The crystallization and preliminary X-ray diffraction analysis of MraZ, formerly known as hypothetical protein YabB, from Escherichia coli K-12 is presented.
Keywords: MraZ, YabB, cell-wall biosynthesis, Escherichia coli, structural genomics
Abstract
The MraZ family of proteins, also referred to as the UPF0040 family, are highly conserved in bacteria and are thought to play a role in cell-wall biosynthesis and cell division. The murein region A (mra) gene cluster encodes MraZ proteins along with a number of other proteins involved in this complex process. To date, there has been no clear functional assignment provided for MraZ proteins and the structure of a homologue from Mycoplasma pneumoniae, MPN314, failed to suggest a molecular function. The b0081 gene from Escherichia coli that encodes the MraZ protein was cloned and the protein was overexpressed, purified and crystallized. This data is presented along with evidence that the E. coli homologue exists in a different oligomeric state to the MPN314 protein.
1. Introduction
The cell wall of microorganisms is composed of a giant polymer of peptidoglycan that confers both shape and rigidity on the cell. In Gram-negative organisms, the cell wall contains an additional layer made of lipopolysaccharide, which forms a second lipid bilayer containing both polysaccharide and protein. When a cell enlarges during cell division, new cell-wall synthesis involving the addition of new material to the pre-existing wall must take place without compromising the structural integrity of the cell. The processes of cell-wall biosynthesis and cell division are therefore tightly coupled.
Cell-wall biosynthesis by Gram-negative organisms is a complex process that involves numerous proteins and has both cytoplasmic and membrane components (van Heijenoort, 2001 ▶). In Escherichia coli, protein products of the mra or murein region A gene cluster are responsible for both cell-wall biosynthesis (murEFDGC, mraWY and DdlB) and cell division (ftsLIWQAZ; Fig. 1 ▶; Mengin-Lecreulx et al., 1998 ▶). Also located in this cluster is the gene mraZ, which was formerly referred to as yabB. The mraZ gene product is a protein of unknown function that owing to its genomic location is thought to play a role in cell-wall biosynthesis and/or cell division. Members of the MraZ protein family are very highly conserved in bacteria and although there are inconsistencies in the literature, some groups have reported that expression of MraZ proteins is essential for the growth and survival of their respective organisms (Arigoni et al., 1998 ▶; Carrion et al., 1999 ▶; Hutchison et al., 1999 ▶). However, other studies have provided evidence that the MraZ homologues are not essential genes (Daniel et al., 1996 ▶; Dassain et al., 1999 ▶; Merlin et al., 2002 ▶). There is also little functional information available for these proteins and their exact contribution to cell-wall biosynthesis and/or cell division remains unclear.
Figure 1.
(a) Organization of the mra gene cluster. (b) Sequence alignment of the E. coli MraZ protein with MPN314. For simplicity, the purification tags are not shown for either protein. Conserved residues are denoted with an asterisk.
Recently, the three-dimensional crystal structure of MPN314 from Mycoplasma pneumoniae was reported (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1n0g; Chen et al., 2004 ▶). This protein displays 28% sequence identity and good overall similarity with the E. coli homologue (Fig. 1 ▶). All members of this protein family are composed of two sequence domains, each belonging to the 70-residue UPF0040 protein family. The overall structure of MPN314 is an octameric ring arranged to form a pseudo-eightfold symmetry. Each monomer has two subdomains related by a pseudo-twofold axis and is a single-domain α + β protein with a roll-type fold consisting mainly of α-helices wrapped around a core of β-strands. Structural similarity searches using the programs DALI (Holm & Sander, 1996 ▶) and CE (Shindyalov & Bourne, 1998 ▶) failed to identify any structural homologues in the Protein Data Bank. The structure also did not reveal any potential active site or ligand-binding site. Electrostatic surface-potential calculations revealed that the protein is highly negatively charged on the inner ring surface, while the outer ring surface is mainly positively charged. Although this structure strongly suggests that this protein functions as an octameric ring, it has not provided any clear molecular function.
