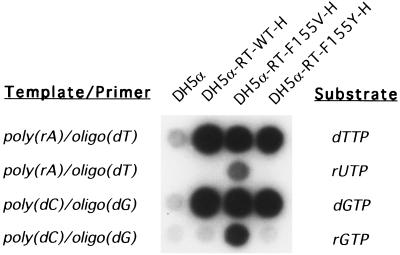
Examination of the active site of a high-resolution structure of the MMLV RT and modeling of template-primer and substrate into the structure resulted in a strong prediction that the 2′OH of an incoming rNTP would overlap with the bulky sidegroup of Phe-155 (14). To test the structural prediction, a mutant MMLV RT was constructed carrying a substitution of Phe-155 with valine in the hope that the smaller side chain would open the “door” of the enzyme to ribonucleotides (see Methods). As a control, a mutant was also generated in which Phe-155 was replaced by tyrosine, found at the corresponding position in HIV-RT and thought to play the same role as the phenylalanine in MMLV-RT. In anticipation of potential cleavages of the RNA products by the RNase H activity in MMLV-RT, the substitutions were introduced into an RNase H-defective MMLV-RT backbone (designated RT-WT-H), which contains a mutation from Asp-524 to Asn (28). RT-WT-H lacks almost all RNase H activity but retains DNA polymerase activity comparable to the wild-type MMLV-RT (28). Each mutant enzyme was expressed in bacteria, partially purified, and assayed on homopolymer templates with various substrates. As predicted, the substitution of Phe-155 with valine rendered the enzyme (designated RT-F155V-H) capable of incorporating ribonucleotides into the products from two different templates (Fig. 2). The wild-type enzyme was unable to use ribonucleotides, and the tyrosine substitution (designated RT-F155Y-H) did not alter the enzyme’s behavior. Of 20 substitution mutants with changes of unrelated residues also located near the active site, none showed similar effects (data not shown).
Figure 2.
RT-F155V-H can incorporate rUTP or rGTP into products, using poly(rA)/oligo(dT) or poly(dC)/oligo(dG) as template/primer. Reactions were performed with the indicated template/primer, enzymes, and labeled substrates, and elongated products were assayed by spotting on DE81 paper, washing, and autoradiography. Mutant RT-F155V-H was uniquely able to incorporate ribonucleotides.
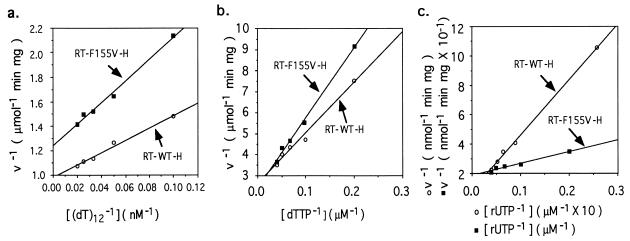
The effect of valine substitution was further analyzed by kinetic measurements of the enzyme activity using purified RTs and poly(rA)/oligo(dT) as template/primer (Fig. 3). The results are summarized in Table 1. RT-F155V-H was similar to RT-WT-H with respect to rate constants (Km) for oligo(dT) and dTTP, as well as maximum velocity (Vmax) using dTTP as a substrate. These similarities between RT-WT-H and RT-F155V-H in these parameters, along with their comparable enzymatic activities on various template/primers under different conditions (data not shown), indicate that the overall structure of RT-F155V-H was hardly, if at all, changed by the substitution. However, the affinity of RT-F155V-H for rUTP was dramatically increased. Whereas the Km of the wild-type enzyme for rUTP was almost 50-fold higher than for dTTP, mutant RT-F155V-H displayed comparable Km values for rUTP and dTTP (Table 1). These results strongly imply that Phe-155 is a key amino acid that dictates the selective binding of deoxyribonucleotides to the enzyme. Interestingly, the Vmax of RT-F155V-H for rUTP was only marginally changed compared with the wild-type, remaining ≈100-fold less than for dTTP.
Figure 3.
