The purification and structure determination of the murine constitutive androstane receptor bound to its inverse agonist/antagonist androstenol is described.
Keywords: nuclear receptor, constitutive androstane receptor, CAR, androstenol
Abstract
The constitutive androstane receptor (CAR) is a member of the nuclear receptor superfamily. In contrast to classical nuclear receptors, which possess small-molecule ligand-inducible activity, CAR exhibits constitutive transcriptional activity in the apparent absence of ligand. CAR is among the most important transcription factors; it coordinately regulates the expression of microsomal cytochrome P450 genes and other drug-metabolizing enzymes. The murine CAR ligand-binding domain (LBD) was coexpressed with the steroid receptor coactivator protein (SRC-1) receptor-interacting domain (RID) in Escherichia coli. The mCAR LBD subunit was purified away from SRC-1 by affinity, anion-exchange and size-exclusion chromatography, crystallized with androstenol and the structure of the complex determined by molecular replacement.
1. Introduction
The constitutive androstane receptor (CAR) is a member of the nuclear receptor superfamily that exhibits constitutive or ligand-independent activity. CAR acts as a transcriptional factor by forming a heterodimer with the nuclear receptor RXR and by binding to specific DNA sequences in the promoters of genes that encode cytochrome P450 enzymes, xenobiotic transporters and various phase II xenobiotic conjugating enzymes. From studies performed in mice, CAR has been observed to play a central role in the clearance of bilirubin (Huang et al., 2003 ▶; Xie et al., 2003 ▶) and bile acids (Saini et al., 2004 ▶) and in the clearance or activation of xenobiotic drugs such as zoxazolamine, cocaine (Wei et al., 2000 ▶) and acetaminophen (Zhang et al., 2002 ▶).
Although mouse CAR (mCAR) displays transactivation in the apparent absence of ligand, there are two classes of small-molecule ligands that alter CAR activity. The first, exemplified by 5α-androst-16-en-3α-ol (androstenol; Forman et al., 1998 ▶), suppresses CAR activity. The second, 1,4-bis[2-(3–5-dichloropyridyloxy)]benzene (TCPOBOP; Tzameli et al., 2000 ▶), can both increase constitutive mCAR activity and reverse the inhibitory affect of androstenol.
The unique pharmacological significance of this protein in addition to its distinctive constitutive activity makes it a prime target for structural studies. In this paper, we describe the preparation of X-ray diffraction-quality crystals and the structure-determination process of the murine CAR ligand-binding domain–androstenol complex.
2. Protein expression and purification
Recombinant mCAR ligand-binding domain (LBD) encompassing amino acids Lys111–Ser358 (accession No. O35627) was produced as an N-terminal hexahistidine-tagged protein in Escherichia coli BL21(DE3) Gold cells (Novagen Inc.) using the NdeI and BamHI restriction sites of the pET-15b vector (Novagen). To facilitate the overexpression of the mCAR LBD, a human SRC1 peptide containing all three receptor-interaction domains (RID 1–3; Asp 617–Asp769; accession No. U59302) was coproduced in the same E. coli cell utilizing the NcoI and BamHI restriction sites of the pACYC184 vector (New England Biolabs). Therefore, the mCAR LBD protein is expressed as an mCAR LBD–SRC-1 RID protein–protein complex.
