In order to clarify the structural basis of the pathogenesis-related-1 domain, Na-ASP-1, the first multi-domain ASP from the human hookworm parasite N. americanus, has been crystallized. 2.2 Å resolution data have been collected from a crystal belonging to the monoclinic space group P21.
Keywords: pathogenesis-related proteins, hookworm, ASP, Necator americanus, ancylostoma, vaccines
Abstract
Human hookworm infection is a major cause of anemia and malnutrition in the developing world. In an effort to control hookworm infection, the Human Hookworm Vaccine Initiative has identified candidate vaccine antigens from the infective larval stage (L3) of the parasite, including a family of pathogenesis-related-1 (PR-1) proteins known as the ancylostoma-secreted proteins (ASPs). The functions of the ASPs are unknown. In addition, it is unclear why some ASPs have one while others have multiple PR-1 domains. There are no known structures of a multi-domain ASP and in an effort to remedy this situation, recombinant Na-ASP-1 has been expressed, purified and crystallized. Na-ASP-1 is a 406-amino-acid multi-domain ASP from the prevalent human hookworm parasite Necator americanus. Useful X-ray data to 2.2 Å have been collected from a crystal that belongs to the monoclinic space group P21 with unit-cell parameters a = 67.7, b = 74.27, c = 84.60 Å, β = 112.12°. An initial molecular-replacement solution has been obtained with one monomer in the asymmetric unit.
1. Introduction
Hookworms are one of the most predominant parasites of humans: they cause morbidity in almost one billion people worldwide (Hotez et al., 2004 ▶). Hookworm disease is prevalent wherever rural poverty occurs, including the tropics and subtropics, placing a serious burden on those who can least afford it. The nematodes Necator americanus and Ancylostoma duodenale produce injury through intestinal hemorrhage as they feed on host hemoglobin, resulting in blood loss, protein malnutrition, anemia and other complications. Current control methods for hookworm infection include barrier foot protection, sanitation (construction of pit latrines) and anthelminthic chemotherapy primarily aimed at deworming school-age children. The foot barrier protection is an ineffective control method because Ancylostoma is orally infective and Necator infects through the upper and lower extremeties (Bethony et al., 2002 ▶). Anthelminthic chemotherapy using benzimidazoles is only a temporary solution, as rapid reinfection occurs within four to six months (World Health Organization, 2002 ▶).
As an alternative or complementary approach to hookworm control, an international effort has been initiated to develop an anti-hookworm vaccine aimed at reducing worm burdens and intensity (Hotez et al., 2003 ▶). The selection of antigens for hookworm vaccine development is based partly on the early observation that the infective stages of the parasite, the third-stage larvae (L3), are immunogenic (Miller, 1971 ▶) and that radiation-attenuated living L3 have been successfully used as a veterinary vaccine for canine hookworm infection in the late 1970s (Miller, 1978 ▶). A major goal of vaccine development for humans is to reproduce the effect of live attenuated L3 vaccines by substituting with L3-derived antigens in recombinant form (Hotez et al., 2003 ▶). It was subsequently determined that there are two major antigens secreted by L3 upon host entry: ancylostoma-secreted protein-1 (ASP-1) and ASP-2. ASPs are nematode-specific proteins that belong to the pathogenesis-related (PR) protein superfamily. PR proteins were initially isolated from plants and their name is related to the elevated expression of these proteins upon injuries inflicted by pathogens or by other stress factors (Van Loon & Van Strien, 1999 ▶).
ASPs are members of the PR-1 subfamily, which includes the mammalian cysteine-rich secreted protein (CRISP) family as well as a diversity of proteins from the animal, plant and fungal kingdoms. Specific members include mammalian testis-specific protein (Kasahara et al., 1989 ▶), human brain tumor protein GliPR (Mochca-Morales et al., 1990 ▶) and antigen 5 from Vespula vilgaris venom (Lu et al., 1993 ▶). While the function of this subfamily is unknown, its members are characterized by a highly conserved cysteine-rich domain of about 15–16 kDa molecular weight and have been implicated in conditions requiring cellular defense or proliferation such as plant pathogenesis and brain-tumor growth. Two types of ASP have been isolated from larval nematodes and they either possess a single PR-1 domain (Na-ASP-2) or two PR-1 domains (Na-ASP-1). Na-ASP-1 is a 406-amino-acid protein that has a molecular weight of 43.9 kDa and is approximately twice the size of the 197-amino-acid 21.3 kDa Na-ASP-2.