MPN314 is currently the only representative structure of this protein family and although this individual structure failed to directly suggest molecular function, a better understanding of residue and molecular-surface conservation may provide insights crucial to understanding the involvement of MraZ proteins in cell-wall biosynthesis and/or cell division. In this paper, we present the cloning, expression, purification and crystallization of the E. coli MraZ homologue. Additionally, we provide both preliminary crystallographic and light-scattering evidence for the oligomerization of the E. coli MraZ protein as a dodecamer. It appears that there is substantial variability in this protein family and additional information concerning homologues of MPN314 may be important for the functional characterization of this protein family.
2. Materials and methods
2.1. Cloning procedure, expression and purification
The mraZ open reading frame was amplified by the polymerase chain reaction using BL21 (DE3) E. coli genomic DNA as a template and the forward and reverse primers 5′-CGGATCCATGTTTGGTAAAGGCGGTCT-3′ and 5′-CGCGGCCGCTGAACGGCATCTTAAAGCCAG-3′, respectively. The amplified DNA was inserted into the pET21b plasmid (Novogen, Madison, WI, USA) to yield the pET21b-mraZ-His6 construct. The recombinant protein produced from this construct contains the amino-acid sequence GRVEHHHHHH added to the C-terminus. The plasmid was sequenced to ensure that no mutations had been introduced during the amplification reaction.
E. coli BL21 (DE3) transformants containing the pET21b-mraZ-His6 construct were grown in 5 ml aliquiots and then subcultured into 1 l fresh Terrific Broth (Bioshop Canada Inc.) supplemented with ampicillin (200 µg ml−1) and grown to an A 600 of 0.650 at 310 K. Protein expression was then induced for 6 h with 0.4 mM isopropyl-β-thiogalactoside and cells were harvested by centrifugation (20 min, 3297g). The cell pellet was then resuspended in 50 mM sodium phosphate pH 8.0, 0.1% Triton X-100 and 0.3 M NaCl and cells were disrupted by sonication. Following centrifugation (40 min, 20 070g) at 277 K, the supernatant was incubated with 5 ml Ni–NTA resin (Qiagen) pre-equilibrated with 50 mM sodium phosphate pH 8.0 and 0.3 M NaCl for 90 min at 277 K. The supernatant–resin slurry was then loaded into a 25 ml column and the flowthrough was collected. A washing step with ten column volumes of buffer containing 50 mM sodium phosphate pH 8.0 and 0.3 M NaCl was performed. Further elution of non-specifically bound protein was accomplished by washing with 50 mM sodium phosphate pH 8.0, 0.3 M NaCl and 0.1 M imidazole. The protein was eluted from the column with 50 mM sodium phosphate pH 8.0, 0.3 M NaCl and 300 mM imidazole.
At this stage, recombinant MraZ protein was pure as judged by SDS–polyacrylamide gel electrophoresis followed by Coomassie Brilliant Blue R-250 staining. The yield was estimated to be 50 mg per litre of culture. The observed monomeric weight was approximately 19 kDa as observed by SDS–PAGE. The protein was dialyzed into 50 mM Tris–HCl pH 8.0 and 500 mM NaCl.
2.2. Dynamic light scattering of MraZ
The oligomeric state of MraZ in the crystallization buffer (50 mM Tris–HCl pH 8.0 and 500 mM NaCl) was assessed via dynamic light scattering using a DynaPro MSX/TC from Protein Solutions. Oligomerization was assessed at both 1 and 2 mg ml−1. Measurements were taken at 277, 293 and 310 K.
2.3. Crystallization
The purified protein was concentrated using a Centricon (Amicon/Millipore) with a 10 kDa molecular-weight cutoff (277 K, 997g). Crystallization experiments were performed using the hanging-drop vapour-diffusion method at room temperature. Drops were prepared by mixing 2 µl protein solution with an equal volume of reservoir solution and were suspended over 1 ml reservoir solution. Initial crystallization screens were carried out at a protein concentration of 27 mg ml−1 in 0.1 M Tris–HCl pH 8.0 and 500 mM NaCl using Crystal Screens 1 and 2 (Hampton Research). The initial crystallization conditions were further refined.