The transformed E. coli strain was plated onto LB-agar plates supplemented with 100 µg ml−1 ampicillin (Amp) and 25 µg ml−1 chloramphenicol (CamR). The bacterial LB-agar AMP/CamR plates were grown for 11 h at 310 K. Pilot cultures of 35 ml were grown overnight for approximately 12 h in LB media with 200 µg ml−1 Amp and 35 µg ml−1 CamR. Preparative-scale cultures of 2 l were prepared by diluting the pilot culture 100-fold with LB including 100 µg ml−1 Amp and 25 µg ml−1 CamR. This culture was grown at 310 K to an optical density of 0.7–0.8 (λ = 600 nm). Synthesis of protein was induced with 0.5 mM isopropylthiogalactopyranoside (IPTG) at 303 K for 9 h. The cells were harvested by centrifugation at 5000g at 277 K. Cells were either lysed immediately for protein purification or stored at 193 K until later use. Cell lysis was carried out at 300 K. The cell pellet, which yielded 20 mg of recombinant protein per litre of medium, was resuspended in ice-cold resuspension buffer (20 mM Tris–HCl pH 8.0 at 277 K, 300 mM NaCl, 10% glycerol) containing one Complete EDTA-free tablet (Roche), 0.5 mM PMSF, 10 mM β-mercaptoethanol (βME), 10 µl DNAse (Roche) and 10 mM MgSO4. Cells were lysed using a French press at a pressure of 10 MPa. The cell lysate was clarified by centrifugation at 30 000g for 1 h. The pH of the clarified lysate was adjusted to 8.0 and the lysate was applied onto a 1 ml Ni2+-Sepharose affinity matrix (Qiagen). This protein-bound Ni2+-Sepharose column was washed with 20 column volumes of ice-cold resuspension buffer as described above. This was followed by washing the column with thrombin cleavage buffer (Novagen). The protein-bound column was incubated with 1 U thrombin protease (Novagen) per milligram of protein at room temperature for 2 h and then incubated overnight at 277 K. The mCAR LBD–SRC1 liberated from the hexahistidine-tagged protein was then washed off the column with the appropriate amount of wash buffer. The mCAR LBD–SRC RID was subsequently purified by anion-exchange chromatography by diluting the protein solution tenfold in anion-exchange buffer A (50 mM Tris–HCl pH 8.0 at 277 K, 10% glycerol, 1 mM DTT). The protein mixture was then loaded onto a Poros HQ anion-exchange column (Perseptive Biosystems) at 5.0 ml min−1. Bound protein was eluted at 120 mM NaCl with an increasing salt concentration with buffer B (50 mM Tris–HCl pH 8.0 at 277 K, 500 mM NaCl, 10% glycerol, 1 mM DTT). The mCAR LBD–SRC-1 RID complex was disassociated by incubating the protein with 20 µM androstenol. The mCAR LBD was separated from any undissociated mCAR LBD–SRC-1 RID complex by gel-filtration chromatography. The protein solution was concentrated to a volume of 2 ml and applied onto a pre-equilibrated (30 mM Tris–HCl, 100 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 20% glycerol) Superdex-75 High-Load 16/60 column (Amersham Biosciences). The protein was eluted with the same buffer. Protein fractions were analyzed by SDS–PAGE. The mCAR LBD-containing fractions were pooled and concentrated to 6–8 mg ml−1. This protein sample was utilized for crystallization.
3. Preparation and crystallization of the mCAR LBD–androstenol complex
The mCAR LBD–androstenol complex was prepared by introducing a threefold molar excess of androstenol to purified mCAR LBD and the mixture was incubated at room temperature for 1 h. Initial conditions for crystallization of the mCAR LBD–androstenol complex were obtained from preliminary screens using the sparse-matrix crystal screening kits of Hampton Research (Laguna Niguel, CA, USA) and Jena Biosciences (Jena, Germany). All experiments were performed using the vapor-diffusion technique with hanging drops in Linbro plates. This was performed by mixing 1 µl 6–8 mg ml−1 protein with 1 µl mother liquor. Crystalline precipitate appeared in condition No. 11 of Crystal Screen 1 (Hampton Research) in approximately 14 d. This condition was further optimized to 18% PEG 400, 0.2 M CaCl2, 0.1 M HEPES pH 7.2 to yield single crystals at 287 K. Diffraction-grade crystals were obtained by the addition of 0.01 M l-cysteine.
4. Data collection
A 2.9 Å native data set was recorded at the Advanced Photon Source, DuPont–Northwestern–Dow Collaborative Access Team Sector 5ID-B, utilizing a MAR Mosaic CCD225 detector. Crystals were briefly dipped into cryoprotectant solution (20% PEG 400, 0.2 M CaCl2, 0.1 M HEPES pH 7.2, 5 µM androstenol, 5 mM DTT, 25% glycerol) and cryocooled in liquid nitrogen.