Kinetic analysis of RT-WT-H and RT-F155V-H. (a) Various concentrations of oligo(dT)12 primer were annealed to 15 μg/ml poly(rA) template in a reaction buffer containing 60 mM Tris·HCl (pH 8.0), 75 mM NaCl, 0.7 mM MgCl2, 5 mM DTT, 100 μM dTTP, and 20 μCi/ml [32P]dTTP. Reactions were initiated by adding 800 ng/ml purified enzyme. Data points were taken at 20-sec intervals. (b) Reactions were performed in the presence of 15 μg/ml poly(rA), 7.5 μg/ml oligo(dT), 800 ng/ml enzyme, and various concentrations of dTTP and [32P]dTTP. Data points were taken at 20-sec intervals. (c) Reactions were performed in the presence of 15 μg/ml poly(rA), 7.5 μg/ml oligo(dT), 8 μg/ml enzyme, and various concentrations of rUTP and [32P]rUTP. Data points were taken at intervals of 2 min (for RT-WT-H) or 20 sec (for RT-F155V-H).
Table 1.
Summary of kinetic parameters
| Enzyme | (dT)12
|
dTTP
|
rUTP
|
|||
|---|---|---|---|---|---|---|
| Vmax, μmol·min−1·mg−1 | Km, nM | Vmax, μmol·min−1·mg−1 | Km, μM | Vmax, nmol·min−1·mg−1 | Km, μM | |
| RT-WT-H | 1.02 ± 0.05 | 5.27 ± 1.95 | 0.38 ± 0.07 | 9.2 ± 3.4 | 1.17 ± 0.51 | 443 ± 221 |
| RT-F155V-H | 0.81 ± 0.06 | 8.27 ± 3.44 | 0.41 ± 0.04 | 13.6 ± 2.4 | 5.45 ± 0.53 | 4.34 ± 1.71 |
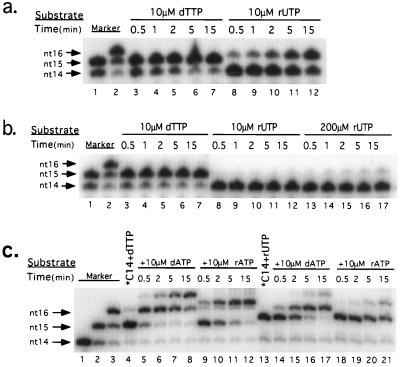
The lower catalytic rate of RT-F155V-H for ribonucleotide substrates could be accounted for by either an inherently lower ribonucleotide incorporation rate or a lower extension rate of a ribonucleotide-containing primer, or both. To distinguish between these possibilities, a single nucleotide extension assay was performed. As shown in Fig. 4a, the mutant extended ≈75% of the primer by addition of dTTP within 0.5 min, while even after 15 min, only ≈50% of the primer was extended by rUTP. In comparison, RT-WT-H incorporated barely detectable amount of rUTP, even at a concentration as high as 200 μM (Fig. 4b). Furthermore, the incorporation of rUTP slowed the catalysis of the following nucleotide, either dATP or rATP, ≈5-fold (Fig. 4c). We note that products with a 3′ terminal ribonucleotide migrated slightly more slowly than those with a deoxyribonucleotide.
Figure 4.
Both incorporation and extension of incorporated ribonucleotides by RT-F155V-H are slower than for deoxyribonucleotides. 5′ end radiolabeled 14-mer DNA primer was annealed to a 17-mer DNA template at a 1:1 ratio. (a) The primer was extended by RT-F155V-H using either 10 μM dTTP or rUTP as a substrate. At the indicated time points, an aliquot of the reaction was removed and analyzed by gel electrophoresis. (b) The primer was extended by RT-WT-H in the presence of 10 μM dTTP, 10 μM rUTP, or 200 μM rUTP. (c) The 14-mer primer was first extended by either 10 μM dTTP or 10 μM rUTP to completion. The extended 15-mer primers were then further extended by adding either 10 μM dATP or 10 μM rATP to the reaction. Markers were generated by extending *C14 with ddTTP (nt15) or dTTP plus ddATP (nt16). Products with ribonucleotides at the 3′ terminus migrated more slowly than those with deoxyribonucleotides at the terminus. The bands migrating at the position of nt17 (lanes 5–8 and 15–17) presumably resulted from untemplated extension of dATP at the last nucleotide of the template.