2.4. Data collection and diffraction measurements
MraZ crystals suitable for X-ray data collection were mounted using the crystallization condition with 50% glycerol added as a cryoprotectant at cryogenic temperature using an Oxford Cryosystems Cryostream. Native X-ray data were collected using the in-house facility equipped with a Rigaku rotating-anode generator operated at 50 kV and 100 mA and a MAR Research imaging plate. The crystal-to-detector distance was set to 350 mm and the crystal oscillation was set to 1° with 1500 s exposure time per image. All data sets were indexed and integrated with DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
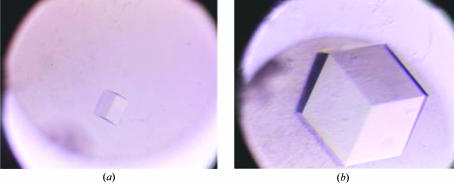
Recombinant MraZ protein was purified to homogeneity using a one-step procedure. The nickel-affinity column produced protein of very high purity that was sufficient for crystallization trials. Initial crystallization conditions were obtained from condition No. 34 of Crystal Screen 1 from Hampton Research (Fig. 2 ▶). The optimal crystallization conditions were refined to 0.1 M sodium acetate trihydrate pH 4.5 and 1.7 M sodium formate using a starting protein concentration of 30 mg ml−1 in 0.1 M Tris–HCl pH 8.0 and 500 mM NaCl (Fig. 2 ▶). The crystals typically reached dimensions of 0.4 × 0.4 × 0.4 mm.
Figure 2.
Crystallization of MraZ from E. coli. Initial crystallization conditions were obtained from Crystal Screen 1 condition 34 containing 0.1 M sodium acetate trihydrate pH 4.6 and 2.0 M sodium formate (a). Crystallization conditions were optimized to 0.1 M sodium acetate trihydrate pH 4.5 and 1.7 M sodium formate (b).
The diffraction pattern obtained using the in-house facility was consistent with the cubic P4132 symmetry, with unit-cell parameters a = b = c = 231.83 Å. A total of 126 902 reflections were collected in the resolution range 50.0–4.2 Å (Table 1 ▶). The average mosaicity was 0.5°. A reasonable Matthews coefficient of 2.28 Å3 Da−1 was obtained for 12 molecules in the asymmetric unit. The resulting calculated solvent content is 45.6%. The large unit cell required data to be collected with a longer crystal-to-detector distance to allow improved spot separation and indexing. Crystals typically provided low-resolution data of good quality; however, we have not been able to generate crystals capable of diffracting to high resolution in this space group. No differences in diffraction quality or resolution of the crystals were observed between capillary-mounted crystals at room temperature and crystals collected under cryogenic temperatures, regardless of the cryoprotectant solution employed.
Table 1. Diffraction data for native MraZ crystals.
Values in parentheses are for the high-resolution shell.
| Space group | P4132 |
| Unit-cell parameters (Å) | a = b = c = 231.83 |
| Temperature (K) | 100 |
| Resolution (Å) | 50–4.2 (4.35–4.2) |
| No. of observations/No. of unique observations | 126902/16121 |
| Completeness (%) | 99.8 (91.6) |
| 〈I/σ(I)〉 | 22.7 (2.77) |
| Rsym† (%) | 6.9 (35.8) |
R
sym = 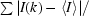
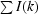 , where I(k) and 〈I〉 represent the diffraction intensity values of the individual measurements and the corresponding mean values, respectively. The summation is over all measurements.
, where I(k) and 〈I〉 represent the diffraction intensity values of the individual measurements and the corresponding mean values, respectively. The summation is over all measurements.