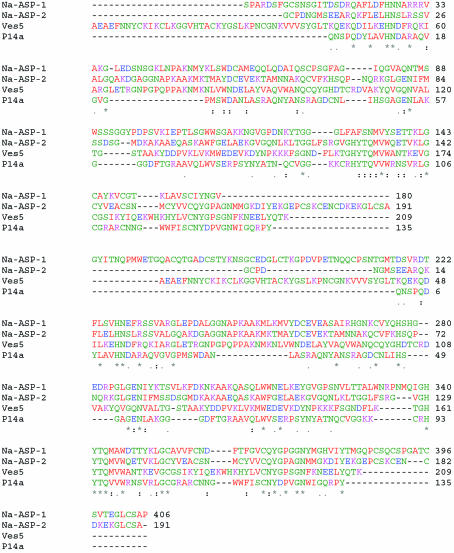
There are no known structures of multi-domain PR-1 proteins and only three structures of single-domain PR-1 proteins have been reported. The three reported structures are the X-ray structure of Ves v 5 (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1qnx; Henriksen et al., 2001 ▶), the NMR structure of p14a, a PR-1 protein from tomato (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1cfe; Fernandez et al., 1997 ▶), and the high-resolution (1.68 Å) structure of Na-ASP-2 (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1u53; Asojo et al., 2005 ▶), in which the protein exists in the form of a crystallographic dimer. The dimeric nature of Na-ASP-2 was also confirmed by SEC-MALS analysis (Asojo et al., 2005 ▶). The sequence identity between Na-ASP-1 and Ves v 5, p14a and Na-ASP-2 are 19, 17 and 27%, respectively. Despite limited sequence identity, there is a significant conservation and alignment within the conserved PR-1 domain (Fig. 1 ▶). In order to clarify the structural basis of multi-domain PR-1 proteins and their associated antigenic properties, we have expressed, purified and crystallized recombinant Na-ASP-1. Here, we present our preliminary crystallographic analysis.
Figure 1.
CLUSTALW alignment of PR-1 proteins of known structure with Na-ASP-1. Two monomers of the single-domain PR-1 proteins Na-ASP-2, Ves5 and P14a are aligned with a monomer of Na-ASP-1. Na-ASP-2, L3 ASP from N. americanus (AY288089); Ves5, antigen Ves v 5 from Vespula vilgaris (Q05110); P14a is from cherry tomato (P04284). PUBMED protein ID codes are shown in parentheses (http://www.pubmed.org). The PDB codes are http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1u53, http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1qnx and http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1cfe for Na-ASP-2, Ves5 and P14a, respectively. Amino acids are colored as follows: red for small and hydrophobic amino acids (AVFPMILW), blue for acidic amino acids (DE), magenta for basic amino acids (RHK) and green for hydroxyl, amine and basic (STYHCNGQ). ‘*’ indicate sites in all aligned sequences with identical residues, while ‘.’ and ‘:’ indicate conservatively mutated positions (there is higher conservation for the latter than the former).
2. Expression and purification of Na-ASP-1
2.1. Cloning
A cDNA library of infective third-stage larvae (L3) of N. americanus was constructed as described previously (Hawdon et al., 1996 ▶; Zhan et al., 1999 ▶). The entire coding sequence minus the N-terminal signal peptides of Na-asp-1 was PCR amplified from the first-strand cDNA of N. americanus L3 with Na-asp-1 gene-specific primers: 5′-CTCTCGAGAAGAGATCTCCAGCAAGAGACAGCTTC-3′ and 5′-GGGGTACCTTAAGGAGCGCTGCACAAGCC-3′. The primers introduced XhoI and KpnI sites (bold) on the 5′ and 3′ ends of the gene, respectively. The PCR product was subcloned into the expression vector pPICZαA partially digested with XhoI and KpnI restriction enzymes. The Na-asp-1 gene was fused to the pre-pro leader sequence of Saccharomyces cerevisiae α-mating factor under the control of the AOX1 promoter. The construct sequence was confirmed by sequencing the insert. pPICZαNa-asp-1 was linearized before transforming into Pichia pastoris KM71H (muts: methanol utilization slow). Transformants were selected on YPD plates containing Zeocin (100 mg l−1).