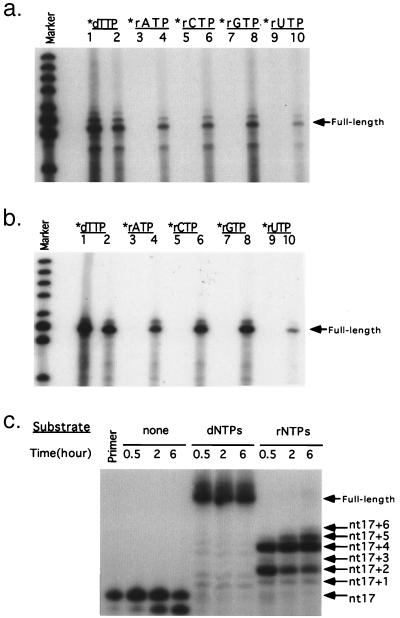
To expand these kinetics analysis results, RT-WT-H and RT-F155V-H were used to copy single-stranded DNA or RNA templates using dNTPs or rNTPs as substrates (Fig. 5). RT-F155V-H incorporated all four ribonucleotides into long products from a mixture of rNTPs and dNTPs, using either DNA (Fig. 5a) or RNA (Fig. 5b) as a template. In contrast, RT-WT-H could barely incorporate detectable ribonucleotides into the product, though it could synthesize DNA products efficiently. That RT-F155V-H had little preference for any particular ribonucleotide substrate supports the specific function of Phe-155 to distinguish ribonucleotides from deoxyribonucleotides.
Figure 5.
RT-F155V-H incorporates ribonucleotides into products. (a) A short primer was extended by RT-WT-H (lanes 1, 3, 5, 7, and 9) or RT-F155V-H (lanes 2, 4, 6, 8, and 10) on a long single-stranded RNA template in the presence of a mixture of four dNTPs and radiolabeled nucleotides as indicated. (b) Similar experiment on a single-stranded DNA template. (c) End-labeled P17 oligonucleotide primer was extended by RT-F155V-H on T28 oligonucleotide template using either four dNTPs or rNTPs as substrates.
To further evaluate the ability of the enzyme to synthesize RNA, we used only ribonucleotides as substrates (Fig. 5c). Despite the slow ribonucleotide polymerization rate, which is consistent with the kinetics results, RT-F155V-H was able to make an extended product of pure RNA of at least 6 nt under the given conditions. It is worth noting again that the ribonucleotide products migrated slower than the corresponding deoxyribonucleotide ones, indicating that the ribonucleotide products did not come from possible dNTPs contamination in the rNTP preparations.
The results presented above provide direct evidence supporting the common stepwise mechanism of nucleic acid polymerization underlying all nucleotide polymerases catalysis. Such a common catalysis mechanism, along with structural similarities among the polymerases (3, 11, 29), suggests that all nucleotide polymerases might have evolved from the same ancestor. Indeed, this notion is supported by the observation that point mutations in T7 RNA polymerase can result in the reciprocal change in specificity to that observed here; these mutations rendered the enzyme, normally specific for ribonucleotides, capable of incorporating deoxyribonucleotides (16, 30). The mechanism by which these mutations act is unknown. Our results also provide insight into how a DNA polymerase selectively uses deoxyribonucleotides as opposed to ribonucleotides as substrates. Multiple devices are employed by the enzyme to prevent incorporation of ribonucleotides, which might be lethal to organisms. In MMLV RT, Phe-155 serves as a door to preclude ribonucleotides’ binding to the enzyme; and ribonucleotides, once bound to the enzyme, are further discriminated against by catalytic machinery for both incorporation and extension. It would be interesting to identify sequences governing the catalytic rate of the enzyme to use ribonucleotide substrates, by structure-based mutagenesis or by colony screening following random mutagenesis (31). RT-F155V-H may prove to be a powerful tool to serve this purpose.
Acknowledgments
We thank Alice Telesnitsky, Hengyin Yang, Bruce Strober, and Jason Gonsky for useful discussions. This work was supported in part by National Institutes of Health Grant R01 CA 30488. G.G. is an Associate and W.H. and S.P.G. are Investigators of the Howard Hughes Medical Institute.