The diffraction pattern revealed the unit-cell parameters to be a = 60.35, b = 155.04, c = 134.61 Å, α = β = γ = 90°. Systematic absences of reflections confirmed that the space group was C2221. The crystallographic asymmetric unit contains two molecules of mCAR LBD–androstenol with a solvent content of 54.3% and a Matthews coefficient of 2.7 Å3 Da−1. The data were indexed and integrated with the MOSFLM (Leslie, 1992 ▶) and XDS (Kabsch, 1993 ▶) packages and scaled using SCALA (Collaborative Computational Project, Number 4, 1994 ▶).
5. Structure determination by molecular replacement
The structure of the mCAR LBD–androstenol complex was determined by molecular replacement using AMoRe (Collaborative Computational Project, Number 4, 1994 ▶). The search model was based on the PXR LBD (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1ilh) structure (Watkins et al., 2001 ▶). The two proteins have an approximately 35.1% sequence identity. However, in the successful search model, only helices 3–9 were used. The β-strands were deleted and the loop regions were given a B factor of 100 Å2.
6. Results and discussion
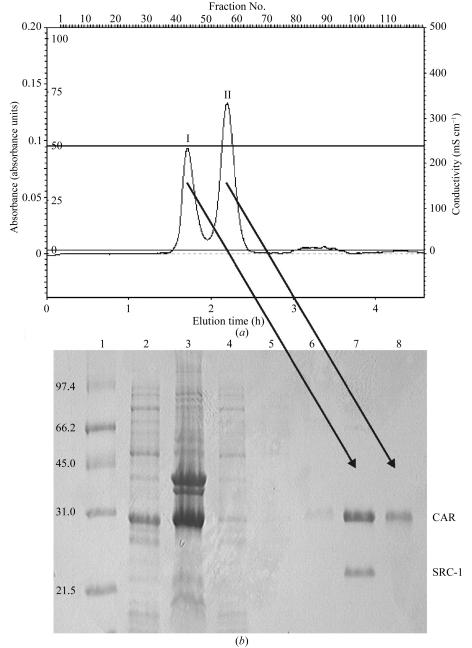
The size-exclusion step results in a separation of the mCAR LBD from the undissociated mCAR LBD–SRC-1 RID complex (Fig. 1 ▶ a). The compositions of the eluted fractions were observed by SDS–PAGE (Fig. 1 ▶ b). The concentrated mCAR LBD–androstenol complex was used for crystallization. Initially, a microcrystalline precipitate was observed upon mixing the protein solution with the reservoir. However, crystals grew from within this precipitate, appearing in 2–3 d and growing to maximum dimensions of 200 × 50 × 10 µm within 5 d (Fig. 2 ▶). Diffraction data were >99% complete with an overall R merge of 10% (Table 1 ▶). Several search models were examined; however, other nuclear receptor LBD structures failed as search models. The only model that gave a meaningful solution to the search was the modified PXR structure described above. The final solution was obtained after rigid-body refinement of the initial solution. The final rotation, translation, correlation coefficient and R factor are listed in Table 2 ▶. The current mCAR LBD–androstenol model is currently being refined to generate an accurate representation of the structure of this protein–ligand complex.
Figure 1.
(a) Gel-filtration chromatogram of mCAR LBD–SRC1 complex and mCAR LBD. (b) SDS–PAGE, 15% gel. Lane 1, molecular-weight markers. Lane 2, soluble extract from E. coli. Lane 3, pelleted fraction from E. coli. Lane 4, Ni–NTA flowthrough upon loading of mCAR LBD and mCAR LBD–SRC1. Lane 5, Ni–NTA flowthrough post-wash (20 mM Tris, 150 mM NaCl, 2.5 mM CaCl2 pH 8.4 at room temperature) of mCAR LBD–SRC and mCAR LBD-bound Ni–NTA resin. Lane 6, Ni–NTA column post-protein elution. Lane 7, fraction 42 of gel-filtration chromatogram containing mCAR LBD–SRC1 complex. Lane 8, fraction 58 of gel-filtration chromatogram containing purified mCAR LBD.
Figure 2.