2.2. Fermentation
A single colony was grown in BMGY medium (1% yeast extract, 2% peptone, 0.1 M potassium phosphate buffer pH 6.0, 1.3% yeast nitrogen base and 1.2% glycerol) in shaker flasks. The culture was grown for approximately 48 h to an optical density of 4–5; 150 ml of the seed culture was aseptically transferred to 2 l fermentation medium. Fermentations were performed in a 5 l Bioflo III/3000 stirred-tank fermentor (New Brunswick Scientific, Edison, GA, USA) with a 2 l starting volume interfaced with a computer using Biocommand 32 software (New Brunswick Scientific) for data acquisition and control. Methanol was fed into the fermentor using a closed-loop control system with a methanol feed pump (Model 101 U/R, Watson-Marlow, UK) and a balance (Model PG 12001-S, Mettler Toledo, Switzerland) for weighing the methanol bottle interfaced with the computer using Biocommand 32 software. The methanol concentration in the off-gas was analyzed with a methanol concentration monitor and controller (Model MC-168, PTI Instruments, Kathleen, GA, USA). Fermentations were performed at 303 K. The pH during growth (glycerol batch and fed-batch phase) was maintained at 5.0 and then ramped up to 6.0 over a 2 h period starting from induction with methanol (beginning of the transition phase). Saturated aqueous ammonium hydroxide was used to control the pH at 6.0 until the end of fermentation. FM22 medium (Stratton et al., 1998 ▶) was employed for growth and recombinant protein production.
The fermentation procedure was divided into three phases: batch phase, fed-batch phase and induction phase. Glycerol (40 g l−1) was also added during the batch phase, which lasted 20–24 h after inoculation. The fed-batch phase began with the addition of 50% glycerol (20 g l−1) and lasted until the wet cell weight was about 300 g l−1. The induction phase began with the addition of methanol (1 ml l−1), upon which the glycerol feed was slowly ramped down. Once the cells were adapted to methanol, a methanol feed containing 2 ml l−1 PTM1 salts was started. The methanol sensor was used to control the methanol concentration, maintaining about 1 g l−1 throughout the induction period, which lasted 60 h. 20% peptone was also added to reduce the proteolytic activity in fermentors.
2.3. Purification
The fermentation broth was harvested at 6000g for 20 min at 277 K. PMSF and EDTA were immediately added to final concentrations of 2 and 5 mM, respectively. The broth was diafiltered into 50 mM Tris, 5 mM EDTA pH 8.3 using a 3 kDa regenerated cellulose membrane at 206 kPa at 277–285 K. The retentate was loaded onto a ∼1 l Q-Sepharose FF column. After the column had been equilibrated with 50 mM Tris, 5 mM EDTA pH 8.3, the diafiltered ASP-1 was loaded onto the column. The product was collected in the Q column flowthrough. The product from the Q column flowthrough was concentrated from a volume of 3 l down to 10 ml. A bench-top ultrafiltration unit (regenerated cellulose membrane, 10 kDa molecular-weight cutoff) was used for the initial concentration. An Amicon Stirred Cell (Omega membrane, 5 kDa molecular-weight cutoff) was then used to reach the required final volume and concentration. Size-exclusion chromatography using Sephacryl S-100 High Resolution resin was used as the final polishing step. The product was eluted with PBS buffer pH 7.2. Purity was confirmed by SDS–PAGE analysis and prior to crystallization, protein was desalted and concentrated with a Microcon (Millipore, 30 kDa molecular-weight cutoff) concentrator.