Footnotes
Abbreviations: RT, reverse transcriptase; MMLV, Moloney murine leukemia virus.
References
- 1.Kornberg A. DNA Synthesis. San Francisco: Freeman; 1974. [Google Scholar]
- 2.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1991. [Google Scholar]
- 3.Delarue M, Poch O, Tordo N, Moras D, Argos P. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 4.McAllister W T, Raskin C A. Mol Microbiol. 1993;10:1–6. doi: 10.1111/j.1365-2958.1993.tb00897.x. [DOI] [PubMed] [Google Scholar]
- 5.Arnold E, Ding J, Hughes S H, Hostomsky Z. Curr Opin Struct Biol. 1995;5:27–38. doi: 10.1016/0959-440x(95)80006-m. [DOI] [PubMed] [Google Scholar]
- 6.Ollis D L, Brick P, Hamlin R, Xuong N G, Steitz T A. Nature (London) 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 7.Arnold E, Jacobo-Molina A, Nanni R G, Williams R L, Lu X, Ding J, Clark A D J, Zhang A, Ferris A L, Clark P, Hizi A, Hughes S H. Nature (London) 1992;357:85–89. doi: 10.1038/357085a0. [DOI] [PubMed] [Google Scholar]
- 8.Kohlstaedt L A, Wang J, Rice P A, Friedman J M, Steitz T A. In: Reverse Transcriptase. Skalka A M, Goff S P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 223–250. [Google Scholar]
- 9.Davies J F I, Almassy R J, Hostomsky Z, Ferre R A, Hostomsky Z. Cell. 1994;76:1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- 10.Sousa R, Chung Y J, Rose J P, Wang B C. Nature (London) 1993;364:593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- 11.Pelletier H, Sawaya M R, Kuman A, Wilson S H, Kraut J. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 12.Unge T, Knight S, Bhikhabhai R, Lovgren S, Dauter Z, Wilson K, Strandberg B. Structure (London) 1994;2:953–961. doi: 10.1016/s0969-2126(94)00097-2. [DOI] [PubMed] [Google Scholar]
- 13.Sawaya M R, Pelletier H, Kumar A, Wilson S H, Kraut J. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- 14.Georgiadis M M, Jessen S M, Ogata C M, Telesnitsky A, Goff S P, Hendrickson W A. Structure (London) 1995;3:879–892. doi: 10.1016/S0969-2126(01)00223-4. [DOI] [PubMed] [Google Scholar]
- 15.Rodgers D W, Gamblin S J, Harris B A, Ray S, Culp J S, Hellmig B, Woolf D J, Debouck C, Harrison S C. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joyce C M, Steitz T A. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 17.Johnson K A. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 18.Erie D A, Hajiseyedjavadi O, Young M C, von Hippel P H. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 19.Baltimore D. Nature (London) 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 20.Temin H, Mizutani S. Nature (London) 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 21.Skalka A M, Goff S P. Reverse Transcriptase. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [PubMed] [Google Scholar]
- 22.Telesnitsky A, Goff S P. In: Reverse Transcriptase. Skalka A M, Goff S P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 49–84. [Google Scholar]
- 23.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A J, Arnold E. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 24.Patel P H, Jacobo-Molina A, Ding J, Tantillo C, Clark A D J, Rag R, Nanni R G, Hughes S H, Arnold E. Biochemistry. 1995;34:5351–5363. doi: 10.1021/bi00016a006. [DOI] [PubMed] [Google Scholar]
- 25.Blain S W, Goff S P. J Biol Chem. 1993;268:23585–23592. [PubMed] [Google Scholar]
- 26.Telesnitsky A, Goff S P. Proc Natl Acad Sci USA. 1993;90:1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goff S P, Traktman P, Baltimore D. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blain S W, Goff S P. J Virol. 1995;69:4440–4452. doi: 10.1128/jvi.69.7.4440-4452.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steitz T A, Smerdon S J, Jäger J, Joyce C M. Science. 1994;266:2022–2025. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- 30.Sousa R, Padilla R. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad V R, Goff S P. J Biol Chem. 1989;264:16689–16693. [PubMed] [Google Scholar]