MraZ from E. coli displays 28% sequence identity and good overall sequence similarity to MPN314 from M. pneumoniae, the structure of which has previously been reported. Consequently, a molecular-replacement search using a monomeric MPN314 model was attempted using a variety of molecular-replacement programs. However, to date no solutions have been found. This is not surprising given the somewhat low level of sequence identity combined with differences in oligomeric states. We have therefore generated selenomethionine-labelled recombinant MraZ protein and are currently optimizing the crystallization conditions for subsequent multiple anomalous dispersion data collection and structure solution. Additionally, we are working to obtain an alternative crystal form capable of providing higher resolution data.
In an attempt to improve crystal quality, the oligomeric state in solution for the E. coli MraZ protein was characterized by dynamic light scattering. At both 1 and 2 mg ml−1 protein concentrations, the E. coli homologue was demonstrated to form a dodecamer that was stable across the temperature range 277–313 K. This complex could not be dissociated by the addition of a variety of detergents or through dramatic alterations in salt concentration or buffer pH. The samples were generally monodisperse, with only minor contaminating species of very high molecular weight (>1 × 108 Da), which are likely to be dust particles or non-functional protein aggregates. The dodecameric state of this MraZ homologue is supported by the Matthews coefficient calculations and is a major difference between this protein and the M. pneumoniae homologue. The large weight of protein (>180 kDa) contained within the asymmetric unit is likely to contribute substantially to the poor diffraction of these crystals. Further attempts will be made to obtain an alternate crystal form with improved internal order. Given the relatively low sequence identity, as well as the differences in oligomerization, significant functional information may possibly be garnered from the structure of E. coli MraZ that was not possible from the previously reported MPN314 structure. Additionally, future exploration of the conserved features of the tertiary and quaternary structures of E. coli MraZ and MPN314 may suggest a more precise role for these proteins in cell-wall biosynthesis and/or cell division.
Acknowledgments
The authors would like to thank Drs M. Cygler and A. Matte for their support as part of the Montreal–Kingston Bacterial Structural Genomics Initiative. MA is supported by a Postgraduate Doctoral Scholarship from NSERC and this research is funded by the CIHR. ZJ is a Canada Research Chair in Structural Biology and recipient of the Steacie Fellowship.
References
- Arigoni, F., Talabot, F., Peitsch, M., Edgerton, M. D., Meldrum, E., Allet, E., Fish, R., Jamotte, T., Curchod, M. L. & Loferer, H. (1998). Nature Biotechnol.16, 851–856. [DOI] [PubMed]
- Carrion, M., Gomez, M. J., Merchante-Schubert, R., Dongarra, S. & Ayala, J. A. (1999). Biochimie, 81, 879–888. [DOI] [PubMed] [Google Scholar]
- Chen, S., Jancrick, J., Yokota, H., Kim, R. & Kim, S.-H. (2004). Proteins, 55, 785–791. [DOI] [PubMed] [Google Scholar]
- Daniel, R. A., Williams, A. M. & Errington, J. (1996). J. Bacteriol.178, 2343–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassain, M., Leroy, A., Colosetti, L., Carole, S. & Bouche, J. P. (1999). Biochimie, 81, 889–895. [DOI] [PubMed] [Google Scholar]
- Heijenoort, J. van (2001). Glycobiology, 11, 25R–36R. [DOI] [PubMed]
- Holm, L. & Sander, C. (1996). Science, 273, 595–603. [DOI] [PubMed] [Google Scholar]
- Hutchison, C. A., Peterson, S. N., Gill, S. R., Cline, R. T., White, O., Fraser, C. M., Smith, H. O. & Venter, J. C. (1999). Science, 286, 2165–2169. [DOI] [PubMed] [Google Scholar]
- Mengin-Lecreulx, D., Ayala, J., Bouhss, A., van Heijenoort, J., Parquet, C. & Hara, H. (1998). J. Bacteriol.180, 4406–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlin, C., McAteer, S. & Masters, M. (2002). J. Bacteriol.184, 4573–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Shindyalov, I. N. & Bourne, P. E. (1998). Protein Eng.11, 739–747. [DOI] [PubMed] [Google Scholar]