3. Crystallization
Initial crystallization screens were carried out on samples of Na-ASP-1 using sparse-matrix screens from Emerald Biostructures and Hampton Research, including Wizard Screens, Crystal Screens I and II and all Cryo Screens. These experiments were carried out at three different temperatures: 293, 285 and 277 K. Similar behavior was observed regardless of growth temperature. Crystals were grown by vapor diffusion in hanging drops which were equilibrated against 1 ml crystallization solution. Drops were prepared by mixing 2 µl of protein solution with an equal volume of crystallization solution. All protein solutions for crystallization experiments consisted of 15 mg ml−1 Na-ASP-1 in 10 mM Tris–HCl pH 7.5. The initial protein concentration was confirmed spectrophotometrically prior to setting up crystallization experiments. Several conditions from the Cryo Screens yielded promising leads and gave small crystals or showers of crystals. These conditions were characterized by a high concentration [between 20 and 40%(w/v)] of polyethylene glycol (6000 or 8000) and a low concentration of buffer (less than 200 mM). The pH did not seem to be important to nucleation, as plate-like crystals were obtained at pH values ranging from 5.1 (citrate) to 9.3 (CHES). Large plate-like crystals were obtained upon optimization of the crystallization conditions. Despite the relative ease of crystal growth, crystal cracking and degradation after four weeks (at 298 and 277 K) has been observed. Consequently, the diffraction patterns from older crystals were significantly poorer than those from fresh crystals. The protein solution degraded over time and samples that initially crystallized would not crystallize after three weeks at 277 K.
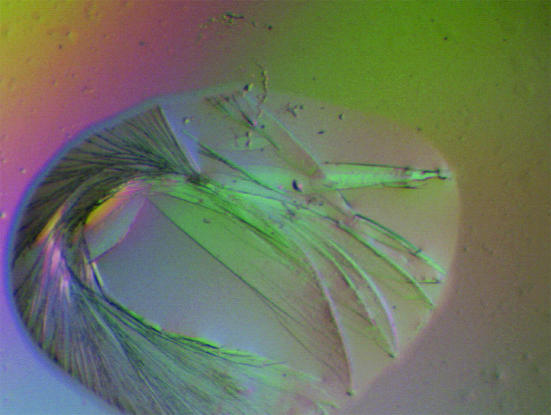
The largest crystals were obtained overnight by vapor diffusion from hanging drops containing a mixture of 3 µl (7.5 mg ml−1) protein and 1.5 µl reservoir solution, with a reservoir containing 21%(w/v) PEG 6000 and 200 mM sodium citrate pH 6.0. Similar crystals were also obtained from a reservoir solution containing 30%(w/v) PEG 8000 and 100 mM CHES pH 9.3. These plate-like crystals grew to dimensions of 0.3 × 0.5 × 0.05 mm on the smallest face (Fig. 2 ▶) and degraded rapidly over a four-week period.
Figure 2.
Crystals of Na-ASP-1 are thin flat plates of less than 0.05 mm on the smallest face.
4. Diffraction experiments and structure determination
Since crystals grew in solutions that contained adequate cryoprotectant, the crystals were flash-cooled in a stream of N2 gas at 113 K prior to collecting diffraction data on a MAR 345 detector (MAR Research). The X-ray source was a Rigaku RU-200 rotating-anode generator operating at 50 kV and 100 mA, with a double focusing-mirror system. A complete data set was collected from a single crystal using a crystal-to-detector distance of 150 mm and exposure times of 20 min for 1.0° oscillations. All X-ray diffraction data sets were processed using the programs DENZO (Otwinowski, 1993a ▶) and SCALEPACK (Otwinowski, 1993b ▶). Crystallographic data is shown in Table 1 ▶. With the volume of the unit cell (P2 or P21) being 394 122 Å3 and the solvent content being between 40 and 80% of the unit-cell volume (with respective Matthews coefficients between 2.1 and 5.4 Å3 Da−1), the asymmetric unit of those crystals was expected to contain either a monomer or dimer of Na-ASP-1. These molecular packings would correspond to Matthews coefficients of 4.48 Å3 Da−1 (72.6% solvent) and 2.24 Å3 Da−1 (45.1% solvent) for a monomer and dimer in the asymmetric unit, respectively.
Table 1. Data-collection and reduction statistics.
Values in parentheses are for the highest resolution shell.
| Space group | P21 |
| Unit-cell parameters (Å, °) | a = 67.7, b = 74.27, c = 84.60, β = 112.12 |
| Resolution limits (Å) | 50–2.18 (2.18–2.26) |
| 〈I/σ(I)〉 | 13.5 (2.68) |
| No. of reflections | 324988 |
| No. of unique reflections | 39175 |
| Redundancy | 8.3 (7.4) |
| Rmerge† (%) | 10.6 (49.7) |
| Completeness (%) | 96.1 (93.9) |
R
merge = 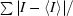
 , where I is the observed intensity and 〈I〉 is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
, where I is the observed intensity and 〈I〉 is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
Using residues 12–180 of the full structure of Na-ASP-2 as the search model and the program AMoRe (Navaza, 1994 ▶), a satisfying molecular-replacement solution was found in space group P21 but not in P2. The correlation coefficient and initial R factors were 32.1 and 47.8% for a monomer of Na-ASP-1 in the asymmetric unit. The resulting packing is reasonable and negates the possibility of a dimeric form of Na-ASP-1. Model building and refinement are in progress.