Crystals of mCAR LBD–androstenol complex obtained by the hanging-drop method. The crystals are approximately 200 µm in length.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Space group | C2221 |
| Unit-cell parameters | |
| a (Å) | 60.3 |
| b (Å) | 155.04 |
| c (Å) | 134.61 |
| α = β = γ (°) | 90.0 |
| Matthews coefficient (Å3 Da−1) | 2.7 |
| Solvent content (%) | 54.3 |
| No. molecules per AU | 2 |
| Resolution range (Å) | 30–2.9 (3.0–2.9) |
| Total reflections | 157834 |
| Unique reflections | 14399 |
| Redundancy | 11.0 (11.1) |
| 〈I〉/〈σ(I)〉 | 21.84 (6.94) |
| Data completeness (%) | 99.8 (100.0) |
| Rsym† (%) | 10.8 (45.1) |
R
sym = 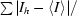
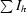 , where Ih is the integrated intensity of a given reflection and 〈Ih〉 is the average intensity over symmetry equivalents.
, where Ih is the integrated intensity of a given reflection and 〈Ih〉 is the average intensity over symmetry equivalents.
Table 2. Molecular-replacement solution.
Model: PXR, amino acids 150–413.
| θ1 | θ2 | θ3 | x | y | z | CC | R factor | |
|---|---|---|---|---|---|---|---|---|
| Initial solution | ||||||||
| 1 | 101.9 | 27.9 | 221.8 | 0.1241 | 0.3164 | 0.1172 | 37.8 | 52.4 |
| 2 | 35.2 | 58.6 | 297.4 | 0.8341 | 0.9156 | 0.6177 | 41.7 | 51.0 |
| Final solution | ||||||||
| 1 | 100.6 | 27.0 | 221.5 | −36.0 | 1.4 | 31.0 | — | — |
| 2 | 35.6 | 57.7 | 295.6 | 5.8 | 22.5 | 132.7 | 45.5 | 49.5 |
Acknowledgments
We thank Chris Dealwis, Elias Lolis and Jim Pflugrath for advice and assistance. This work was performed at the DuPont–Northwestern–Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center located at Sector 5 of the Advanced Photon Source. DND-CAT is supported by E. I. DuPont de Nemours & Co., The Dow Chemical Company, the US National Science Foundation through grant DMR-9304725 and the State of Illinois through the Department of Commerce and the Board of Higher Education Grant IBHE HECA NWU 96. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Energy Research under Contract No. W-31-102-Eng-38.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Forman, B. M., Tzameli, I., Choi, H. S., Chen, J., Simha, D., Seol, W., Evans, R. M. & Moore, D. D. (1998). Nature (London), 395, 612–615. [DOI] [PubMed] [Google Scholar]
- Huang, W., Zhang, J., Chua, S. S., Qatanani, M., Han, Y., Granata, R. & Moore, D. D. (2003). Proc. Natl Acad. Sci. USA, 100, 4156–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800. [Google Scholar]
- Leslie, A. G. W. (1992). Jnt CCP4/EACBM Newsl. Protein Crystallogr.26
- Saini, S. P., Sonoda, J., Xu, L., Toma, D., Uppal, H., Mu, Y., Ren, S., Moore, D. D., Evans, R. M. & Xie, W. (2004). Mol. Pharmacol.65, 292–300. [DOI] [PubMed] [Google Scholar]
- Tzameli, I., Pissios, P., Schuetz, E. G. & Moore, D. D. (2000). Mol. Cell. Biol.20, 2951–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, R. E., Wisely, G. B., Moore, L. B., Collins, J. L., Lambert, M. H., Williams, S. P., Willson, T. M., Kliewer, S. A. & Redinbo, M. R. (2001). Science, 292, 2329–2333. [DOI] [PubMed] [Google Scholar]
- Wei, P., Zhang, J., Egan-Hafley, M., Liang, S. & Moore, D. D. (2000). Nature (London), 407, 920–923. [DOI] [PubMed] [Google Scholar]
- Xie, W., Yeuh, M. F., Radominska-Pandya, A., Saini, S. P., Negishi, Y., Bottroff, B. S., Cabrera, G. Y., Tukey, R. H. & Evans, R. M. (2003). Proc. Natl Acad. Sci. USA, 100, 4150–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Huang, W., Chua, S. S., Wei, P. & Moore, D. D. (2002). Science, 298, 422–424. [DOI] [PubMed] [Google Scholar]