Acknowledgments
The Human Hookworm Vaccine Initiative is supported by a grant from The Bill and Melinda Gates Foundation awarded to the Sabin Vaccine Institute and George Washington University Medical Center.
References
- Asojo, O. A., Goud, G. N., Dhar, K., Loukas, A., Zhan, B., Deumic, V., Liu, S. Borgstahl, G. E. O. & Hotez, P. J. (2005). J. Mol. Biol.346, 801–814. [DOI] [PubMed] [Google Scholar]
- Bethony, J., Chen, J., Lin, S., Xiao, S., Zhan, B., Li, S., Xue, H., Xing, F., Humphries, D., Yan, W., Chen, G., Foster, V., Hawdon, J. M. & Hotez, P. J. (2002). Clin. Infect. Dis.35, 1336–1344. [DOI] [PubMed] [Google Scholar]
- Fernandez, C., Szyperski, T., Bruyere, T., Ramage, P., Mosinger, E. & Wuthrich, K. (1997). J. Mol. Biol.266, 576–593. [DOI] [PubMed] [Google Scholar]
- Henriksen, A., King, T. P., Mirza, O., Monsalve, R. I., Meno, K., Ipsen, H., Larsen, J. N., Gajhede, M. & Spangfort, M. D. (2001). Proteins, 45, 438–448. [DOI] [PubMed] [Google Scholar]
- Hotez, P. J., Brooker, S., Bethony, J. M., Bottazzi, M. E., Loukas, A. & Xiao, S. (2004). N. Engl. J. Med.351, 799–807. [DOI] [PubMed] [Google Scholar]
- Hotez, P. J. et al. (2003). Int. J. Parasitol.33, 1245–1258. [DOI] [PubMed] [Google Scholar]
- Hawdon, J. M., Jones, B. F., Hoffman, D. R. & Hotez, P. J. (1996). J. Biol. Chem.271, 6672–6678. [DOI] [PubMed] [Google Scholar]
- Kasahara, M., Gutknecht, J., Brew, K., Spurr, N. & Goodfellow, P. N. (1989). Genomics, 5, 527–534. [DOI] [PubMed] [Google Scholar]
- Lu, G., Villalba, M., Coscia, M. R., Hoffman, D. R. & King, T. P. (1993). J. Immunol.150, 2823–2830. [PubMed] [Google Scholar]
- Miller, T. A. (1971). Adv. Parasitol.9, 153–183. [DOI] [PubMed] [Google Scholar]
- Miller, T. A. (1978). Adv. Parasitol.16, 333–342. [DOI] [PubMed] [Google Scholar]
- Mochca-Morales, J., Martin, B. M. & Possani, L. D. (1990). Toxicon, 28, 299–309. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Otwinowski, Z. (1993a). DENZO: An Oscillation Data Processing Program for Macromolecular Crystallography. Yale University, Connecticut, USA.
- Otwinowski, Z. (1993b). SCALEPACK: Software for the Scaling Together of Integrated Intensities Measured on a Number of Separate Diffraction Images. Yale University, Connecticut, USA.
- Stratton, J., Chiruvolu, V. & Meagher, M. M. (1998). High Cell-Density Fermentation, edited by D. R. Higgins & J. M. Cregg, pp. 107–120. New Jersey: Humana. [DOI] [PubMed]
- Van Loon, L. C. & Van Strien, E. A. (1999). Physiol. Mol. Plant Pathol.55, 85–97.
- WHO (2002). World Health Organ. Tech. Rep. Ser.912, 547–551.
- Zhan, B., Hawdon, J., Qiang, H., Hainan, R., Qiang, H., Wei, H., Shu-Hua, X., Tie-Hua, L., Xing, G., Zheng, F. & Hotez, P. (1999). Mol. Biochem. Parasitol.98, 143–149. [DOI] [PubMed] [Google Scholar]